Open Access Article
Open Access ArticleGenetically encoded self-assembly of large amyloid fibers†
D. M.
Ridgley
,
B. G.
Freedman
,
P. W.
Lee
and
J. R.
Barone
*
Biological Systems Engineering, Virginia Tech, 203 Seitz Hall, Blacksburg, VA 24061, USA. E-mail: jbarone@vt.edu; Tel: (+1 540) 231-0680
First published on 14th January 2014
Abstract
“Functional” amyloids are found throughout nature as robust materials. We have discovered that “template” and “adder” proteins cooperatively self-assemble into micrometer-sized amyloid fibers with a controllable, hierarchical structure. Here, Escherichia coli is genetically engineered to express a template protein, Gd20, that can initiate self-assembly of large amyloid fibrils and fibers. Through atomic force microscopy (AFM) we found that expression of Gd20 produces large amyloid fibrils of 490 nm diameter and 2–15 μm length. Addition of an extracellular adder protein, myoglobin, continues self-assembly to form amyloid tapes with widths of ∼7.5 μm, heights of ∼400 nm, and lengths exceeding 100 μm. Without myoglobin the amyloid fibrils are metastable over time. When myoglobin is present, the amyloid fiber continues self-assembling to a width of ∼18 μm and height of ∼1 μm. Experimental results demonstrate that large amyloid fibers with a tailored stiffness and morphology can be engineered at the DNA level, spanning four orders of magnitude.
Introduction
Amyloid fibrils are usually studied in the context of neurodegenerative diseases.1 “Functional” amyloids used by organisms for survival are being discovered, such as in the barnacle cement of Megabalanus rosa.2–4 The functional nature, high specific modulus, and ability to be self-assembled from a variety of proteins make amyloids interesting designer nanomaterials.5–7 There are many studies of the nanometer-sized amyloid fibril, which displays consistent morphology and properties when produced from a variety of proteins.8–10 The amyloid fibril is composed of high strand density β-sheets that are oriented perpendicular to the fibril axis.11,12 Recent research has demonstrated that it is possible to grow large amyloid fibers with tailored morphology (circular or rectangular cross-sections) and modulus (0.1–2.5 GPa) in vitro by utilizing a “template” and “adder” protein mixture.13 The large amyloid fibers form from nanometer fibrils that continue to interact to the micrometer scale.14 Different amyloid fibers can be self-assembled by altering solution conditions, template to adder protein molar ratios, or adder protein length and sequence.13,15 Template proteins are capable of conformation change on their own but adder proteins are not. The predominantly hydrophobic template protein assumes the β-sheet as the lowest energy state to hide most hydrophobic groups between the sheets. For a population of template proteins, most hydrophobic groups are hidden inside the β-sheets but the template still has hydrophobic faces. Hydrophobic groups on the α-helices of predominantly hydrophilic adder proteins prefer the exposed hydrophobic groups on the template, undergo an α to β transition, and “add” into the structure to form larger β-sheets and fibrils. It is the “addition” event that allows further self-assembly beyond the stable nanometer template scale and is what makes this system unique. The conformational changes resulting from addition allow a large entropy gain by the water relative to template formation alone. Further assembly involves a competition between hydrophobic and hydrogen bonding interactions that differentiates morphology and properties based on adder protein properties.Observation of a controllable, hierarchical protein self-assembly process fostered the notion that large structures of varying shape and modulus could be encoded at the genetic level. For instance, a cylindrical fiber 20 μm across and 104 μm long with a 1 GPa modulus could be built simply by inserting the DNA of the correct template and adder proteins into a cellular expression system. The fiber could be constructed into a composite by expressing a third, non-assembling protein or polymer to act as a matrix. Silk-like copolymers and recombinant collagen have been produced by expressing the desired protein(s) in E. coli or P. pastoris.16,17 Unfortunately, the target protein(s) must undergo substantial post-expression processing to assemble the protein fiber.18,19 Scheibel et al. used Saccharomyces cerevisiae to express the Sup35p prion determinant protein that self-assembled into amyloid fibril templates to form conducting nanowires.20 Here we investigate the potential for genetically encoded self-assembly through the expression of our template protein, Gd20, in E. coli with and without the extracellular addition of the My adder protein.
Materials and methods
DNA insertion
All chemicals were obtained from Thermo Fisher Scientific unless otherwise specified. All bacteria were grown on LB media supplemented with 100 mg L−1 ampicillin or 15 g L−1 agar where appropriate.21 All Escherichia coli strains, plasmids, and enzymes were purchased from New England BioLabs (Ipswich, MA, USA). The oligopeptide sequence of Gd20 (Start-TFLILALLAIVATTATTAVR-Stop-Stop) was optimized for expression in E. coli strain K12 using the JCat program22 and the resulting double-stranded DNA was ordered from Integrated DNA Technologies (Coralville, IA). The gene fragment was amplified by PCR (Eppendorf, Hauppauge, NY) with oligonucleotide primers Forward (5′-GGTGGTCATATGAC CTTCCTGATCCTGGC-3′) and Reverse (5′-GTGGTTGCTCTTCCGCATTATTAACGAACA GCGGTGGT-3′) which included NdeI and SapI restriction sites for entry into vector pTXB1 (New England BioLabs). The resulting plasmid, pTXGd20, was transformed into E. coli NEB5-alpha by standard protocols21 and verified by PCR. The plasmid DNA was purified by miniprep (Gerard BioTec, Oxford, OH, USA) and transformed into the protein expression E. coli cell line ER2566.Gd20 expression
Cells containing the Gd20 plasmid were grown up in LB-Amp to an optical density at 600 nm (0D600) of 0.5. Isopropyl β-D-1-thiogalactopyranoside (IPTG) was added at a concentration of 0.4 mM to X and XMy to induce expression of Gd20. Myoglobin (My, from equine skeletal muscle, Sigma-Aldrich, St. Louis, MO, UniProt P68082) was added to NXMy and XMy at a concentration of 12.5 mg ml−1, which was simply the concentration used previously and was expected to be sufficient to allow addition into any Gd20 template produced.13Fourier-transform infrared (FT-IR) spectroscopy
The four cell cultures were dried on Teflon coated aluminum foil after 43 and 72 hours of incubation. Spectra of the dried cultures were acquired on a Thermo Fisher Scientific, Inc. (Waltham, MA) 6700 FT-IR with a Smart Orbit diamond attenuated total reflectance (ATR) accessory. Spectra were acquired using Omnic v8.0 software in the same manner as previously reported.13,15 The spectral region from 1720–1580 cm−1 was isolated and manually smoothed with the Savitzky–Golay algorithm using 9–13 points. The second derivative of the Amide I spectral region was taken without filtering to identify the individual Amide I components.Atomic force microscopy (AFM)
AFM was performed on an Innova AFM (Bruker, Santa Barbara, CA) after 37 hours (X, NX, XMy) and 72 hours (X, NX, XMy and NXMy) with samples prepared, imaged, and enhanced in the same manner as previously reported.14 Nanoindentation of the large fibrils (X) and fibers (XMy) was performed in contact mode with a 0.01–0.025 Ohm cm antimony-doped Si probe (Bruker, part: MPP-31123-10, R: 8 nm and k: 0.9 N m−1). The deflection-displacement curves were obtained with NanoDrive v8.01 software using the Point Spectroscopy mode with 512 points taken at a 0.5 μm s−1 approach/retreat rate. The curves were converted to a force-displacement curve utilizing the probe's spring constant according to the manufacturer.23 Young's modulus was extrapolated according to Guo et al. with Poisson's ratio taken as 0.3, which has been used in previous amyloid studies.24,25 11 indentations were performed on 2 XMy fibers (XMy) and 6 X large fibrils (X) each at 37 hours. 7 indentations were performed on 1 XMy fiber and 4 X large fibrils each at 72 hours. The averages ± the standard errors are reported.Results and discussion
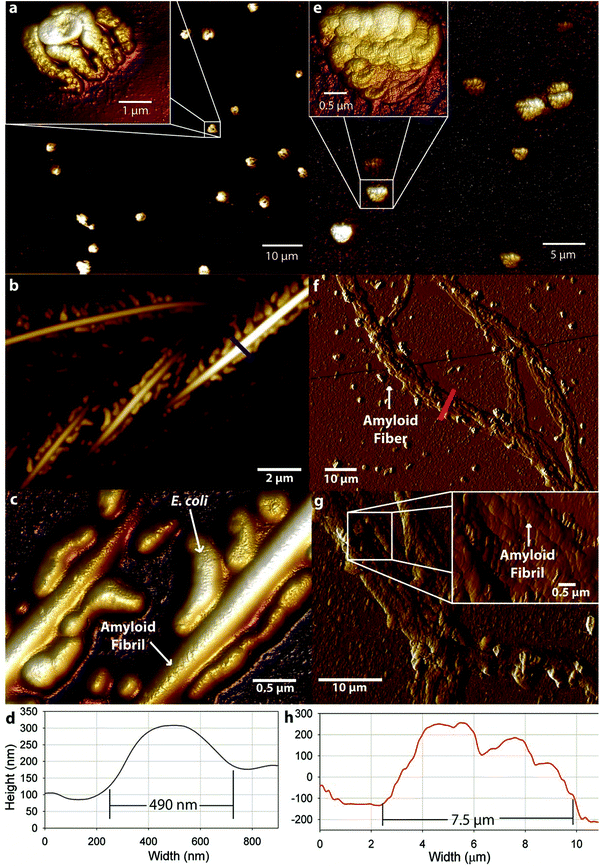
Four cell cultures are analyzed with atomic force microscopy (AFM) and Fourier transform-infrared (FT-IR) spectroscopy to determine the presence of amyloid fibrils or fibers and their structure: (1) cells without expression of Gd20 (NX), (2) cells without expression of Gd20 but My addition (NXMy), (3) cells with expression of Gd20 (X), and (4) cells with expression of Gd20 and My addition (XMy).The four cell cultures (NX, NXMy, X, XMy) are analyzed with AFM to assess fiber formation in the presence or absence of Gd20 or My. The NX culture shows coalesced, partially lysed E. coli with no fibrous structures (Fig. 1a). When Gd20 is expressed in the X culture several fibrous structures appear throughout the solution. X produces large amyloid fibrils of 490 nm diameter and 2–15 μm length (Fig. 1b–d) similar to the large fibrils observed in vitro previously.14 Closer examination of the fibrils reveals that E. coli is attached to the surface (Fig. 1c). The small molecular weight and high hydrophobicity of Gd20 (ESI Table 1† and Fig. 1) may cause some of it to embed in the cell lipid bilayer and become partially exposed outside the cell surface, acting as a bridge between the cell and the self-assembled amyloid fibril (Scheme 1).
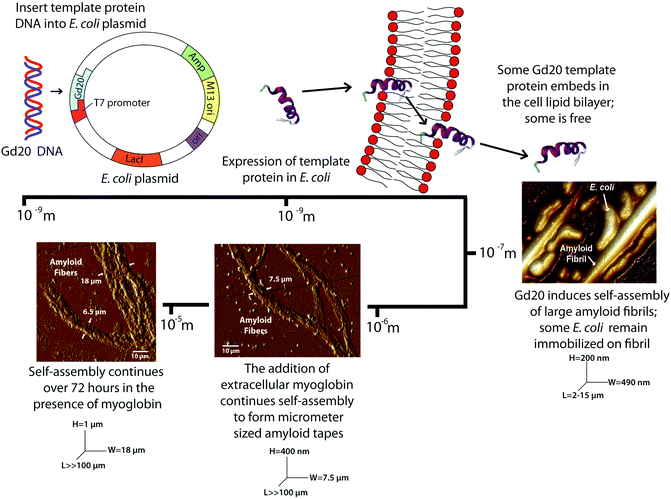 | ||
| Scheme 1 Genetic encoding of the Gd20 template protein into an E. coli plasmid results in the self-assembly of amyloid fibrils and fibers spanning four orders of magnitude. | ||
Previous studies have indicated that a template and an adder protein are necessary for the formation of large amyloid fibers at near physiological conditions.13 Extracellular My is added to the cultures to determine how a proven adder protein affects the formation of amyloid fibers. The NXMy culture tests how My interacts with E. coli in the absence of Gd20. NXMy does not form fibrous structures and the My appears to coalesce along with E. coli, similar to what is observed in the NX culture upon spin coating (Fig. 1e). On the other hand, XMy self-assembles into large amyloid tapes with width of ∼7.5 μm, height of ∼400 nm, and lengths exceeding 100 μm (Fig. 1f–h). The tape is composed of large fibrils of widths 400–900 nm (Fig. 1g) that resemble those in Fig. 1b and c from the X culture and previous studies.14 This process largely mimics the self-assembly of protein mixtures in vitro where Gd20 and My aggregate into (a) ∼30 nm wide protofibrils, (b) protofibrils aggregate into ∼100 nm wide fibrils, (c) fibrils aggregate into 400–900 nm wide large fibrils of elliptical cross-section that then (d) aggregate extensively laterally and limitedly vertically into ∼7.5 μm wide tapes of rectangular cross-section.13–15 Protofibrils have been shown to have an intrinsic twist due to the left-handed chirality of the amino acids within the high density β-sheet core.26 This was confirmed with WG and Gd:My template and adder systems in vitro with AFM.14 Spectroscopic studies have indicated that hydrophobic interactions promote self-assembly of smaller structures into larger structures, which is what drives aggregation to higher scales here as well (Scheme 1 and Fig. 3).13,27 For Gd:My, the twist at the protofibril stage does not persist to higher stages.
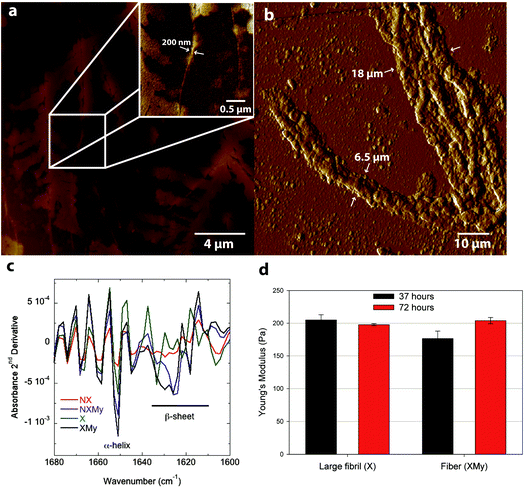
X and XMy cultures are imaged after 72 hours to determine if the amyloid structures continue to self-assemble over time. Fibrils from the X culture are of smaller width, ∼200 nm (Fig. 2a and ESI Fig. 2†), than at 37 hours, ∼490 nm (Fig. 1b–d). The decrease in width suggests that the 490 nm large fibril is metastable or not fully compacted at early times. Indeed, few studies have been able to surpass the ∼100 nm width barrier for amyloid fibrils or tapes formed after long incubation times.28,29 So the metastability of a larger structure may be intrinsic to certain amyloid forming proteins, especially systems that have only 1 assembling protein. After 72 hours XMy assembles into a tape similar to early times but with a larger width of ∼18 μm and height of ∼1 μm (Fig. 2b and ESI Fig. 2†) suggesting that unassembled proteins in a mixture of template and adder proteins continue to interact as time progresses to grow toward an equilibrium structure.
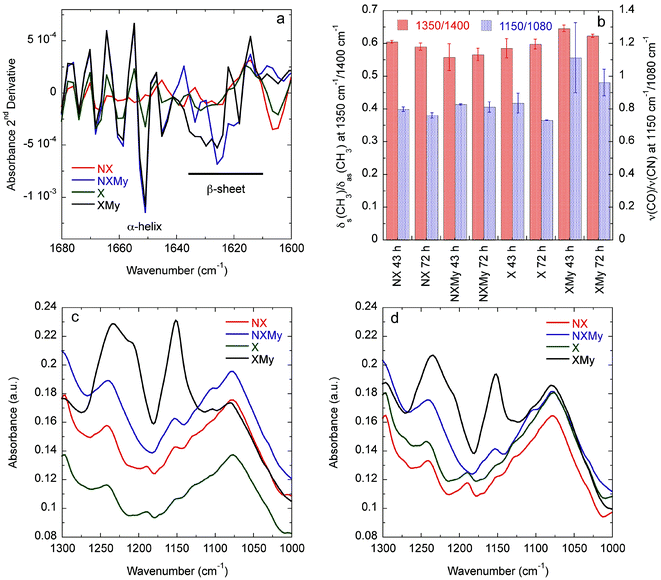
Analysis of the Amide I absorbance obtained from Fourier transform-infrared (FT-IR) spectroscopy can be used to determine protein secondary structure.30,31 NX does not contain any significant structure (Fig. 2c and 3a). Expression of Gd20 (X) shows the presence of α-helices (∼1650 cm−1), the predominant structure of Gd20 as predicted with PSIPRED,32 and β-sheets (1610–1630 cm−1) as the template forms, including high strand density β-sheets found in amyloids at <1625 cm−1.33 X β-sheets appear metastable as the ratio of the area of the β-sheet region to the α-helix region decreases from 2.1 at 43 hours to 1.8 at 72 hours (Fig. 3a). The addition of My is evident from increased α-helical content, the predominant My secondary structure. β-sheet formation occurs when My is exposed to E. coli (NXMy) or E. coli that express Gd20 (XMy) at times coincident with the AFM data in Fig. 1. Native My is 79% α-helix with no β-sheet content. We have observed consistent α to β transitions in our in vitro fiber-forming systems with My as the adder protein and this would appear to be the case with NXMy and XMy. NXMy does not show fibril or fiber formation but My can self-assemble into amyloid fibrils under the right conditions.34 Here, My forms β-sheets, the elementary amyloid structure, probably on the hydrophobic cell surface, which mimics our template protein characteristics to allow the α to β transition but not fibril formation. Proteins have been shown to undergo α to β transitions on hydrophobic surfaces.13,35 XMy displays the largest β-sheet content and consistent large fiber formation indicating that the Gd20 template can influence β-sheet formation and is important to fiber formation. XMy has the highest ratio of δs(CH3)/δas(CH3) at 1350 cm−1/1400 cm−1, which we term “hydrophobic packing” and describes the interdigitation of alanine (A), isoleucine (I), leucine (L), and valine (V) amino acid side groups as one driving force for self-assembly of large amyloid fibrils and fibers (Fig. 3b).15 More importantly, XMy has dramatic changes in the 1250–1050 cm−1 region, which is quantified by the ratio of ν(CO)/ν(CN) at 1150 cm−1/1080 cm−1 (Fig. 3b–d).36 Gd20 contains 4 threonine (T) and My contains a significant amount of amino acids with CO and CN in the side groups (ESI Fig. 1†) so the profound increase in ν(CO) and ν(CN) would indicate these amino acids release from My α-helices and aggregate with each other.
Modulus results for X large fibrils and XMy tapes, obtained from AFM nanoindentation in contact mode, support the conclusion that XMy continues self-assembly into a stable large tape with an increasing modulus, while X forms a metastable large fibril as evidenced by a decreasing modulus over time (Fig. 2d). These moduli are much lower than previously reported results for fibrils, fibers, and tapes.13,15,37 The in vivo formed XMy tapes (Fig. 2b) show that the large fibrils are not fully aggregated together and thus do not produce as cohesive a tape as observed from in vitro systems.13–15 This could be for several reasons: (1) some portion of the Gd20 template protein remains embedded in the cell membrane while the remainder allows for limited self-assembly with itself or My (Scheme 1) and (2) the molar concentration of fully excreted Gd20 may be sufficient to initiate My conformation change and addition into the self-assembling structure but that addition is limited due to a limited Gd20 concentration because the template![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) adder molar ratio has been shown to play a role in the self-assembly of large amyloid fibers.13
adder molar ratio has been shown to play a role in the self-assembly of large amyloid fibers.13
Conclusion
By utilizing E. coli to express an amyloid-forming template protein it is possible to grow micrometer sized amyloid tapes upon addition of a suitable adder protein. XMy forms tapes of the same size, morphology, and fibrillar hierarchy as observed in vitro.14 These tapes can vary in modulus and can be coaxed to twist into cylinders to change morphology and properties.15 By understanding self-assembly from the molecular to the macroscopic scale, it is possible to build fibers of predictable cross section, from rectangular to circular, and modulus, from soft to rigid, by controlling the type and amount of template and adder proteins. Since protein amino acid sequence can be controlled at the genetic level, it is then also possible to encode the properties of a macroscopic structure at the genetic level in a predictable and controllable manner. It is envisioned that large-scale structures for use in engineering applications could be encoded at the genetic level. The scheme also shows that it is possible to program cells to build their own fibrous scaffold, making the self-assembly process a unique tissue engineering motif. Furthermore, this method could be used to reconstruct the cell cytoskeleton or to recompartmentalize cells to make more robust organisms for industrial bioprocessing. It is hopeful that this study will provide the basis for future genetic engineering of spontaneously forming macroscopic biomaterials.Acknowledgements
Generous funding through NSF-CMMI-0856262 and the USDA funded Virginia Tech Biodesign and Bioprocessing Research Center is gratefully acknowledged.Notes and references
- C. M. Dobson, Trends Biochem. Sci., 1999, 24, 329–332 CrossRef CAS.
- D. M. Fowler, A. V. Koulov, W. E. Balch and J. W. Kelly, Trends Biochem. Sci., 2007, 32, 217–224 CrossRef CAS PubMed.
- M. F. B. G. Gebbink, D. Claessen, B. Bouma, L. Dijkhuizen and H. A. B. Wosten, Nat. Rev. Microbiol., 2005, 3, 333–341 CrossRef CAS PubMed.
- D. E. Barlow, G. H. Dickinson, B. Orihuela, J. L. Kulp III, D. Rittschof and K. J. Wahl, Langmuir, 2010, 26, 6549–6556 CrossRef CAS PubMed.
- S. Keten, Z. Xu, B. Ihle and M. J. Buehler, Nat. Mater., 2010, 9, 359–367 CrossRef CAS PubMed.
- T. P. J. Knowles and M. J. Buehler, Nat. Nanotechnol., 2011, 6, 469–479 CrossRef CAS PubMed.
- C. E. MacPhee and C. M. Dobson, J. Am. Chem. Soc., 2000, 122, 12707–12713 CrossRef CAS.
- M. Bouchard, J. Zurdo, E. J. Nettleton, C. M. Dobson and C. V. Robinson, Protein Sci., 2000, 9, 1960–1967 CrossRef CAS PubMed.
- T. P. Knowles, A. W. Fitzpatrick, S. Meehan, H. R. Mott, M. Vendruscolo, C. M. Dobson and M. E. Welland, Science, 2007, 318, 1900–1903 CrossRef CAS PubMed.
- J. Adamcik, J.-M. Jung, J. Flakowski, P. De Los Rios, G. Dietler and R. Mezzenga, Nat. Nanotechnol., 2010, 5, 423–428 CrossRef CAS PubMed.
- M. Bouchard, J. Zurdo, E. J. Nettleton, C. M. Dobson and C. V. Robinson, Protein Sci., 2000, 9, 1960–1967 CrossRef CAS PubMed.
- L. Nielsen, S. Frokjaer, J. F. Carpenter and J. Brange, J. Pharm. Sci., 2001, 90, 29–37 CrossRef CAS.
- D. M. Ridgley, K. C. Ebanks and J. R. Barone, Biomacromolecules, 2011, 12, 3770–3779 CrossRef CAS PubMed.
- D. M. Ridgley and J. R. Barone, ACS Nano, 2013, 7, 1006–1015 CrossRef CAS PubMed.
- D. M. Ridgley, E. C. Claunch and J. R. Barone, Soft Matter, 2012, 8, 10298–10306 RSC.
- A. Vuorela, J. Myllyharju, R. Nissi, T. Pihlajaniemi and K. I. Kivirikko, EMBO J., 1997, 16, 6702–6712 CrossRef CAS PubMed.
- J. Cappello, J. Crissman, M. Dorman, M. Mikolajczak, G. Textor, M. Marquet and F. Ferrari, Biotechnol. Prog., 1990, 6, 198–202 CrossRef CAS PubMed.
- D. L. Kaplan, Nat. Biotechnol., 2002, 20, 239–240 CrossRef CAS PubMed.
- S. T. Krishnaji, G. Bratzel, M. E. Kinahan, J. A. Kluge, C. Staii, J. Y. Wong, M. J. Buehler and D. L. Kaplan, Adv. Funct. Mater., 2013, 23, 241–253 CrossRef CAS.
- T. Scheibel, R. Parthasarathy, G. Sawicki, X.-M. Lin, H. Jaeger and S. L. Lindquist, Proc. Natl. Acad. Sci. U. S. A., 2003, 100, 4527–4532 CrossRef CAS PubMed.
- J. Sambrook and D. W. Russell, Molecular Cloning: A Laboratory Manual, 2001 Search PubMed.
- A. Grote, K. Hiller, M. Scheer, R. Münch, B. Nörtemann, D. C. Hempel and D. Jahn, Nucleic Acids Res., 2005, 33, W526–W531 CrossRef CAS PubMed.
- Innova AFM User Manual, Bruker Instruments, 2013 Search PubMed.
- S. Guo and B. B. Akhremitchev, Biomacromolecules, 2006, 7, 1630–1636 CrossRef CAS PubMed.
- L. L. del Mercato, G. Maruccio, P. P. Pompa, B. Bochicchio, A. M. Tamburro, R. Cingolani and R. Rinaldi, Biomacromolecules, 2008, 9, 796–803 CrossRef CAS PubMed.
- A. W. P. Fitzpatrick, G. T. Debelouchina, M. J. Bayro, D. K. Clare, M. A. Caporini, V. S. Bajaj, C. P. Jaroniec, L. Wang, V. Ladizhansky, S. A. Müller, C. E. MacPhee, C. A. Waudby, H. R. Mott, A. De Simone, T. P. J. Knowles, H. R. Saibil, M. Vendruscolo, E. V. Orlova, R. G. Griffin and C. M. Dobson, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 5468–5473 CrossRef CAS PubMed.
- D. M. Ridgley, E. C. Claunch and J. R. Barone, Appl. Spectrosc., 2013, 57, 1417–1426 CrossRef PubMed.
- C. C. Lara, J. Adamcik, S. Jordens and R. Mezzenga, Biomacromolecules, 2011, 12, 1868–1875 CrossRef CAS PubMed.
- J. L. Jiménez, E. J. Nettleton, M. Bouchard, C. V. Robinson, C. M. Dobson and H. R. Saibil, Proc. Natl. Acad. Sci. U. S. A., 2002, 99, 9196–9201 CrossRef PubMed.
- S. Krimm and J. Bandekar, in Advances in Protein Chemistry, ed. C. B. Anfinsen, J. T. Edsall and F. M. Richards, Academic Press, Inc., Orlando, 1986, vol. 38, pp. 181–364 Search PubMed.
- A. Dong, P. Huang and W. S. Caughey, Biochemistry, 1990, 29, 3303–3308 CrossRef CAS.
- D. W. A. Buchan, S. M. Ward, A. E. Lobley, T. C. O. Nugent, K. Bryson and D. T. Jones, Nucleic Acids Res., 2010, 38, W563–W568 CrossRef CAS PubMed.
- G. Zandomeneghi, M. R. H. Krebs, M. G. McCammon and M. Fändrich, Protein Sci., 2004, 13, 3314–3321 CrossRef CAS PubMed.
- M. Fandrich, M. A. Fletcher and C. M. Dobson, Nature, 2001, 410, 165–166 CrossRef CAS PubMed.
- A. Sethuraman, G. Vedantham, T. Imoto, T. Przybycien and G. Belfort, Proteins: Struct., Funct., Bioinf., 2004, 56, 669–678 CrossRef CAS PubMed.
- A. Barth, Prog. Biophys. Mol. Biol., 2000, 74, 141–173 CrossRef CAS.
- J. F. Smith, T. P. Knowles, C. M. Dobson, C. E. MacPhee and M. E. Welland, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 15806–15811 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: A table of the intrinsic properties of Gd20 and myoglobin as well as their respective amino acid sequences, cross-sectional measurements of the X fibril and XMy fiber at 72 hours. See DOI: 10.1039/c3bm60223k |
| This journal is © The Royal Society of Chemistry 2014 |