An ultra-sensitive colorimetric detection of tetracyclines using the shortest aptamer with highly enhanced affinity†
Young Seop
Kwon‡
,
Nurul Hanun
Ahmad Raston‡
and
Man Bock
Gu
*
School of Life Sciences and Biotechnology, Korea University, Anam-dong, Seongbuk-Gu, Seoul 136-713, Republic of Korea. E-mail: mbgu@korea.ac.kr; Fax: +82-2-9286050; Tel: +82-2-3290-3417
First published on 17th October 2013
Abstract
A shortened 8-mer ssDNA aptamer was successfully truncated for four different tetracyclines with high affinity. The ultrasensitive colorimetric detection of oxytetracycline using this shortened aptamer was possible, which was about 500-fold enhanced compared to that obtained using the original 76-mer aptamer.
The production of aptamers from the well-known Systematic Evolution of Ligands by Exponential Enrichment (SELEX) method1 has some inevitable shortcomings that restrict the maximum interactions between the single-stranded DNA or RNA molecules with targets during the selection process. This issue is mainly due to the primer regions required on a random library of single-stranded DNA or RNA and long sequences for their diversity, which cause the interference of unnecessary sequence regions which are not involved in binding reactions. In efforts to increase the aptamer affinity to its target, only a few simple or heuristic approaches towards the truncation of aptamers were reported,2–4 in addition to an effort to optimize the length of the random library used in the SELEX process.5 Engineering of aptamers is crucially needed to reveal the core sequence region which participates in binding interactions and remove the unnecessary sequence region. There are several studies reported on aptamer engineering approaches, which focused on the G-quadruplex region,6 secondary structure (stem-loop),7 and stem-loop multiplication since the stem loop is known to be the binding motif for most proteins.8 However, since the binding motifs of the aptamers for the small molecules have not been clearly ascertained, these binding motif based approaches are not eligible.
Tetracyclines (TCs) are the most common antibiotics that have been extensively used for the treatment of animal diseases and were found to accumulate in dairy food, livestock, and soil, posing serious risks to human health.9–12 Thus, the detection of TCs has captured our attention to enhance the sensitivity of detection by using a simple colorimetric assay. This flat poly-cyclic molecule with one aromatic ring enabled one face of the TC molecules to make ionic interactions with the charged RNA backbone of 74 bp in size, while the other face allows either hydrophobic or stacking interactions.13
Several studies on TC-targeted aptamers were done using 74 and 60 nucleotides RNA aptamers to explain the formation of RNA–ligand complexes in various crystallographic models and fluorescence spectroscopic approaches, such as NMR, stopped-flow fluorescence and fluorescent ligand studies.14–19 However, we found that the shortened ssDNA aptamer obtained in this study, neither long nor RNA aptamer, interacts with the TCs, yielding a new molecular interaction complex between the aptamer and the ligand, which is completely different from the previous case.
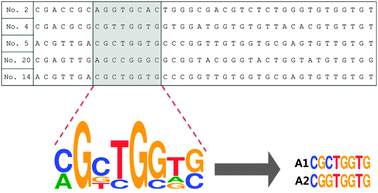
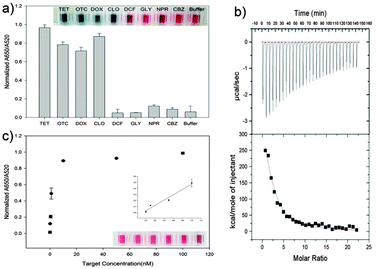
We have heuristically truncated the 76-mer ssDNA aptamers that bind to the OTC,20 to 8 mer, by utilizing the conserved sequences with high homogeneity that exist in the selected aptamers as shown in Scheme 1. In this step, the variable regions composed of 40-nucleotides (N40), in total 76-mer of the five original aptamer sequences (aptamers No. 2, No. 4, No. 5, No. 20 and No. 14) were subjected to further analysis (Table S1, ESI†). In fact, three aptamers denoted as No. 4, No. 5 and No. 20 shared strong affinities with a Kd value of 9.61, 12.08 and 56.84 nM, respectively, while the selectivity for OTC was 72–76%. The design of the shortened aptamers has been directly implemented in this heuristic approach by selecting nucleotide bases which exhibited high homogeneity in accordance to their conserved regions, after these five sequences of the original aptamers were aligned (please see the details of this heuristic approach in the materials and methods and Fig. S1, ESI†). A1 and A2 of 8-mer sequences heuristically selected were successfully conceived and found to exhibit high affinity and specificity to TCs, according to a simple well-known unmodified gold nanoparticle-based colorimetric method.21,22 The A2 aptamer which evinced stronger affinity to TCs compared to the A1 aptamer was chosen for further analysis (Fig. S2, ESI†). Fig. 1a shows the specific interaction of the A2 aptamer with TCs [tetracycline (TET), oxytetracycline (OTC), doxycycline (DOX) and chlortetracycline (CHLOR)], along with diclofenac (DIC), glyphosate (GLY), naproxen (NPX) and carbamazepine (CBZ) as counter targets by using colorimetric assay. As can be seen in Fig. 1a, the A2 aptamer was found to show good selectivity for all tested TCs rather than only for OTC as reported in the previous work with the original 76-mer aptamer, while still discriminating other counter targets (DIC, GLY, NPX and CBZ).23
To evaluate the binding strength between these truncated A2 aptamer and the targets, we used ultrasensitive Isothermal Titration Calorimetry (VP-ITC)24 analysis for measurement of the Kd value. VP-ITC measures heat generated or absorbed due to the binding of the aptamer to the target. The binding curve obtained from the VP-ITC was then analyzed using the single-site binding model25 to accurately measure the association constant. In this case, Ka (9.366 × 105 mM) between A2-TET was measured, which corresponds to an apparent Kd value of 1.067 nM (Fig. 1b). This shortened A2 aptamer which exhibited the highest affinity to TET and the lowest Kd value, compared to other TC derivatives, enables the ultrasensitive detection of TET down to 0.1 nM, as shown in Fig. 1c.
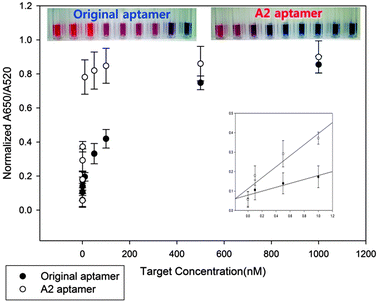
Since the original 76-mer aptamer was found not to interact with TET, only OTC should be used for comparing the detection limits between A2 and the original 76-mer aptamers. Therefore, we performed dose dependent experiments of OTC for both A2 and original 76-mer aptamers based on AuNP-based colorimetric assay. The absorbance ratios of A2 and original 76-mer aptamers at 650/520 nm were plotted against OTC concentrations as shown in Fig. 2. The detection limit of the A2 aptamer to OTC was enhanced down to 0.1 nM, which is about 500-fold improved, compared to that of the original 76-mer aptamer. It also showed that even the color changes at 10 nM of OTC can be observed by the naked eye. The Kd value between A2 aptamer and OTC was also measured to be 1.104 nM by ITC assay, while no detectable binding reaction was observed for the original 76-mer aptamer under the same assay conditions (Fig. S3, ESI†). This result is not only another confirmation of this shortened 8-mer A2 aptamer interacting with TCs with highly enhanced affinity, but also an example of how this shortened aptamer can be utilized in the detection of TCs as a diagnostic tool.
In addition, we performed molecular dynamics (MD) simulations26 for further observations on the binding mechanism and found that the binding interaction of the A2 aptamer to TET, OTC and DOX was almost in the same fashion (Fig. S4, ESI†). According to the simulation, the first two bases of A2 aptamer, 5′-CG, worked as a binding pocket conferring the stacking interaction, similar to phi–phi stacking, with the C11–C15 carbon ring of TCs. Each of the TCs stacked between C and G is found to be separated from both nucleotide rings by 3.5 and 3.7 Å, respectively, which are measured from C14 of TCs to the CG binding pocket of the A2 aptamer. Furthermore, each of the TCs alternatively formed hydrogen bonding with phosphate backbones of G5–G7 and the G2 base of the A2 aptamer, while it was consistently stacked in the CG pocket. This association of the carbon rings of TCs to the A2 aptamer which resulted in highly specific binding mediated by a combination of stacking interactions and hydrogen bonding was also supported by the previous studies.27–29 Considering that there are no interactions found in both gold nanoparticle-based colorimetric and ITC assays for all the possible sequence variants of the A2 aptamer (Fig. S5, S6 and Table S2, ESI†), this shortened A2 aptamer seemed to possess a unique base combination to interact with TCs. In addition, according to the MD simulation, the binding mechanism is the same for all of the tested TCs, and the A2 aptamer highly recognizes four rings of the basic TCs. These findings suggest that, since the original 76-mer aptamer contains unnecessary sequences not involved in binding interactions, some sequences in the original 76-mer aptamer might still contribute to different selectivities to different TCs. Accordingly, based on these findings, we believe that the length of the truncated aptamer could be related to the length of the conserved regions with high homogeneity located on the original aptamers. In addition, this heuristic truncation approach can be applied for any aptamers with conserved regions and high homogeneity amongst several aptamer sequences available for a target.
In conclusion, we have found that 76 mer ssDNA aptamers with high affinity and selectivity for OTC can be truncated to a unique shortened 8-mer ssDNA aptamer that conserved only the original stacking pocket and 6 additional specific bases while exhibiting higher binding affinity (Kd 1.067 nM for tetracycline). The ultra-sensitive detection of tetracyclines down to 0.1 nM was possible by using this shortened ssDNA aptamer, which is about 500-fold enhanced. To the best of our knowledge, this comprehensive molecular interaction study for this shortened 8-mer aptamer, which is the shortest ssDNA aptamer ever with highly enhanced affinity, was successfully conducted for the first time. These new findings are expected to promote studies on the construction and development of productive aptamer science and technology.
This research was supported by the National Research Foundation of Korea (NRF) grant funded by the Korean government (MEST) (No. 2012R1A2A2A01011056).
Notes and references
- C. Tuerk and L. Gold, Science, 1990, 249, 505–510 CAS
.
- W. M. Rockey, F. J. Hernandez, S. Y. Huang, S. Cao, C. A. Howell, G. S. Thomas, X. Y. Liu, N. Lapteva, D. M. Spencer, J. O. McNamara, X. Zou, S. J. Chen and P. H. Giangrande, Nucleic Acid Ther., 2011, 21, 299–314 CrossRef CAS PubMed
.
- P. C. Anderson and S. Mecozzi, J. Am. Chem. Soc., 2005, 127, 5290–5291 CrossRef CAS PubMed
.
- M. G. Wallis, U. von Ahsen, R. Schroeder and M. Famulok, Chem. Biol., 1995, 2, 543–552 CrossRef CAS
.
- S. K. Silverman, Chem. Commun., 2008, 3467–3485 RSC
.
- D. M. Tasset, M. F. Kubik and W. Steiner, J. Mol. Biol., 1997, 272, 688–698 CrossRef CAS PubMed
.
- H. Kaur and L. Y. Yung, PLoS One, 2012, 7, e31196 CAS
.
- P. R. Mallikaratchy, A. Ruggiero, J. R. Gardner, V. Kuryavyi, W. F. Maguire, M. L. Heaney, M. R. McDevitt, D. J. Patel and D. A. Scheinberg, Nucleic Acids Res., 2011, 39, 2458–2469 CrossRef CAS PubMed
.
- D. A. Silvestrini, G. W. Anderson and E. S. Snyder, Poult. Sci., 1958, 37, 1243 CrossRef
.
- I. Chopra, T. G. B. Howe, A. H. Linton, K. B. Linton, M. H. Richmond and D. C. E. Speller, J. Antimicrob. Chemother., 1981, 8, 5–21 CrossRef CAS PubMed
.
- J. J. Mcfarland, Ear, Nose Throat J., 1981, 60, 226–227 Search PubMed
.
- M. L. Nelson and S. B. Levy, Ann. N. Y. Acad. Sci., 2011, 1241, 17–32 CrossRef CAS PubMed
.
- C. Berens, A. Thain and R. Schroeder, Bioorg. Med. Chem., 2001, 9, 2549–2556 CrossRef CAS
.
- M. Pioletti, F. Schlunzen, J. Harms, R. Zarivach, M. Gluhmann, H. Avila, A. Bashan, H. Bartels, T. Auerbach, C. Jacobi, T. Hartsch, A. Yonath and F. Franceschi, EMBO J., 2001, 20, 1829–1839 CrossRef CAS PubMed
.
- S. Hanson, G. Bauer, B. Fink and B. Suess, RNA, 2005, 11, 503–511 CrossRef CAS PubMed
.
- H. Xiao, T. E. Edwards and A. R. Ferre-D'Amare, Chem. Biol., 2008, 15, 1125–1137 CrossRef CAS PubMed
.
- D. Wunnicke, D. Strohbach, J. E. Weigand, B. Appel, E. Feresin, B. Suess, S. Muller and H. J. Steinhoff, RNA, 2011, 17, 182–188 CrossRef CAS PubMed
.
- M. Muller, J. E. Weigand, O. Weichenrieder and B. Suess, Nucleic Acids Res., 2006, 34, 2607–2617 CrossRef PubMed
.
- U. Forster, J. E. Weigand, P. Trojanowski, B. Suess and J. Wachtveitl, Nucleic Acids Res., 2012, 40, 1807–1817 CrossRef PubMed
.
- J. H. Niazi, S. J. Lee and M. B. Gu, Bioorg. Med. Chem., 2008, 16, 7245–7253 CrossRef CAS PubMed
.
- H. Li and L. Rothberg, Proc. Natl. Acad. Sci. U. S. A., 2004, 101, 14036–14039 CrossRef CAS PubMed
.
- F. Xia, X. Zuo, R. Yang, Y. Xiao, D. Kang, A. Vallee-Belisle, X. Gong, J. D. Yuen, B. B. Hsu, A. J. Heeger and K. W. Plaxco, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 10837–10841 CrossRef CAS PubMed
.
- Y. S. Kim, J. H. Kim, I. A. Kim, S. J. Lee, J. Jurng and M. B. Gu, Biosens. Bioelectron., 2010, 26, 1644–1649 CrossRef CAS PubMed
.
- M. W. Freyer and E. A. Lewis, Methods Cell Biol., 2008, 84, 79–113 CrossRef CAS
.
- T. Egawa, A. Tsuneshige, M. Suematsu and T. Yonetani, Anal. Chem., 2007, 79, 2972–2978 CrossRef CAS PubMed
.
- Fh. Stilling and A. Rahman, J. Chem. Phys., 1974, 60, 1545–1557 CrossRef
.
- G. R. Zimmermann, R. D. Jenison, C. L. Wick, J. P. Simorre and A. Pardi, Nat. Struct. Biol., 1997, 4, 644–649 CrossRef CAS PubMed
.
- M. Sassanfar and J. W. Szostak, Nature, 1993, 364, 550–553 CrossRef CAS PubMed
.
- D. E. Huizenga and J. W. Szostak, Biochemistry, 1995, 34, 656–665 CrossRef CAS
.
Footnotes |
| † Electronic supplementary information (ESI) available: Fig. S1–S4 and Tables S1 and S2, and materials and methods. See DOI: 10.1039/c3cc47108j |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2014 |