Gold nanoparticle-based colorimetric chiral discrimination of histidine: application to determining the enantiomeric excess of histidine†
Seong Hyeok
Seo
,
Sudeok
Kim
and
Min Su
Han
*
Department of Chemistry, Chung-Ang University, Seoul 156-756, Republic of Korea. E-mail: mshan@cau.ac.kr; Fax: +82 2 825 4736; Tel: +82 2 820 5198
First published on 5th November 2013
Abstract
The proposed colorimetric assay system consisting of L-Pro group-functionalized gold nanoparticles (L-Pro–AuNPs) and Cu2+ ions can discriminate the absolute configuration of His and determine the enantiomeric excess of His.
Chirality is a unique and significant property in the fields of pharmaceutics, biochemistry, and asymmetric catalyst development.1 Most bioactive materials of in vivo systems are mainly composed of enantiomers, such as carbohydrates, peptides, and amino acids, which play key roles in metabolism, toxicity, and biochemical activities. The chiralities of these materials demonstrate very different pharmaceutical and biochemical effects.2 Amino acid chirality is particularly important because amino acids have been intensively used as chiral building blocks for various drug intermediates.3 The development of a method for determining the chirality and enantiomeric excess of amino acids is valuable in the fields of pharmaceutical science and biotechnology.4 Many methods have already been developed to discriminate the chirality of amino acids, including chiral high-performance liquid chromatography (HPLC), gas chromatography (GC), and capillary electrophoresis (CE).5 Although these methods are useful in analyzing the chirality of amino acids, most of them require a laborious setup process and are impractical for real time analysis. Therefore, it is desirable to develop a simple and rapid method that discriminates the chirality of amino acids. Many chiral sensors of amino acids have been developed based on fluorometry, colorimetry, electrochemistry, and circular dichroism (CD).6 These tools provide simple and quick discrimination of amino acid chirality.
Nanoparticles are a desirable platform for developing colorimetric chemosensors because they have unique optical properties, such as higher extinction coefficients and distance-dependent surface plasmon resonance.7 Many nanoparticle-based colorimetric chemosensors have been developed to detect small molecules, metal ions, DNA, and proteins.8 However, only a few nanoparticle-based colorimetric sensing systems have been reported for the detection of chiral compounds.9 Recently, N-acetyl-L-cysteine-modified gold nanoparticles, nucleotide-capped silver nanoparticles, and zinc tetraphenylporphyrin-modified silver nanoparticle cluster-bearing L-arginine have been developed as colorimetric chiral recognition systems for amino acids.10 Although some nanoparticle-based colorimetric chiral recognition systems for amino acids have been developed, it is still challenging to develop a colorimetric sensing system that visually discriminates chirality and determines the enantiomeric excess in amino acids. The chiral recognition of amino acids is of tremendous pharmaceutical value.
Proline (Pro) is the only cyclic α-amino acid, and has a more rigid structure than other amino acids. For this reason, it is widely used as a chiral auxiliary for synthesis and as the stationary phase for chiral separation in chromatography.11 In the presence of Cu2+ ions, proline and other amino acids readily combine to construct diastereomers, with stability determined by the amino acid chirality. In general, a combination of the same enantiomers has a more stable structure because the stability of ternary complexes depends on the steric hindrance and trans effect of the ligand.12 Therefore, the L-Pro–Cu2+ complex with the L-enantiomer has a stronger binding affinity than with the D-enantiomer, which has been intensively used to separate enantiomers in chromatography.11f–m Based on this information, we expected that amino acid–Cu2+ binds to L-Pro on gold nanoparticles (AuNPs) and forms amino acid–Cu2+–L-Pro complexes on AuNPs. The binding affinity of amino acid–Cu2+ to L-Pro on the AuNP is determined by the chirality of amino acids. Therefore, as shown in Scheme 1, only L-amino acid can construct the amino acid–Cu2+–L-Pro complex on gold nanoparticles under certain conditions, which results in the aggregation of AuNPs in assay solution because the stability of AuNPs is affected by any modification of ligands on these AuNPs.8,13
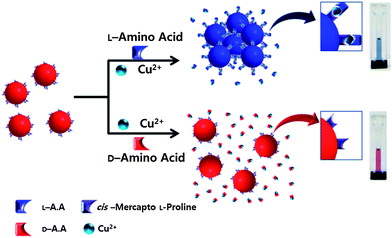 | ||
| Scheme 1 Schematic representation of the colorimetric sensor for the discrimination of the enantiomer. | ||
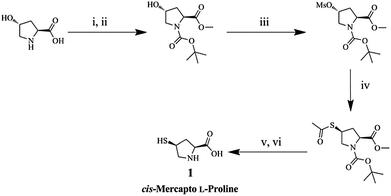
Chiral ligand (1) was synthesized from trans-hydroxy L-Pro including a mercapto moiety to build a chiral environment on the AuNP surface, as shown in Scheme 2. Hydroxyproline was protected by N-Boc and a methyl ester moiety. The protected proline was converted to the corresponding acetylthio proline by simple substitution, and 1 was obtained by deprotection reactions. The AuNPs functionalized with 1 (L-Pro–AuNPs) were prepared as chiral amino acid probes by mixing with citric acid capped-AuNPs.
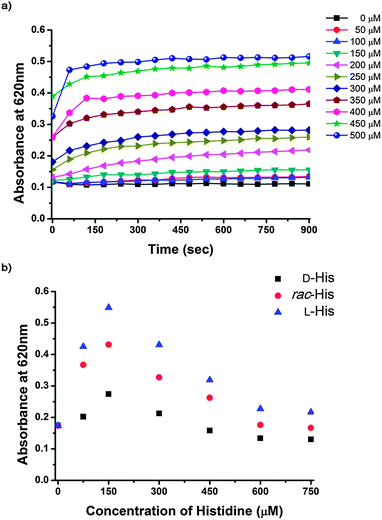
L-Pro–AuNPs can aggregate in the presence of Cu2+, which is one component of this assay system, because L-Pro–AuNPs can interconnect with Cu2+ ions. The stability of L-Pro–AuNPs decreased with increasing concentrations of Cu2+ ions. If a low concentration of Cu2+ ions was used in this assay system to avoid aggregation, the assay system may not respond to amino acids. To enhance the amino acid sensitivity of this assay, it was necessary to determine the highest concentration of Cu2+ that did not affect the stability of L-Pro–AuNPs within a given detection time. The optimal concentration of Cu2+ in this study was determined by measuring absorbance changes in L-Pro–AuNPs in the presence of various concentrations of Cu2+. As shown in Fig. 1a, in the presence of Cu2+ concentrations lower than 150 μM, the absorbance of L-Pro–AuNPs at 620 nm barely changed in 15 minutes. Therefore, colorimetric chiral assay systems for amino acids in this study were prepared by combining L-Pro–AuNPs (3 nM) with Cu2+ (150 μM). To determine the optimal concentration of amino acids in this system, UV-Vis spectra of the assay solution were recorded 15 min after adding various concentrations of enantiomerically pure L-His, D-His, and rac-His because His has a high binding affinity to Cu2+ in some amino acids. Absorbance changes in the assay mixtures were recorded at 620 nm and are shown in Fig. 1b. These samples showed distinct UV-Vis absorbance values and the differences were as large as ∼0.2 and correlated well with the enantiomeric excess of His at 300 μM of the analyte. However, when His concentration exceeded 450 μM, the difference decreased. The difference was very small at 750 μM, which is likely due to the fact that high concentrations of His prevent Cu2+-mediated interactions between His and L-Pro in L-Pro–AuNPs by masking Cu2+ ions.
The responses of chiral assay systems to enantiomerically pure Phe, Asp, Lys, and His were evaluated because these amino acids have representative side chains including hydrophobic, carboxylic, amine, and imidazole groups, respectively. In a typical experiment, a stock solution of amino acid was added to the assay mixture containing L-Pro–AuNPs (3 nM) and Cu2+ ions (150 μM) so that the final concentration was 300 μM.
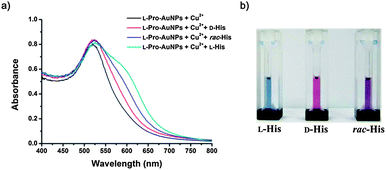
Absorbance changes of the assay solution were recorded 15 min after adding enantiomerically pure amino acid. The absorbance difference between the assay solutions in the presence of the His enantiomer was distinct, as shown in Fig. 2a, while the differences were small for other amino acids (see the ESI†). An assay system that can discriminate the chirality of enantiomers without a spectrometer is of particular interest due to its convenience, but only a few such systems have been reported. The use of the present assay system for this purpose is demonstrated in Fig. 2b. This chiral assay system was blue in the presence of L-His, red with D-His, and purple with rac-His.
The enantiomeric excess is defined as the absolute difference between the mole fractions of each enantiomer. The determination of enantiomeric excess is important in many fields, such as chiral drug production and asymmetric catalyst discovery.4,14 Therefore, this system is applied to determine the enantiomeric excess of His. The relationship between the enantiomeric excess of His and absorbance was obtained at constant His concentration (300 μM) for this assay system. As shown in Fig. 3, the observed absorbance intensities at 620 nm were linearly proportional to the enantiomeric excess of L-His. In addition, it is possible that this system can be applied to separate L-His from racemic His because these presented AuNPs can recognize only L-His under certain conditions. However, this separation is impractical because the number of chiral ligands on AuNPs is very smaller than total His under these chiral assay conditions.
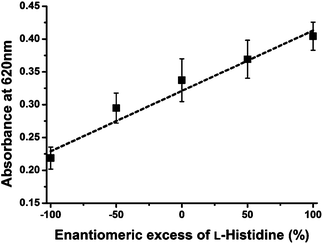 | ||
| Fig. 3 Plot of assay solution absorbance intensities at 620 nm versus the enantiomeric excess of L-His (300 μM) combined with Cu2+ (150 μM). | ||
In conclusion, a AuNP-based colorimetric chiral discrimination system was developed for histidine using AuNPs and a simple synthesizable proline derivative as a chiral ligand. This system allows the measurement of enantiomeric excess and chirality of His by the naked eye and provides a more rapid analysis time than traditional methods such as GC, HPLC, CD, and NMR.
This work was supported by a National Research Foundation of Korea Grant funded by the Korean Government (NRF-2013R1A2A2A01007000) and the Priority Research Centers Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology (2009-0093817).
Notes and references
- (a) N. M. Davies and X. W. Teng, Advances in Pharmacy, 2003, 1, 242 Search PubMed; (b) J. M. Hawkins and T. J. N. Watson, Angew. Chem., Int. Ed., 2004, 43, 3224 CrossRef CAS PubMed; (c) M. Diéguez, O. Pàmies and C. Claver, Chem. Rev., 2004, 104, 3189 CrossRef PubMed.
- (a) I. Agranat and H. Caner, Drug Discovery Today, 1999, 4, 313 CrossRef CAS; (b) D. J. Cordato, L. E. Mather and G. K. Herkes, J. Clin. Neurosci., 2003, 10, 649 CrossRef CAS PubMed; (c) K. Mori, Bioorg. Med. Chem., 2007, 15, 7505 CrossRef CAS PubMed.
- (a) S. Bera, D. Mondal, M. Singh and R. K. Kale, Tetrahedron, 2013, 69, 969 CrossRef CAS PubMed; (b) C. Lamberth, Tetrahedron, 2010, 66, 7239 CrossRef CAS PubMed; (c) T. Darbre and J.-L. Reymond, Acc. Chem. Res., 2006, 39, 925 CrossRef CAS PubMed; (d) S. Hanessian and L. Auzzas, Acc. Chem. Res., 2008, 41, 1241 CrossRef CAS PubMed; (e) X. Li, Y.-D. Wu and D. Yang, Acc. Chem. Res., 2008, 41, 1428 CrossRef CAS PubMed.
- (a) H. Pellissier, Tetrahedron, 2008, 64, 1563 CrossRef CAS PubMed; (b) R. N. Patel, Biomol. Eng., 2001, 17, 167 CrossRef CAS.
- (a) I. Ilisz, A. Aranyi, Z. Pataj and A. Péter, J. Chromatogr., A, 2012, 1269, 94 CrossRef CAS PubMed; (b) V. Schurig, J. Chromatogr., A, 2001, 906, 275 CrossRef CAS; (c) V. Schurig, J. Chromatogr., B: Anal. Technol. Biomed. Life Sci., 2011, 879, 3122 CrossRef CAS PubMed; (d) M. Kato and T. Toyo'oka, Chromatography, 2001, 22, 159 CAS; (e) E. Gassmmar, J. E. Kuo and R. N. Zare, Science, 1985, 230, 813 Search PubMed; (f) P. Gozel, E. Gassmann, H. Michelsen and R. N. Zare, Anal. Chem., 1987, 59, 44 CrossRef CAS; (g) T. J. Ward, Anal. Chem., 2002, 74, 2863 CrossRef CAS; (h) P. M. P. Nazareth and O. A. C. Antunes, J. Braz. Chem. Soc., 2002, 13, 658 CrossRef CAS PubMed; (i) A. Dobashi and M. Hamada, Electrophoresis, 1999, 20, 2761 CrossRef CAS.
- (a) Y. Zhou and J. Yoon, Chem. Soc. Rev., 2012, 41, 52 RSC; (b) T. Kassa, L. P. Undesser, H. Hofstetter and O. Hofstetter, Analyst, 2011, 136, 1113 RSC; (c) L. Zhu and E. V. Anslyn, J. Am. Chem. Soc., 2004, 126, 3676 CrossRef CAS PubMed; (d) L. Zhu, Z. Zhong and E. V. Anslyn, J. Am. Chem. Soc., 2005, 127, 4260 CrossRef CAS PubMed; (e) J. F. Folmer-Andersen, V. M. Lynch and E. V. Anslyn, J. Am. Chem. Soc., 2005, 127, 7986 CrossRef CAS PubMed; (f) D. Leung, J. F. Folmer-Andersen, V. M. Lynch and E. V. Anslyn, J. Am. Chem. Soc., 2008, 130, 12318 CrossRef CAS PubMed; (g) D. Leung and E. V. Anslyn, J. Am. Chem. Soc., 2008, 130, 12328 CrossRef CAS PubMed; (h) X. Mei and C. Wolf, J. Am. Chem. Soc., 2006, 128, 13326 CrossRef CAS PubMed; (i) Y. Fu, Q. Han, Q. Chen, Y. Wang, J. Zhou and Q. Zhang, Chem. Commun., 2012, 48, 2322 RSC; (j) H. Kim, S. M. So, C. P.-H. Yen, E. Vinhato, A. J. Lough, J.-I. Hong, H.-J. Kim and J. Chin, Angew. Chem., Int. Ed., 2008, 47, 8657 CrossRef CAS PubMed; (k) S. Sambasivan, D.-S. Kim and K. H. Ahn, Chem. Commun., 2010, 46, 541 RSC.
- (a) M. E. Stewart, C. R. Anderton, L. B. Thompson, J. Maria, S. K. Gray, J. A. Rogers and R. G. Nuzzo, Chem. Rev., 2008, 108, 494 CrossRef CAS PubMed; (b) N. J. Halas, S. Lal, W.-S. Chang, S. Link and P. Nordlander, Chem. Rev., 2011, 111, 3913 CrossRef CAS PubMed; (c) P. C. Ray, Chem. Rev., 2010, 110, 5332 CrossRef CAS PubMed.
- (a) N. L. Rosi and C. A. Mirkin, Chem. Rev., 2005, 105, 1547 CrossRef CAS PubMed; (b) Y. Lu and J. Liu, Acc. Chem. Res., 2007, 40, 315 CrossRef CAS PubMed; (c) S. K. Ghosh and T. Pal, Chem. Rev., 2007, 107, 4797 CrossRef CAS PubMed; (d) C. Burda, X. Chen, R. Narayanan and M. A. El-Sayed, Chem. Rev., 2005, 105, 1025 CrossRef CAS PubMed.
- (a) C. Gautier and T. Bürgi, ChemPhysChem, 2009, 10, 483 CrossRef CAS PubMed; (b) D. Balogh, Z. Zhang, A. Cecconello, J. Vavra, L. Severa, F. Teply and I. Willner, Nano Lett., 2012, 12, 5835 CrossRef CAS PubMed; (c) Z. An and M. Yamaguchi, Chem. Commun., 2012, 48, 7383 RSC; (d) W. Wei, L. Wu, C. Xu, J. Ren and X. Qu, Chem. Sci., 2013, 4, 1156 RSC.
- (a) H. Su, Q. Zheng and H. Li, J. Mater. Chem., 2012, 22, 6546 RSC; (b) Y. Sun, L. Zhang and H. Li, New J. Chem., 2012, 36, 1442 RSC; (c) M. Zhang and B.-C. Ye, Anal. Chem., 2011, 83, 1504 CrossRef CAS PubMed.
- (a) H. Pellissier, Tetrahedron, 2006, 62, 1619 CrossRef CAS PubMed; (b) L. M. Geary and P. G. Hultin, Tetrahedron: Asymmetry, 2009, 20, 131 CrossRef CAS PubMed; (c) C. W. Y. Chung and P. H. Toy, Tetrahedron: Asymmetry, 2004, 15, 387 CrossRef CAS PubMed; (d) G. Guillena and D. J. Ramón, Tetrahedron: Asymmetry, 2006, 17, 1465 CrossRef CAS PubMed; (e) G. Guillena, C. Nájera and D. J. Ramón, Tetrahedron: Asymmetry, 2007, 18, 2249 CrossRef CAS PubMed; (f) S. H. Lee, T. S. Oh and H. W. Lee, Bull. Korean Chem. Soc., 1992, 13, 280 CAS; (g) M. G. Schmid and G. Gübitz, Maced. J. Chem. Chem. Eng., 2011, 30, 127 CAS; (h) S. Zhao and Y.-M. Liu, Anal. Chim. Acta, 2001, 426, 65 CrossRef CAS; (i) X. Mu, L. Qi, H. Zhang, Y. Shen, J. Qiao and H. Ma, Talanta, 2012, 97, 349 CrossRef CAS PubMed; (j) Z. Zhang, G. Malikin and S. Lam, J. Chromatogr., A, 1992, 603, 279 CrossRef CAS; (k) C. D. Haurou, G. Declercq, P. Ramiandrasoa and J. L. Millet, J. Chromatogr., A, 1991, 547, 31 CrossRef CAS; (l) S. Lam and G. Malikin, J. Chromatogr., A, 1986, 368, 413 CrossRef CAS; (m) S. Lam, J. Chromatogr., A, 1986, 355, 157 CrossRef CAS.
- (a) S. H. Laurie, Chem. Commun., 1967, 155 RSC; (b) J. Sabolović, C. S. Tautermann, T. Loerting and K. R. Liedl, Inorg. Chem., 2003, 42, 2268 CrossRef PubMed; (c) Z. Chen, J. Lin, K. Uchiyama and T. Hobo, Anal. Sci., 2000, 16, 131 CrossRef CAS.
- (a) J. Nam, W.-G. La, S. Hwang, Y. S. Ha, N. Park, N. Won, S. Jung, S. H. Bhang, Y.-J. Ma, Y.-M. Cho, M. Jin, J. Han, J.-Y. Shin, E. K. Wang, S. G. Kim, S.-H. Cho, J. Yoo, B.-S. Kim and S. Kim, ACS Nano, 2013, 7, 3388 CrossRef CAS PubMed; (b) J. Nam, N. Won, H. Jin, H. Chung and S. Kim, J. Am. Chem. Soc., 2009, 131, 13639 CrossRef CAS PubMed; (c) A. Laromaine, L. Koh, M. Murugesan, R. V. Ulijn and M. M. Stevens, J. Am. Chem. Soc., 2007, 129, 4156 CrossRef CAS PubMed; (d) Y. Choi, N.-H. Ho and C.-H. Tung, Angew. Chem., Int. Ed., 2007, 46, 707 CrossRef CAS PubMed; (e) D. Li, Q. He, Y. Cui and J. Li, Chem. Mater., 2007, 19, 412 CrossRef CAS; (f) W. Zhao, W. Chiuman, J. C. F. Lam, M. A. Brook and Y. Li, Chem. Commun., 2007, 3729 RSC.
- R. E. Gawley, J. Org. Chem., 2006, 71, 2411 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Synthetic procedures of compound 1 and experimental details for colorimetric assay for the enantiomeric excess of histidine and discrimination of various chiral amino acids. See DOI: 10.1039/c3ay41735b |
| This journal is © The Royal Society of Chemistry 2014 |

![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
: