Fluorescence modification of the AAAA (4A) loop: toward a probe of the structural dynamics of the i-motif of the retinoblastoma gene†
Jung Woo
Park
a,
Young Jun
Seo
*b and
Byeang Hyean
Kim
*a
aDepartment of Chemistry, BK School of Molecular Science, Pohang University of Science and Technology, Pohang 790-784, South Korea. E-mail: bhkim@postech.ac.kr; Fax: +82-54-279-3399; Tel: +82-54-279-2115
bDepartment of Chemistry, Chonbuk National University, Jeonju 561-756, South Korea. E-mail: yseo@jbnu.ac.kr; Fax: +82-63-270-3408; Tel: +82-63-270-3417
First published on 22nd October 2013
Abstract
Systematic modification of the 4A loop region of the Rb gene with PyA fluorophore units allows discrimination of the fluorescence signals corresponding to structural dynamics from single-stranded to i-motif structures.
The retinoblastoma (Rb) gene encodes retinoblastoma, a tumor suppressor protein that is dysfunctional in several major cancers.1 Alleles of this gene induce mutation of the protein that is inactivated, resulting in the development of retinoblastoma cancer. The Rb gene is extremely rich in G and C strands at the 5′ position, potentially forming several secondary structures, including G-quadruplex, i-motif, and Z-DNA species.2 Among these sequences, C-rich strands induce i-motif structures with hemiprotonated C·C+ base pairs and their loops at low pH.3 In addition, i-motif sequences are discovered frequently in eukaryotic genomes,4 especially in the regulatory regions of myeloid-specific genes and human c-ki-ras.5 In these regions, several proteins, including helicase, single-stranded binding protein, and DNA polymerase, bind selectively to the C-rich strands.6 Thus, the development of new systems for probing i-motif structures of the Rb gene should benefit cancer therapeutics and the study of oncogenes, and also provide insight into the dynamics of the conformational transitions of C-rich single-stranded DNA.
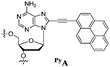
Our goal in this study was to develop a simple and efficient exciplex signaling system for probing the i-motif of the Rb gene sequence 5′-GCCGCCCAAAACCCCCCG and to use fluorescence to understand its structural dynamics.7 We focused on the 4A loop region, hoping that we could induce variations in observable signals representing conformational changes from single-stranded and duplex forms to the i-motif structure without disturbing the hemiprotonated C·C+ base pairs at low pH. We chose a pyrene-modified deoxyadenosine (PyA) as a fluorophore unit, anticipating that it would be sensitive to its local environment and could be incorporated into several DNA structures.8 We expected the incorporation of one or more PyA residues into the 4A loop region to result in signals that would depend on both the distance between the PyA residues and the shapes of the various secondary structures. We synthesized various oligodeoxynucleotides (ODNs, Table 1) to systematically probe the structural transitions of the i-motif, incorporating pyrene units at the 1 (ODN i1), 1,2 (ODN i2), 1,3 (ODN i3), and 1,4 (ODN i4) positions of the 4A loop (Scheme 1).
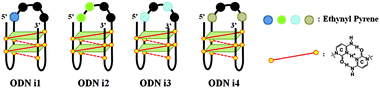 | ||
| Scheme 1 Four different fluorescence signals from i-motifs at a single excitation wavelength (386 nm), depending on the distance between the pyrene units in the 4A loop structure. | ||
We recorded circular dichroism (CD) spectra to investigate the conformational transitions of the natural i-motif strand ODN N and the PyA-containing i-motif strands ODNs i1–i4 (Fig. S1, ESI†). The spectrum of each of these ODNs featured a strong positive band near 285 nm and a negative band at 260 nm under acidic conditions (pH 4.0), characteristic of an i-motif structure.9 After adding the G-rich strand ODN G to form a duplex, in each case we observed a positive band near 270 nm and a negative band near 240 nm, typical of B-form duplex DNA.10 These CD spectra suggest that the incorporation of one or two PyA units in the 4A loop does not disturb the conformational changes of the i-motif structures.
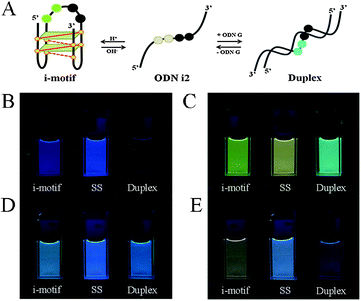
Next, we investigated the fluorescence properties of the various structures: the i-motif structures at pH 4, the single-stranded structures at pH 7.2, and the duplex structures formed with ODN G at pH 7.2. All of our modified ODNs exhibited various colors depending on their structures (i.e., i-motif, single strand, or duplex). Interestingly, for the i-motif structures with substitution at the 1, 1,2, 1,3 and 1,4 positions, we observed, at pH 4, four different colors—blue, green, bluish, and reddish, respectively—depending on the number of and distance between the pyrene units, at a constant excitation wavelength of 386 nm (Fig. 1 and Fig. S2, ESI†). In the case of ODN i1, we observed a strong change in fluorescence intensity depending on its structure: the blue signal (at 450 nm) of the i-motif structure was stronger than that of its duplex, but weaker than that of its randomly coiled single strand. In the case of ODN i2, we observed dramatic color changes depending on each conformational transition. At pH 7.2 (randomly coiled single strand), the emission maximum appeared at 492 nm, arising from a pyrene exciplex, and the sample displayed green fluorescence. Interestingly, when the structure transformed to the i-motif at pH 4.0, the emission maximum shifted to 507 nm and the sample displayed a yellowish green color; in the duplex state ODN i2·ODN G, the emission maximum shifted again to 476 nm and the sample displayed a blue emission. In the case of ODN i3, we observed only slight color changes depending on its structure; the i-motif structure provided a greenish blue solution (emission maximum at 470 nm), while the other two structures provided blue solutions (emission maxima at 453 nm). Finally, in the case of ODN i4, we observed a very dramatic broad, red-shifted band near 550 nm that resulted in the solution of the i-motif structure exhibiting a reddish color; such a signal is not common, even for a pyrene excimer. Solutions containing the other two structures of ODN i4 exhibited blue fluorescence (maximum at 465 nm). Because ODNs i2 and i4 provided the most dramatic changes in fluorescence upon their conformational transitions, we used these systems to probe i-motif structures with visible color changes.
We also recorded fluorescence and CD spectra of our modified ODNs i1–i4 at pH ranging from 9.0 to 4.0 (Fig. S3, ESI†) to comprehend the structural dynamics of the Rb gene sequence. The changes in fluorescence at each value of pH were consistent with those in the CD spectra and corresponded to the conformational changes. The most significant conformational change in each of our modified ODNs i1–i4 was that from the single strand to the i-motif, which occurred in the pH range 6.0–5.0.
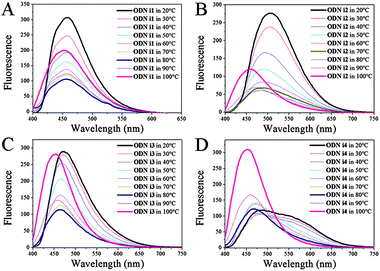
To elucidate further details of the conformational transitions, we investigated the fluorescence melting of the i-motif structures (Fig. 2). Interestingly, the conformational transitions of all of the modified ODNs i1–i4 from i-motif to single strand were not simple one-stage transitions; indeed, from the fluorescence melting data, we observed three turning points.
At temperatures below their melting point ranges, all of our ODNs exhibited the original fluorescence of their i-motif structures. When the temperature was in the melting temperature region (30–70 °C), the fluorescence maxima shifted to those of the randomly coiled single strands (Fig. 2; Table S2, ESI†). Interestingly, when the temperature was above 70 °C, we observed a new blue-shifted band, the fluorescence intensity of which increased upon increasing the temperature, for all of our modified ODNs. This behavior suggested three stages of unfolding: an i-motif structure at temperatures below the melting range; a randomly coiled single strand at temperatures within the melting range; and another, unknown, single-stranded structure at higher temperatures. Thus, we investigated the dynamics of the duplex structures formed from these i-motif sequences; their structures were also different depending on the temperature. When melted to a single strand from a duplex, the emission in each case approached that of the randomly coiled single strand, much like that observed after unfolding of the i-motif. We did not, however, detect any fluorescence representing an additional unfolding stage similar to that found upon unfolding of the i-motif structure; we observed only two stages of emissions corresponding to the duplex and the randomly coiled single strand (Table S2; Fig. S4, ESI†). Thus, the new blue-shifted emissions during the fluorescence melting of ODNs i1–i4 were related only to the unfolding dynamics of their i-motifs at pH 4. To determine the structural differences among the three states during the i-motif fluorescence melting process, we recorded the CD spectra over the same temperature range (Fig. S5, ESI†). Unfortunately, we could distinguish only two states representing the randomly coiled single strands and i-motifs. Although CD might not be the best method for discerning between individual states, because it is not highly sensitive to the conformational changes of DNA,11 our fluorescence data suggested the existence of another structural possibility during the conformational transitions of the i-motifs; in other words, the structural dynamics of the i-motif might not simply involve two states.
Next, we measured UV absorptions to clarify the origins of the signals resulting from each structural state (i-motif, randomly coiled single strand, duplex). The UV spectrum of each modified ODNs i1–i4 featured a strong absorption band near 420 nm, characteristic of pyrene. At pH 4 (i-motif structure), this signal underwent a red-shift relative to that of the randomly coiled single-strand and duplex structures (Fig. S6, ESI†). These red-shifts for the i-motif structures imply an interaction between adjacent pyrene moieties or between the pyrene unit and a neighboring base (e.g., G). The high intensities of the signals for the i-motif structures of ODNs i1 and i3 at 420 nm were characteristic of the original PyA monomer and provided solutions exhibiting blue emissions; for the ODNs i2 and i4, however, we observed substantial peak broadening at 420 nm, implying interactions between the pyrene units. The adjacent pyrene moieties in ODN i2 are sufficiently close for them to interact, while ODN i4 may have suitable conformational flexibility for its pyrene units to do so as well. A staggered or oblique geometry with electrostatic stacking interactions between the two PyA moieties of ODNs i2 and i4 might explain the unique red-shifted emission signal.12 As expected, at pH 7 (randomly coiled single strand), the spectra of ODNs i1, i3, and i4 (but not ODN i2) exhibited similar pyrene absorption patterns at 420 nm; the greater intensity and blue-shift of these bands, relative to that of ODN i2, explain why the blue emissions of these solutions were similar to that of the original PyA monomer. In ODN i2, however, interactions still existed between the two PyA units, resulting in a broad band at 420 nm and a red-shifted band and, hence, a reddish emission. In the duplex state with ODN G, all of the ODNs, even ODN i2, recovered the characteristic emission of the PyA monomer, with increased intensity at 420 nm and a blue-shifted pattern relative to those of their i-motif structures and, hence, blue emissions. These UV spectroscopic data suggest that the unique red-shifted emission signals of the i-motif structures of ODNs i2 and i4 arose from the stacking of two pyrene units.12
Finally, to confirm the stacking structures of the pyrene units on the loop, we determined the melting temperatures (Tm) of our i-motif and duplex structures (Fig. S8; Table S3, ESI†). Unexpectedly, the melting temperatures for the i-motifs of all of our pyrene-attached ODNs i1–i4 were slightly lower or similar to that of the natural i-motif of ODN N; we had suspected that pyrene stacking would increase the structural stability of the i-motif structure. The melting temperature of ODN i1, containing one PyA unit, was lower than that of ODN N. The stability of ODN i2 was also lower than that of ODN N; its two pyrene units are so close that they may destabilize the i-motif structure through steric interactions. The ODNs i3 and i4, however, exhibited stabilities similar to that of the natural sequence, consistent with their pyrene units being positioned at a sufficient distance to avoid such steric clashes. Accordingly, we suspect that the two pyrene units on the loops of ODNs i2 and i4 are not optimally π-stacked to stabilize their i-motif structures; rather, they may have staggered or oblique geometric relationships on the outer surfaces of the i-motif structures.
We are systematically developing a probing system to monitor the i-motif structures, focusing on the 4A-loop of the Rb gene. In terms of fluorescence signals, i-motif structures modified with fluorophores at the 1,2 (ODN i2) and 1,4 (ODN i4) positions of the 4A loop provide the most dramatic fluorescence changes at a single excitation wavelength upon conformational transitions from single-stranded to duplex to i-motif structures, respectively. These probing systems are unique in their ability to discriminate among these three structural states, through fluorescence emissions at three different colors, upon excitation at a single wavelength. Although we are not in a position to identify the exact structures and all of the structural dynamics that occur upon the transitions from single strands to i-motifs, we believe that the three different emission patterns observed upon the unfolding of the i-motif structures during fluorescence melting at pH 4 reflect the existence of an additional single strand stage. We hope that our results will lead to further biological studies of the i-motif structure of the Rb gene and increase its range of applications such as monitoring gene structure as well as understanding the structural dynamics, which is related to gene regulation.
This study was supported by the EPB Center program (2008-0061892) and the NRF (2012R1A2A2A01047069). YJS thanks to the “Leaders-Industry-University Cooperation” Project, funded by the Ministry of Education, Science and Technology (MEST).
Notes and references
- A. L. Murphree and W. F. Benedict, Science, 1984, 223, 1028 CAS.
- S. H. Friend, R. Bernards, S. Rogeli, R. A. Weinberg, J. M. Rapaport, D. M. Alberts and T. P. Dryja, Nature, 1986, 323, 643 CrossRef CAS PubMed.
- K. Gehring, J. L. Leroy and M. Gueron, Nature, 1993, 363, 561 CrossRef CAS PubMed.
- P. Catasti, X. Chen, S. V. Mariappan, E. M. Bradbury and G. Gupta, Genetica, 1999, 106, 15 CrossRef CAS.
- E. H. Postel, S. J. Berberich, J. W. Rooney and D. M. Kaetzel, J. Bioenerg. Biomembr., 2000, 32, 277 CrossRef CAS; G. Manzini, N. Yathindra and L. E. Xodo, Nucleic Acids Res., 1994, 22, 4634 CrossRef PubMed.
- E. Marsich, A. Piccini, L. E. Xodo and G. Manzini, Nucleic Acids Res., 1996, 24, 4029 CrossRef CAS PubMed; E. Marsich, L. E. Xodo and G. Manzini, Eur. J. Biochem., 1998, 258, 93 Search PubMed; L. Lacroix, H. Lienard, E. Labourier, M. D. Mergny, J. Lacoste, H. Leffers, J. Tazi, C. Helene and J. L. Mergny, Nucleic Acids Res., 2000, 28, 1564 CrossRef PubMed.
- Y. J. Seo, J. H. Ryu and B. H. Kim, Org. Lett., 2005, 7, 4931 CrossRef CAS PubMed; Y. J. Seo and B. H. Kim, Chem. Commun., 2006, 150 RSC; I. J. Lee, J. W. Yi and B. H. Kim, Chem. Commun., 2009, 5383 RSC.
- Y. J. Seo, G. T. Hwang and B. H. Kim, Tetrahedron Lett., 2006, 47, 4037 CrossRef CAS PubMed; Y. J. Seo, H. Rhee, T. Joo and B. H. Kim, J. Am. Chem. Soc., 2007, 129, 5244 CrossRef PubMed; Y. J. Seo, S. Bhuniya and B. H. Kim, Chem. Commun., 2007, 1804 RSC; Y. J. Seo, S. Bhuniya, J. W. Yi and B. H. Kim, Tetrahedron Lett., 2008, 49, 2701 CrossRef PubMed; Y. J. Seo, I. J. Lee and B. H. Kim, Mol. Biosyst., 2009, 5, 235 RSC; I. J. Lee and B. H. Kim, Chem. Commun., 2012, 2074 RSC.
- S. S. Pataskar, D. Dash and S. K. Brahmachari, J. Biomol. Struct. Dyn., 2001, 19, 307 Search PubMed; V. Mathur, A. Verma and S. Maiti, Biochem. Biophys. Res. Commun., 2004, 320, 1220 CrossRef CAS PubMed.
- N. Berova, K. Nakanishi and R. W. Woody, Circular Dichroism: Principles and Application, Wiley, New York, 2nd edn, 2000 Search PubMed.
- M. M. Dailey, M. C. Miller, P. J. Bates, A. N. Lane and J. O. Trent, Nucleic Acids Res., 2010, 38, 4877 CrossRef CAS PubMed.
- T. Kawai, M. Ikegami and T. Arai, Chem. Commun., 2004, 824 RSC; J. N. Wilson, J. Gao and E. T. Kool, Tetrahedron, 2007, 63, 3427 CrossRef CAS PubMed; F. Seela and S. A. Ingale, J. Org. Chem., 2010, 75, 284 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: MALDI-TOF mass spectrometric data; UV absorption, CD, and fluorescence spectra at pH 4.0–9.0. See DOI: 10.1039/c3cc46619a |
| This journal is © The Royal Society of Chemistry 2014 |