Personalized protein coronas: a “key” factor at the nanobiointerface†
Mohammad J.
Hajipour
a,
Sophie
Laurent
bc,
Afsaneh
Aghaie
d,
Farhad
Rezaee‡
*ef and
Morteza
Mahmoudi‡
*ag
aDepartment of Nanotechnology and Nanotechnology Research Center, Faculty of Pharmacy, Tehran University of Medical Sciences, Tehran, Iran. E-mail: mahmoudi@stanford.edu
bDepartment of General, Organic, and Biomedical Chemistry, NMR and Molecular Imaging Laboratory, University of Mons, Avenue Maistriau 19, B-7000 Mons, Belgium
cCenter for Microscopy and Molecular Imaging (CMMI), Rue Adrienne Bolland, 8, B-6041 Gosselies, Belgium
dBlood Transfusion Research Center, High Institute for Research and Education in Transfusion Medicine, Tehran, Iran
eUniversity of Groningen, University Medical Center Groningen, Department Cell Biology, Department Medical Proteomics, Groningen, the Netherlands. E-mail: f.rezaee@med.umcg.nl; f.rezaee@erasmusmc.nl
fDepartment of Gastroenterology and Hepatology, Erasmus Medical Center, Rotterdam, the Netherlands
gBiomaterials & Advanced Drug Delivery Laboratory, School of Medicine, Stanford University, Stanford, CA 94305, USA
First published on 30th May 2014
Abstract
It is now well known that the primary interactions of biological entities (e.g., tissues and cells) with nanoparticles (NPs) are strongly influenced by the protein composition of the “corona” (i.e., the NP surface attached proteins). The composition of the corona strongly depends on the protein source (e.g., human plasma). Because the protein source determines the NP corona, it is reasonable to hypothesize that humans with specific disease(s) may have specific NP coronas. To test this hypothesis, we incubated two different hydrophobic/hydrophilic types of NPs (polystyrene and silica) with plasma from human subjects with different diseases and medical conditions (e.g., breast cancer, diabetes, hypercholesterolemia, rheumatism, fauvism, smoking, hemodialysis, thalassemia, hemophilia A and B, pregnancy, common cold and hypofibrinogenemia). Our results demonstrate that the type of disease has a crucial role in the protein composition of the NP corona. Based on these results, we introduce the concept of the “personalized protein corona” (PPC) as a determinant factor in nano-biomedical science. This study will help researchers rationally design experiments based on the “personalized protein corona” for clinical and biological applications.
1. Introduction
Nanoparticles (NPs) are recognized as promising agents for various biomedical applications. Due to their ultra-small size, NPs are able to reach otherwise inaccessible sites within the human body (e.g., the cell nucleus). Thus, from a medical point of view, the safety of NPs is an important concern.1–3 The first important issue for biological evaluation of NPs is the composition of the protein corona on the surface of a NP, which determines the biological function of the NP in a biological fluid.4–7 The protein corona imparts a unique biological identity to a NP, which is much different from the pristine coated surface.8–10 The protein composition of the corona layer (which can significantly alter the biological fate) is strongly dependent on various factors such as the physicochemical properties of the NP (e.g., the composition, size, shape, charge and hydrophobicity/hydrophilicity),11–16 the temperature,17 the protein concentration,18 the protein gradient,19 and the protein source (e.g., fetal bovine serum (FBS) or human plasma).20The plasma proteomes of patients with different diseases show that each type of disease has significant effects on the concentration/conformation of plasma proteins. In addition, the genetic background, post-translational modification, life style, and geographical origin play an important role in the changes in plasma proteins in healthy individuals.21–23 It is well known that the concentration/structure of some critical proteins present in human plasma can be significantly altered by diseases and medical conditions (e.g., cancer, hemodialysis, obesity, diabetes, hypercholesterolemia, rheumatism, fauvism, hemophilia, smoking, hypofibrinogenemia and pregnancy).24–35 It must be noted that the major cause of type 2 diabetes is obesity. Obesity is associated with many other diseases, including coagulation disorders, dyslipidemia and atherosclerosis, all of which affect the protein composition of plasma.23–25 It should be noted that changed/modified plasma proteins can be considered as potential prognostic or diagnostic biomarkers for predicting/treating specific disorders. For example, compared to short hemodialysis, the long hemodialysis patients have higher plasma vitamin D binding protein and lower clusterin, haptoglobin, hemopexin, apolipoprotein A-IV, complement factor B, and complement factor H. Moreover, the isoforms of alpha 1-antitrypsin and the fibrinogen γ-chain are different in these patients. Albumin is the most abundant protein in the plasma,26–29 where it plays a key role in the formation of the protein corona.15,30 Depending on the physicochemical properties of the NP, albumin tends to be adsorbed on the NP surface after a short incubation due to its high concentration in the human plasma. It is subsequently replaced by other proteins, which have higher affinities for the NP surface.14 It is now well recognized that different diseases/situations affect the plasma homeostasis. For example, plasma protein glycosylation in diabetic patients can increase the catabolism of low-density lipoproteins, which in turn reduces the solubility of albumin. Similarly, there is a linear relationship between fibrinogen levels and the number of cigarettes smoked, age, physical exercise, stress, blood glucose/cholesterol, inflammation and pregnancy.24,27,32,36–45 Fibrinogen, an acute phase protein, and fibrinolysis play an important role in the wound healing process.46–54
Although there are extensive reports on the effect of various factors on the corona composition, the effects of altered human plasma protein composition due to different diseases defined as a protein source, which is a determinant factor in the formation of corona (a crucial feature in nano-biomedical applications), have not been studied extensively. Here, we investigate the effects of various human plasma samples with different disease backgrounds on the corona composition of commercially available NPs (i.e., silica and polystyrene NPs with very narrow size distributions). A range of experimental approaches including one-dimensional sodium dodecyl sulfate polyacrylamide gel electrophoresis (1D-SDS-PAGE), dynamic light scattering (DLS), zeta potential analysis and semi-quantitative densitometry were employed to elucidate the effects of the altered plasma on the corona composition.
2. Experimental section
2.1. Plasma preparation
Blood samples from different human subjects (patients and healthy volunteers) were collected following signed informed consent based on ethical laws (see ESI† for details) in citrate-coated tubes (anti-coagulant tubes) and retained for 5 min. The citrated tubes were then centrifuged at 2500g for 5 min at 4 °C. The supernatants were collected and maintained at −20 °C. The plasma samples were aliquoted into 1.5 ml cryovials to avoid re-thawing and re-freezing of the plasma. In some cases, the plasma was centrifuged again at 2500g for 3 min at 4 °C to completely remove the blood cells.More than ten patients were considered for each group. Preliminary analyses showed that there are no considerable differences among the protein composition of coronas from patients with the same disease/health condition. Indeed, the same plasmas showed more similarity in their hard corona composition. In the next step, the hard corona of the age and sex-matched patients, with different diseases/states, was obtained from three subjects. The intensity of protein bands involved in the hard corona of each disease was calculated using the Image J software and the variance was calculated according to the typical formula.
2.2. Nanoparticles (NPs)
Sulfonated polystyrene NPs with an average size of 100 nm and amorphous silica NPs with an average size of 90 nm were used as received from Invitrogen by Life Technologies (USA) and Kisker-Biotech Inc. (Germany), respectively. According to the company, both nanoparticles must have the same size (100 nm). But our particle size analysis revealed that the silica NPs are slightly smaller. However the aim of the study is to investigate the effect of protein source (i.e. plasma derived from patients with different diseases) on the formation of corona regardless of NPs; neither the NP size in general nor this slight NP size variation affect the interpretation of results. The silica and polystyrene NPs are part of the first materials group investigated for safety at the nanoscale (see this report for example http://www.ehsnanonews.com/nanoarchive/ehsnnv03no4update.pdf) and these NPs are the focus of researchers for the biological and medical application.15,18,39 That is the reason why these NPs were chosen in this study.2.3. Hard corona preparation from the human plasma with various diseases
The plasma from different patients (e.g., cancer, hemodialysis, diabetes, rheumatism, fauvism, hypercholesterolemia, hemophilia, hypofibrinogenemia, common cold, pregnancy and smoking) was diluted with PBS to a final concentration of 50% and incubated at 37 °C for 10 min. The NPs (1 mg ml−1) were diluted with PBS and incubated at 37 °C for 10 min. The NPs were then incubated with the plasma solutions at a final concentration of 100 μg ml−1 at 37 °C for 1 hour. The solutions were then centrifuged for 30 min at 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm (18 krcf) at 15 °C to pellet the NP-protein corona complexes. After removal of the supernatant, the NPs were resuspended in 1 ml of cold PBS (15 °C) and centrifuged again for 20 min at 14
000 rpm (18 krcf) at 15 °C to pellet the NP-protein corona complexes. After removal of the supernatant, the NPs were resuspended in 1 ml of cold PBS (15 °C) and centrifuged again for 20 min at 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm at 15 °C to remove the loosely attached proteins. The washing step was repeated three times to obtain the final pellet (i.e., the NP-hard corona sample). To obtain a final concentration of 100 μg ml−1, 100 μl of NP stock solution (1 mg ml−1) was added to 900 μl of plasma–PBS solution (500 μl of plasma + 400 μl of PBS). The total volume of each reaction mixture was 1000 μl containing 500 μl (of plasma) + 400 μl (of PBS) + 100 μl (of NP stock solution (1 mg ml−1)). Since slight temperature changes affect the adsorption of proteins on the NP-surface, the NPs (100 μl of stock solution) and plasma–PBS solution (900 μl) were pre-incubated for 10 min at 37 °C. Then the NPs and plasma–PBS solution having exactly the same temperature (37 °C) were mixed and the process of protein corona preparation was continued. This strategy increases the accuracy of hard corona preparation and decreases the experimental bias.17,55,56 Of note, the employed procedure for the hard corona preparation is a well-established method, which was introduced by other investigators in the field (in terms of particle concentration and incubation time).8,18,39
000 rpm at 15 °C to remove the loosely attached proteins. The washing step was repeated three times to obtain the final pellet (i.e., the NP-hard corona sample). To obtain a final concentration of 100 μg ml−1, 100 μl of NP stock solution (1 mg ml−1) was added to 900 μl of plasma–PBS solution (500 μl of plasma + 400 μl of PBS). The total volume of each reaction mixture was 1000 μl containing 500 μl (of plasma) + 400 μl (of PBS) + 100 μl (of NP stock solution (1 mg ml−1)). Since slight temperature changes affect the adsorption of proteins on the NP-surface, the NPs (100 μl of stock solution) and plasma–PBS solution (900 μl) were pre-incubated for 10 min at 37 °C. Then the NPs and plasma–PBS solution having exactly the same temperature (37 °C) were mixed and the process of protein corona preparation was continued. This strategy increases the accuracy of hard corona preparation and decreases the experimental bias.17,55,56 Of note, the employed procedure for the hard corona preparation is a well-established method, which was introduced by other investigators in the field (in terms of particle concentration and incubation time).8,18,39
2.4. SDS-PAGE
The hard corona-coated NPs were re-suspended in 500 μl of PBS containing loading buffer and boiled for 10 min at 100 °C. Identical volumes of each sample were loaded on a 15% polyacrylamide gel and run at 120 V, 80 mA for approximately 100 min. Since the aim of the study was to investigate whether plasma as a protein source (patient with different disorder) determines the corona composition, we stain the proteins in SDS-PAGEs with highly sensitive silver. All protein bands in SDS-PAGEs were stained with highly sensitive silver nitrate. The silver staining allows us to observe much more protein bands and in particular low abundant proteins than coomassie blue staining. The limit of the silver staining appeared to be between 0.1 and 0.5 ng for each protein band in a SDS-PAGE. To evaluate the protein content of the plasma derived from different patients, the collected plasmas were diluted (1%) and re-suspended in 10 μl of loading buffer and boiled for 10 minutes at 100 °C (without NPs). Finally, the gel was stained using the coomassie blue method.2.5. Characterization of sulfonated polystyrene (SPS) and SiO2 NPs
The SPS and SiO2 NPs were characterized using a FEI Titan 80-300 transmission electron microscope (TEM) to examine their morphology and size (Table S2 of ESI† and Fig. 1). DLS measurements were performed with a Malvern PCS-4700 instrument equipped with a 256-channel correlator. The 488.0 nm line of a Coherent Innova-70 Ar ion laser was used as the incident beam with a power of 250 mW. The temperature was maintained at 25 °C by means of an external circulator. The zeta potentials were determined using a Malvern Zetasizer 3000 HSa.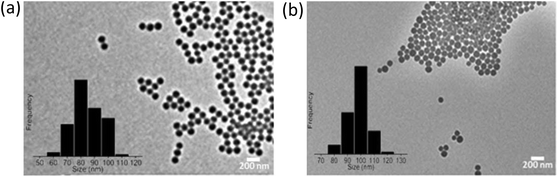 | ||
| Fig. 1 TEM images of (a) hydrophobic sulfonated polystyrene and (b) hydrophilic amorphous silica (SiO2). | ||
2.6. Densitometry analysis of protein bands from the hard coronas of different diseases
A semi-quantitative densitometry analysis was performed to measure the intensity of the protein bands from the hard coronas of different patients. The intensities of the protein bands in various ranges (i.e., >100 kDa, 100–40 kDa and <40 kDa) were measured using the Image J software (Version1.410).3. Results
3.1. The effect of plasma source on NP coronas
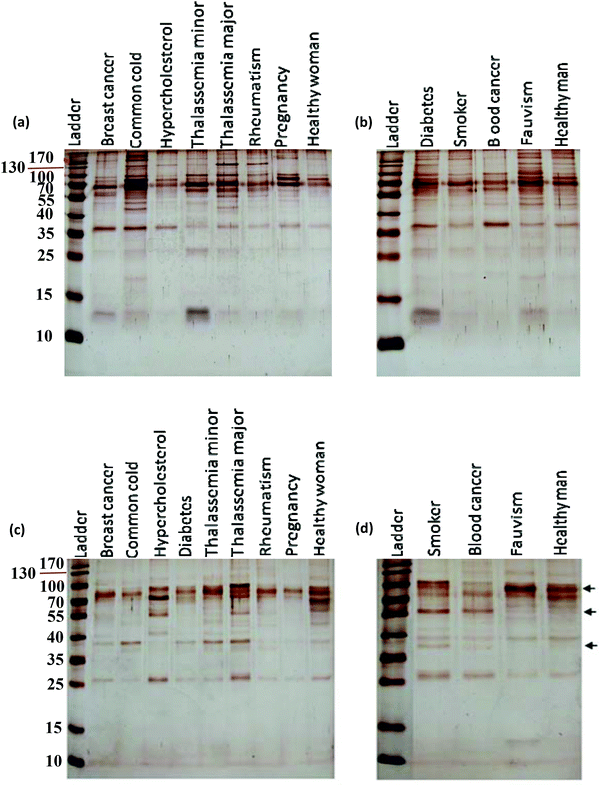
After incubation with the various plasma samples, the hard corona-coated NPs were analyzed under the same conditions (see ESI† for details). Based on the protein patterns of the various hard coronas (Fig. 2), it is clear that the protein compositions of various diseases are completely different. For example, there were significant differences in the composition and band intensities of the hard coronas of rheumatism, thalassemia and blood cancer cases compared to healthy subjects. In contrast, the protein composition of the corona from the fauvism and breast cancer patients showed only a slight difference compared to the control (Fig. 2a and b). As shown in Fig. 2c and d, the protein patterns of the coronas derived from various patients were completely different from each other (for silica NPs). For example, the corona composition of blood cancer patients and smokers was different from that of the healthy man. Of note, the most striking observation was the appearance of two protein bands at ∼37 and ∼57 kDa and the disappearance of the protein band at ∼90 kDa in these cases.
3.2. Plasma source and corona formation
3.3. Densitometry analysis of the protein bands in the coronas of different patients
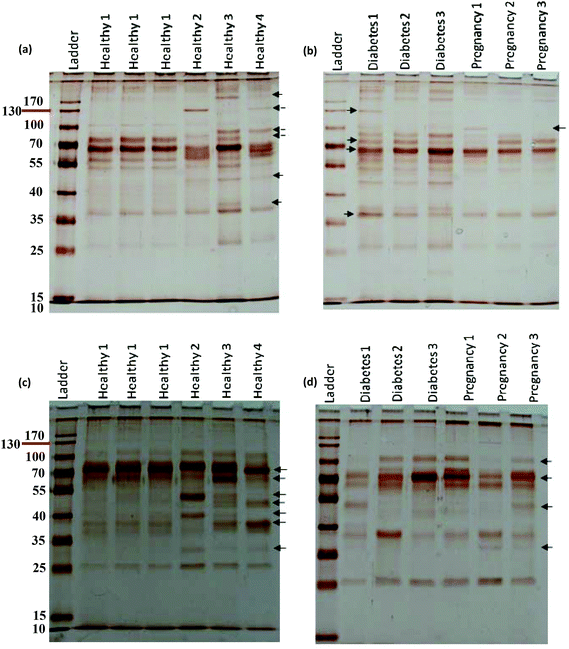
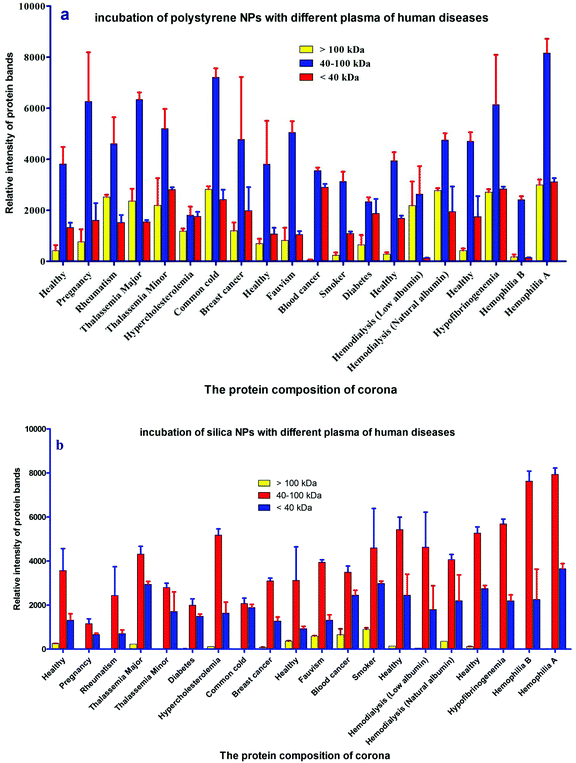
For further analysis, a semi-quantitative densitometry experiment was performed to assess the relative amounts of proteins (based on the protein band intensity), which were classified based on various molecular weight ranges (i.e., >100 kDa, 40–100 kDa, and <40 kDa) of hard coronas from NPs incubated with different plasma samples. The results (see Fig. 5a and 5b) showed variations in the pattern of NP coronas for plasma samples from different diseases. More specifically, there were variations in the intensity of most protein bands (see Fig. S1 and S2 of ESI†), while the intensities of protein bands in the hard coronas of the same subject (diabetes and pregnancy) were similar (see Fig. 5a and 5b). Moreover, the relative amounts of protein in different hard coronas obtained from the same plasma (healthy number 1) were exactly the same (see Fig. S3 and S4 of ESI†). Therefore, it seems that there is no considerable variation in the intensity of proteins in the hard coronas from the plasma of the same subject (Fig. 6).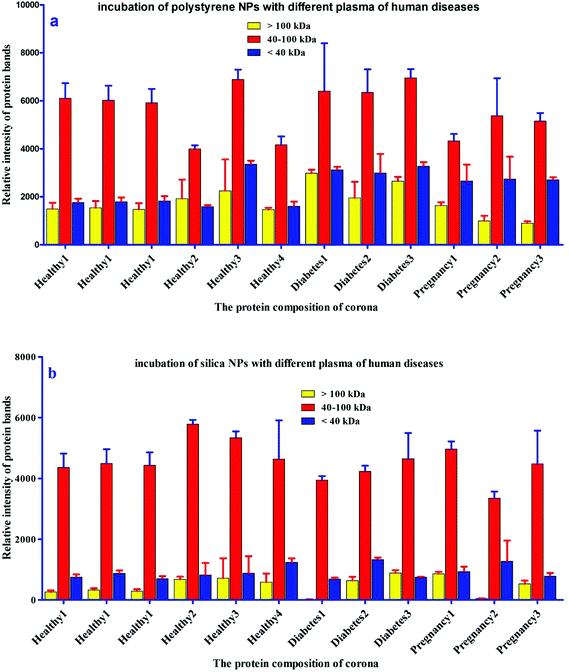 | ||
| Fig. 6 Histogram of the alterations in band intensity during incubation of (a) polystyrene and (b) silica NPs with plasma (50%) from different healthy, pregnant and diabetic cases. | ||
3.4. Dynamic light scattering (DLS) and zeta-potential of the NP hard corona complex for different patients
The protein content of the corona affects the size and surface charge of the NP.18 To investigate whether altered plasmas from different diseases influence the size and surface charge of the NPs, the size and surface charge of NP hard coronas were measured in different patient plasma samples. Dynamic light scattering (DLS) demonstrated that the sizes of the polystyrene/silica NP-protein complexes were higher than for the bare NPs (without hard coronas). On the other hand, the zeta potential for polystyrene/silica NP-protein complexes was lower compared to the bare NPs. As predicted by the SDS-PAGE results, the size distribution and zeta potential values of the hard coronas from various diseases were different from each other, implying that plasma alterations affect the compositions of hard coronas in different patients (Tables 1 and 2).| Disease | ζ-Potential (mV) of hard corona (PS) | ζ-Potential (mV) of hard corona (silica) | DLS (nm) of hard corona (PS) | DLS (nm) of hard corona (silica) |
|---|---|---|---|---|
| PS, polystyrene. | ||||
| Thalassemia minor | 0.54 | −5.43 | 134.42 ± 31.34 | 125.23 ± 35.67 |
| Thalassemia major | −8.47 | −7.68 | 119.8 ± 41.55 | 116.55 ± 39.74 |
| Hemodialysis (low albumin) | −10.50 | −6.78 | 131.34 ± 44.68 | 140.93 ± 33.96 |
| Hemodialysis (natural albumin) | −7.96 | −6.37 | 127.53 ± 36 | 133.64 ± 41.46 |
| Hemophilia B | −9.54 | −8.96 | 135.73 ± 30.55 | 131.73 ± 32.37 |
| Hemophilia A | −10.20 | −9.13 | 132.68 ± 38.93 | 126.49 ± 43.49 |
| Hypofibrinogenemia | −6.44 | −7.76 | 120.92 ± 36.95 | 122.30 ± 47.40 |
| Healthy man | −9.72 | −8.79 | 124.67 ± 40.33 | 135.30 ± 34.78 |
| Fauvism | −10.60 | −9.41 | 143.65 ± 48.71 | 129.43± 46.43 |
| Pregnancy | −7.59 | −5.54 | 132.47 ± 43.149 | 137.72 ± 31.87 |
| Common cold | −8.58 | −9.12 | 130.98 ± 49.52 | 125.53 ± 42.67 |
| Breast cancer | −10.30 | −7.39 | 130.14 ± 43.84 | 143.92 ± 39.44 |
| Rheumatism | −9.76 | −5.93 | 123.89 ± 30.98 | 137.64 ± 36.85 |
| Smoker | −9.58 | −8.27 | 122.816 ± 39.53 | 134.28 ± 47.11 |
| Diabetes | −11.80 | −10.31 | 134.86 ± 42.16 | 119.83 ± 31.92 |
| Hypercholesterolemia | −9.27 | −4.61 | 118.93 ± 37.36 | 132.73 ± 41.83 |
| Blood cancer | −9.56 | −2.94 | 127.84 ± 39.49 | 141.27 ± 33.46 |
| Healthy woman | −17.90 | −6.57 | 132.88 ± 37.38 | 130.53 ± 36.59 |
| Nanoparticle | ζ-Potential (mV) of bare NPs | DLS (nm) of bare NPs |
|---|---|---|
| Polystyrene | 37.4 ± 0.5 | 107.55 ± 30.15 |
| Silica | −24 | 103.14 ± 32.54 |
4. Discussion
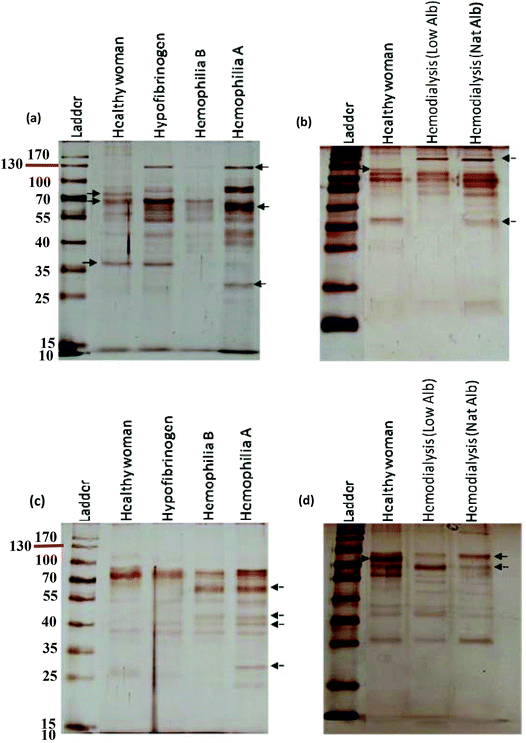
A comprehensive understanding of the interaction of NPs with plasma proteins is important for designing safe NPs with well-regulated biological identities and physiological effects. Because plasma contains various types of proteins, from a personal medicine perspective it may be helpful to establish the concept of personalized NP applications.43–45 Human plasma contains more than 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 proteins of which a very small fraction of proteins are highly abundant (such as albumin, IgG, fibrinogen, apolipoproteins, complement factors, etc.) and the rest of proteins are very low abundant. This small fraction of highly abundant proteins constitutes approximately more than 90% of plasma protein concentrations, which hampers the identification of low abundant proteins in plasma by mass spectrometry analysis.57–62 Previous studies showed that for silica and polystyrene NPs, the hard corona mainly consists of albumin, fibrinogen, immunoglobulin, complement factors and apolipoproteins.15,18 These are the highly abundant proteins of plasma. Of note, the aim of this study is to discover a key factor at the nano-bio interface, which implies that the hard corona of different patients/individuals (protein source) is different from each other and the assumption is not correct. As the images of SDS-PAGE gels strongly approved the significant changes in the hard corona of different patients/individuals in terms of the composition and amount of bound proteins (protein source), and these gels are silver stained, further analyses were not reasonable to perform due to the silver staining, which is highly sensitive and many proteins can be visualized by such staining, but the amount of stained proteins observed in the SDS-band is not enough for mass spectrometry analysis (in this regard, highly abundant proteins are exceptions). Since plasma as a protein source derived from different patients with different diseases (the so-called genetic background) plays an important role in the formation of corona, we applied highly sensitive silver staining to visualize as much as possible protein bands in the SDS gels made from patient plasma with different diseases. However, we measured the intensity of each band to assess the relative amount of each protein present in the fraction derived from each patient. Indeed, our approach makes it possible to observe the clear effect of the plasma source (derived from different genetic backgrounds = patients with different diseases) on the formation of corona. The formation of the protein corona is complex and continuous, whereby many proteins compete to bind to the NP surface as a function of their concentrations, structures and affinities to the NP surface.16 The hard corona consists of strongly adsorbed proteins having a long residence time on the NP surface.44 The formation of hard corona occurred so quickly (during the first seconds of incubation) and the composition of hard corona is constant over different time periods.10 It must be mentioned that in this study, we precisely used the most common method of hard corona preparation. In this method, the NPs should be incubated with plasma for 1 hour.10,15,17,18,39,55 Recently, Tenzer et al.10 showed that the hard corona composition (for different NPs) had no change during different exposure time (0.5–480 minutes), whereas the amount of adsorbed proteins changed significantly over time. They suggested that the adsorption of proteins on the NP-surface cannot be explained by the Vroman effect alone. For the first time, we showed that the diseases/conditions influencing the concentration/conformation of plasma proteins may affect the competitive binding of proteins to the NP-surface. This may lead to significant changes in the hard corona of different patients in terms of the composition and amount of bound proteins. Therefore, we suggest that the adsorption of plasma proteins on the NP-surface is a multi-factorial process in which the protein source (e.g. plasma source) plays an important role in the constitution of corona.
000 proteins of which a very small fraction of proteins are highly abundant (such as albumin, IgG, fibrinogen, apolipoproteins, complement factors, etc.) and the rest of proteins are very low abundant. This small fraction of highly abundant proteins constitutes approximately more than 90% of plasma protein concentrations, which hampers the identification of low abundant proteins in plasma by mass spectrometry analysis.57–62 Previous studies showed that for silica and polystyrene NPs, the hard corona mainly consists of albumin, fibrinogen, immunoglobulin, complement factors and apolipoproteins.15,18 These are the highly abundant proteins of plasma. Of note, the aim of this study is to discover a key factor at the nano-bio interface, which implies that the hard corona of different patients/individuals (protein source) is different from each other and the assumption is not correct. As the images of SDS-PAGE gels strongly approved the significant changes in the hard corona of different patients/individuals in terms of the composition and amount of bound proteins (protein source), and these gels are silver stained, further analyses were not reasonable to perform due to the silver staining, which is highly sensitive and many proteins can be visualized by such staining, but the amount of stained proteins observed in the SDS-band is not enough for mass spectrometry analysis (in this regard, highly abundant proteins are exceptions). Since plasma as a protein source derived from different patients with different diseases (the so-called genetic background) plays an important role in the formation of corona, we applied highly sensitive silver staining to visualize as much as possible protein bands in the SDS gels made from patient plasma with different diseases. However, we measured the intensity of each band to assess the relative amount of each protein present in the fraction derived from each patient. Indeed, our approach makes it possible to observe the clear effect of the plasma source (derived from different genetic backgrounds = patients with different diseases) on the formation of corona. The formation of the protein corona is complex and continuous, whereby many proteins compete to bind to the NP surface as a function of their concentrations, structures and affinities to the NP surface.16 The hard corona consists of strongly adsorbed proteins having a long residence time on the NP surface.44 The formation of hard corona occurred so quickly (during the first seconds of incubation) and the composition of hard corona is constant over different time periods.10 It must be mentioned that in this study, we precisely used the most common method of hard corona preparation. In this method, the NPs should be incubated with plasma for 1 hour.10,15,17,18,39,55 Recently, Tenzer et al.10 showed that the hard corona composition (for different NPs) had no change during different exposure time (0.5–480 minutes), whereas the amount of adsorbed proteins changed significantly over time. They suggested that the adsorption of proteins on the NP-surface cannot be explained by the Vroman effect alone. For the first time, we showed that the diseases/conditions influencing the concentration/conformation of plasma proteins may affect the competitive binding of proteins to the NP-surface. This may lead to significant changes in the hard corona of different patients in terms of the composition and amount of bound proteins. Therefore, we suggest that the adsorption of plasma proteins on the NP-surface is a multi-factorial process in which the protein source (e.g. plasma source) plays an important role in the constitution of corona.
It is well known that each patient has distinct plasma alterations that depend on the heterogeneity, period and severity of the disease, individual genetic variations and environmental factors.20–22 Therefore, we hypothesized that alterations in plasma protein concentrations/structures are mediated by diseases, thus affecting the composition of the hard corona. Intriguingly, changes in the plasma protein composition due to disease alter the NP hard corona composition, which in turn gives the NPs an unpredictable biological identity (compared to the current in vitro data, which do not consider the plasma alteration effect) that determines their biological fate. Notably, the association of multiple disorders can also affect the composition of the hard corona. In addition, the data demonstrate that the composition of the hard corona varies among healthy individuals, as well as among patients with various diseases/medical conditions. This may be due to individual heterogeneity, unspecified disorders, and gene–gene interactions. Thus, the same NPs may have different protein coronas in different individuals. More specifically, depending on the type, period and severity of the disease (which determines the plasma alterations), each patient may have a personalized protein corona; in this case, the patients with the same disease/medical conditions showed more similarity in their protein pattern of corona. It means that precise corona information is needed for safe and high yielding nano-based therapy (see Fig. 7). Therefore, the PPC concept must be considered not only to obtain reliable data on protein corona formation but also to develop high yielding personally designed nanomaterials for therapy.
5. Conclusion
Our results reveal that human plasma samples obtained from different diseases differentially affect the decoration of the protein coronas on the surfaces of NPs. More specifically, we found that changes in plasma protein concentrations and structures (mediated by different disease states) affect the formation of the protein corona on a NP. This study provides an in-depth knowledge on plasma samples from many different diseases and their interactions with NPs, opening new avenues for designing safe and high yielding NPs for clinical and biological applications with respect to personalized nanomedicine.Caution
To collect samples from patients who had only one disorder, we counseled the patients and their doctors. In addition, with permission we checked the medical profiles of the patients. However, it is possible that unrecognized concurrent disorders may occur in patients that could influence the formation of the hard corona.Notes and references
- M. J. Hajipour, K. M. Fromm, A. AkbarAshkarran, D. Jimenez de Aberasturi, I. R. d. Larramendi, T. Rojo, V. Serpooshan, W. J. Parak and M. Mahmoudi, Trends Biotechnol., 2012, 30, 609–610 CrossRef PubMed.
- S. Krol, R. Macrez, F. Docagne, G. Defer, S. Laurent, M. Rahman, M. J. Hajipour, P. G. Kehoe and M. Mahmoudi, Chem. Rev., 2012, 113, 1877–1903 CrossRef PubMed.
- M. Mahmoudi, M. A. Sahraian, M. A. Shokrgozar and S. Laurent, ACS Chem. Neurosci., 2011, 2, 118–140 CrossRef CAS PubMed.
- I. Lynch and K. A. Dawson, Nano Today, 2008, 3, 40–47 CrossRef CAS.
- M. Mahmoudi, S. Sant, B. Wang, S. Laurent and T. Sen, Adv. Drug Delivery Rev., 2011, 63, 24–46 CrossRef CAS PubMed.
- M. F. Engel, A. J. Visser and C. P. van Mierlo, Proc. Natl. Acad. Sci. U. S. A., 2004, 101, 11316–11321 CrossRef CAS PubMed.
- A. E. Nel, L. Mädler, D. Velegol, T. Xia, E. M. Hoek, P. Somasundaran, F. Klaessig, V. Castranova and M. Thompson, Nat. Mater., 2009, 8, 543–557 CrossRef CAS PubMed.
- M. P. Monopoli, C. Åberg, A. Salvati and K. A. Dawson, Nat. Nanotechnol., 2012, 7, 779–786 CrossRef CAS PubMed.
- M. Lundqvist, J. Stigler, T. Cedervall, T. Berggård, M. B. Flanagan, I. Lynch, G. Elia and K. Dawson, ACS Nano, 2011, 5, 7503–7509 CrossRef CAS PubMed.
- S. Tenzer, D. Docter, J. Kuharev, A. Musyanovych, V. Fetz, R. Hecht, F. Schlenk, D. Fischer, K. Kiouptsi and C. Reinhardt, Nat. Nanotechnol., 2013, 8, 772–781 CrossRef CAS PubMed.
- S. Tenzer, D. Docter, S. Rosfa, A. Wlodarski, J. r. Kuharev, A. Rekik, S. K. Knauer, C. Bantz, T. Nawroth and C. Bier, ACS Nano, 2011, 5, 7155–7167 CrossRef CAS PubMed.
- S. Laurent, E. NG, C. Thirifays, L. Lakiss, G. Goupil, S. Mintova, C. Burtea, E. Oveisi, H. Cecile and M. Mahmoudi, Toxicol. Res., 2013, 2, 270–279 RSC.
- M. Mahmoudi, M. P. Monopoli, M. Rezaei, I. Lynch, F. Bertoli, J. J. McManus and K. A. Dawson, ChemBioChem, 2013, 14, 525–525 CrossRef CAS.
- M. S. Ehrenberg, A. E. Friedman, J. N. Finkelstein, G. Oberdörster and J. L. McGrath, Biomaterials, 2009, 30, 603–610 CrossRef CAS PubMed.
- M. Lundqvist, J. Stigler, G. Elia, I. Lynch, T. Cedervall and K. A. Dawson, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 14265–14270 CrossRef CAS PubMed.
- M. Mahmoudi, I. Lynch, M. R. Ejtehadi, M. P. Monopoli, F. B. Bombelli and S. Laurent, Chem. Rev., 2011, 111, 5610 CrossRef CAS PubMed.
- M. Mahmoudi, A. M. Abdelmonem, S. Behzadi, J. H. Clement, S. Dutz, M. R. Ejtehadi, R. Hartmann, K. Kantner, U. Linne, P. Maffre and W. Parak, ACS Nano, 2013, 7, 6555–6562 CrossRef CAS PubMed.
- M. P. Monopoli, D. Walczyk, A. Campbell, G. Elia, I. Lynch, F. Baldelli Bombelli and K. A. Dawson, J. Am. Chem. Soc., 2011, 133, 2525–2534 CrossRef CAS PubMed.
- M. Ghavami, S. Saffar, B. A. Emamy, A. Peirovi, M. A. Shokrgozar, V. Serpooshan and M. Mahmoudi, RSC Adv., 2013, 3, 1119–1126 RSC.
- S. Laurent, C. Burtea, C. Thirifays, F. Rezaee and M. Mahmoudi, J. Colloid Interface Sci., 2013, 392, 431–445 CrossRef CAS PubMed.
- G. P. Chrousos, Nat. Rev. Endocrinol., 2009, 5, 374–381 CrossRef CAS PubMed.
- J. Brewer and O. Gurel, Nanotech. L. & Bus., 2009, 6, 45 Search PubMed.
- D. Nedelkov, U. A. Kiernan, E. E. Niederkofler, K. A. Tubbs and R. W. Nelson, Proc. Natl. Acad. Sci. U. S. A., 2005, 102, 10852–10857 CrossRef CAS PubMed.
- L. Kennedy, T. D. Mehl, E. Elder, M. Varghese and T. J. Merimee, Diabetes, 1982, 31, 52–56 CrossRef CAS.
- K. B. Pandey, N. Mishra and S. I. Rizvi, Clin. Biochem., 2010, 43, 508–511 CrossRef CAS PubMed.
- G. Engström, P. Lind, B. Hedblad, L. Stavenow, L. Janzon and F. Lindgärde, Circulation, 2002, 105, 2632–2637 CrossRef.
- Y.-P. Lin, C.-Y. Yang, C.-C. Liao, W.-C. Yu, C.-W. Chi and C.-H. Lin, PLoS One, 2012, 7, e40232 CAS.
- H. B. Salt, Ann. Rheumat. Dis., 1951, 10, 46 CrossRef CAS.
- S. Acharya and D. Dimichele, Haemophilia, 2008, 14, 1151–1158 CrossRef CAS PubMed.
- J. Joseph, C. Baker, M. Sprang and E. Bermes, Ann. Clin. Lab. Sci., 1978, 8, 130–141 CAS.
- K. S. Kelly-Spratt, S. J. Pitteri, K. E. Gurley, D. Liggitt, A. Chin, J. Kennedy, C.-H. Wong, Q. Zhang, T. B. Buson and H. Wang, PLoS One, 2011, 6, e19721 CAS.
- H. Jin, B.-J. Webb-Robertson, E. S. Peterson, R. Tan, D. J. Bigelow, M. B. Scholand, J. R. Hoidal, J. G. Pounds and R. C. Zangar, Environ. Health Perspect., 2011, 119, 1314 CrossRef CAS PubMed.
- J. E. Frank, Am. Family Phys., 2005, 72, 1277–1282 Search PubMed.
- D. Bowen, Mol. Pathol., 2002, 55, 1–18 CrossRef CAS.
- N. Chu, J. Chang and S. Shieh, Int. J. Obesity, 2003, 27, 735–739 CrossRef CAS PubMed.
- C. P. Riley, X. Zhang, H. Nakshatri, B. Schneider, F. E. Regnier, J. Adamec and C. Buck, J. Trans. Med., 2011, 9, 1–11 CrossRef PubMed.
- S. Alonso-Orgaz, L. Moreno, C. Macaya, L. Rico, P. J. Mateos-Cáceres, D. Sacristán, F. Pérez-Vizcaíno, A. Segura, J. Tamargo and A. López-Farré, J. Proteome Res., 2006, 5, 2301–2308 CrossRef CAS PubMed.
- K. R. Viel, A. Ameri, T. C. Abshire, R. V. Iyer, R. G. Watts, C. Lutcher, C. Channell, S. A. Cole, K. M. Fernstrom and S. Nakaya, N. Engl. J. Med., 2009, 360, 1618–1627 CrossRef CAS PubMed.
- D. Walczyk, F. B. Bombelli, M. P. Monopoli, I. Lynch and K. A. Dawson, J. Am. Chem. Soc., 2010, 132, 5761–5768 CrossRef CAS PubMed.
- B. Furie and B. C. Furie, Cell, 1988, 53, 505–518 CrossRef CAS.
- L. K. Curtiss and J. L. Witztum, Diabetes, 1985, 34, 452–461 CrossRef CAS.
- R. Bucala and A. Cerami, Adv. Pharmacol., 1992, 23, 222 Search PubMed.
- X.-Q. Zhang, X. Xu, N. Bertrand, E. Pridgen, A. Swami and O. C. Farokhzad, Adv. Drug Delivery Rev., 2012, 64, 1363–1384 CrossRef CAS PubMed.
- C. D. Walkey and W. C. Chan, Chem. Soc. Rev., 2012, 41, 2780–2799 RSC.
- H. Mao, W. Chen, S. Laurent, C. Thirifays, C. Burtea, F. Rezaee and M. Mahmoudi, Colloids Surf., B, 2013, 109, 212–218 CrossRef CAS PubMed.
- F. Rezaee, A. Maas, J. H. Verheijen and J. Koopman, Thromb. Haemostasis, 2001, 85, 1025–1030 CAS.
- F. Rezaee, M. J. Gijbels, E. H. Offerman, M. van der Linden, M. P. M. De Maat and J. H. Verheijen, Thromb. Haemostasis, 2002, 88, 329–334 CAS.
- A. A. Gulledge, F. Rezaee, J. H. Verheijen and S. T. Lord, Thromb. Haemostasis, 2001, 86, 511–516 CAS.
- M. Dashty, V. Akbarkhanzadeh, C. J. Zeebregts, C. A. Spek, E. J. Sijbrands, M. P. Peppelenbosch and F. Rezaee, Sci. Rep., 2012, 2, 787 Search PubMed.
- F. Rezaee, A. Maas, M. P. M. De Maat, J. H. Verheijen and J. Koopman, Atherosclerosis, 2002, 164, 37–44 CrossRef CAS.
- J. Koopman, A. Maas, F. Rezaee, J. Havekes, J. Verheijen, M. Gijbels and F. Haverkate, Fibrinolysis Proteolysis, 1997, 11, 19 CrossRef CAS.
- B. Badlou and F. Rezaee, Blood Transfus., 2013, 11, 315 Search PubMed.
- B. Badlou and F. Rezaee, Blood Transfus., 2014, 12(Suppl 1), S245 Search PubMed.
- F. Rezaee, M. J. Gijbels, E. H. Offerman and J. H. Verheijen, Thromb. Haemostasis, 2003, 90, 710–716 CAS.
- M. Mahmoudi, M. A. Shokrgozar and S. Behzadi, Nanoscale, 2013, 5, 3240–3244 RSC.
- A. Lensiak, F. Fenaroli, M. P. Monopoli, C. Åberg, K. A. Dawson and A. Salvati, J. Am. Chem. Soc., 2012, 6, 5845–5857 Search PubMed.
- N. L. Anderson and N. G. Anderson, Mol. Cell Proteomics, 2002, 1, 845–867 CAS.
- M. Dashty, M. Motazacker, H. Levels, M. Mahmoudi, M. Peppelenbosch and F. Rezaee, Thromb. Haemostasis, 2014, 111, 518–530 CrossRef CAS PubMed.
- F. Rezaee, B. Casetta, J. Levels, D. Speijer and J. C. M. Meijers, Proteomics, 2006, 6, 721–730 CrossRef CAS PubMed.
- K. C. S. Queiroz, R. A. Tio, C. Zeebregts, M. F. Bijlsma, F. Zijlstra, C. V. Ferreira, C. A. Spek, B. A. Badlou, M. l de Vries, M. P. Peppelenbosch and F. Rezaee, J. Proteome Res., 2010, 9, 6052–6059 CrossRef CAS PubMed.
- M. Dashti, W. Kulik, F. Hoek, E. Veerman, M. P. Peppelenbosch and F. Rezaee, Sci. Rep., 2011, 1, 139 Search PubMed.
- K. Meijer, M. de Vries, S. Al-Lahham, M. Bruinenberg, D. Weening, M. Dijkstra, N. Kloosterhuis, R. J. van der Leij, H. van der Want, B. J. Kroesen, R. Vonk and F. Rezaee, Sci. Rep., 2011, 6, e17154 CAS.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c4bm00131a |
| ‡ These authors equally contributed to this manuscript. |
| This journal is © The Royal Society of Chemistry 2014 |