Hydrazinonicotinic acid as a novel matrix for highly sensitive and selective MALDI-MS analysis of oligosaccharides†
Jing
Jiao
ab,
Ying
Zhang
ab,
Pengyuan
Yang
b and
Haojie
Lu
*ab
aShanghai Cancer Center, Key Laboratory of Glycoconjugates Research Ministry of Public Health, Fudan University, Shanghai 200032, P. R. China. E-mail: luhaojie@fudan.edu.cn; Fax: +86-21-5423-7961
bDepartment of Chemistry, Institutes of Biomedical Sciences, Fudan University, Shanghai, 200032, P. R. China
First published on 15th October 2014
Abstract
Analysis of oligosaccharides with matrix-assisted laser desorption/ionization mass spectrometry (MALDI-MS) remains challenging due to their low ionization efficiency. The sensitivity achieved by MS for oligosaccharides lags far behind that for proteins/peptides. Here, hydrazinonicotinic acid (HYNIC) is proposed as a new matrix to realize highly sensitive and selective analysis of oligosaccharides in MALDI-MS. The detection limit of maltoheptaose provided by HYNIC is as low as 1 amol, which is five orders of magnitude lower than that provided by the traditional matrix 2,5-dihydroxybenzoic acid (DHB). Interestingly, HYNIC displayed remarkable selectivity for ionization of oligosaccharides, making glycans from glycoproteins become more accessible to be detected even without pre-purification, as demonstrated by the direct detection of the oligosaccharides from human serum without pre-separation of the proteins/peptides. The HYNIC matrix also possessed the virtue of higher homogeneity of crystallization and better salt tolerance (up to 200 mM NaCl, 140 mM urea and 40 mM sulfocarbamide et al.) compared with the traditional matrix DHB. Furthermore, the HYNIC matrix afforded adequate fragmentation, thus providing rich information for the structural elucidation of the oligosaccharide. Therefore, using HYNIC as the matrix to directly analyze oligosaccharides is inherently simple and straightforward.
Introduction
Glycosylation is one of the most common and complicated forms of post-translational modifications (PTMs) in eukaryotic proteins.1 Oligosaccharides, which are structurally diverse, participate in key biological progresses including protein folding, cell–cell recognition, signaling and cancer metastasis in living systems.2–6 Due to its rapidity and accuracy, matrix-assisted laser desorption/ionization mass spectrometry (MALDI-MS) has been the most important technique for the analysis of oligosaccharides.7–9 However, the lack of protonation of the basic groups and the hydrophilic nature of oligosaccharides make them more difficult to be ionized than peptides and proteins. In an attempt to redress the problem, chemical derivatization has frequently been used to improve the limit of detection (LOD) of oligosaccharides in MS.10,11 However, the gain in sensitivity through chemical derivatization is often somewhat counterbalanced by sample loss during the time-consuming sample preparation procedure.12 What is more, the problem resulting from the incomplete derivatization complicates the mass spectrum due to the coexistence of signals from native and derived glycans. Compared with the tedious derivatization methods, analysis of oligosaccharides using a proper matrix is simple and straightforward for giving signals in their native state and reducing the complexity of the spectrum. 2,5-Dihydroxybenzoic acid (DHB) is the most conventional matrix whose weak points are also obvious. First, DHB affords just passable sensitivity with LOD merely at the fmol level as far as we know.13 Second, inhomogeneous needle-shaped crystals of DHB are often formed around the rim of the MALDI spot after drying; as a result, the search for a “sweet spot” where the analytes are ionized is required.14 Therefore, development of novel matrices with high sensitivity has always been a hot research topic.13–16The suppression effect caused by analytes with higher proton affinity (i.e. peptides) leads to another difficulty in detecting glycans.7 The traditional solution to circumvent this problem is to selectively enrich the glycans from the mixtures.15,16 However, necessary manipulation of incubation and elution is a time-consuming and labor-intensive task. Selective improvement of ionization efficiency of oligosaccharides with a proper matrix would undoubtedly facilitate direct and effective mass spectrometry measurements of the trace amount of native carbohydrates from peptide mixtures. Recently, diamond nanoparticles (DNPs) were incorporated as the MALDI matrix to selectively enhance the sensitivity of the mass spectrometer to carbohydrates for the first time.17 However, like other inorganic matrices, DNP with the size of about 500 nm may lead to possible contamination of the MALDI ion source. A matrix with both high sensitivity and selectivity would especially help to discern glycans in complex samples. Herein, a novel matrix 6-hydrazinonicotinic acid (HYNIC) was investigated for highly sensitive and selective analysis of oligosaccharides. We systematically evaluated the advantages of HYNIC as a novel matrix for the analysis of different kinds of oligosaccharides including permethylated oligosaccharides, fucosylated oligosaccharides and sialylated oligosaccharides. Several aspects including the limit of detection for oligosaccharides, ability to selectively ionize oligosaccharides, shot-to-shot reproducibility, salt tolerance and the impact on the oligosaccharides fragmentation pattern were performed for investigating the superiority of the HYNIC matrix. The results showed that HYNIC could be an outstanding matrix for the analysis of oligosaccharides, e.g. enabling highly sensitive detection of maltoheptaose (DP7) as low as 1 amol accompanied by selective enhancement of oligosaccharide ions, high homogeneity of crystals and high tolerance to several kinds of buffers. More than 40 glycans released from the human serum were successfully detected using HYNIC as the matrix.
Experimental section
Materials and reagents
Hydrazinonicotinic acid (HYNIC) and the standard peptide (LSITGTYDLK) were purchased from ChinaPeptides Co., Ltd and the purity of the peptide was more than 99%. Maltoheptaose (DP7, 95%) was purchased from Hayashibara Biochemical Laboratories (Okayama, Japan). NA2F glycan and A1 glycan were purchased from Ludger. 2,5-Dihydroxybenzoic acid (DHB), trifluoroacetic acid (TFA), dithiothreitol (DTT), urea, sulfocarbamide, ammonium bicarbonate (ABC), Dextran 1000 analytical standard (Dextran 1000, 98%), nicotinic acid, trypsin (Proteomics Grade), ribonuclease B (RNase B) and bovine serum albumin (BSA) were obtained from Sigma-Aldrich (St. Louis, MO, USA). Peptide N-glycosidase (PNGase F) was obtained from New England Biolabs (Ipswich, MA, USA). Sodium chloride (NaCl) was purchased from Shanghai Chemical Reagent Company (Shanghai, China). HPLC-grade methanol and acetonitrile (ACN) were purchased from Merck (Darmstadt, Germany). Acetic acid (HAc) of analytical grade was purchased from Sinopharm Chemical Reagent Co., Ltd. (Shanghai, China). Distilled water was purified by using a Milli-Q system (Milford, MA, USA).Sample preparation
HYNIC was dissolved in methanol containing 2.0% HAc. The DHB matrix solution was prepared at the concentration of 10 mg mL−1 in ACN/water (50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50, v/v) containing 0.1% TFA. All oligosaccharides were dissolved in water in a series of concentrations and stored at 4 °C for further use. In order to generate peptide mixtures, BSA was dissolved in 50 mM ABC and denatured by boiling for 10 min; trypsin was then added into the protein solution at an enzyme/substrate ratio of 1
50, v/v) containing 0.1% TFA. All oligosaccharides were dissolved in water in a series of concentrations and stored at 4 °C for further use. In order to generate peptide mixtures, BSA was dissolved in 50 mM ABC and denatured by boiling for 10 min; trypsin was then added into the protein solution at an enzyme/substrate ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50 (w/w) for 12 h at 37 °C to produce peptides. The tryptic peptide mixtures were stored at −20 °C until further use. RNase B (0.5 mg) was dissolved in 50 mM ABC buffer (pH 8.0) and denatured by the same method. After incubating with trypsin for 12 h, PNGase F solution (0.5 μL) was added and the enzymatic deglycosylation reaction was carried out at 37 °C for 24 h. The samples were directly stored at −20 °C without separation.
50 (w/w) for 12 h at 37 °C to produce peptides. The tryptic peptide mixtures were stored at −20 °C until further use. RNase B (0.5 mg) was dissolved in 50 mM ABC buffer (pH 8.0) and denatured by the same method. After incubating with trypsin for 12 h, PNGase F solution (0.5 μL) was added and the enzymatic deglycosylation reaction was carried out at 37 °C for 24 h. The samples were directly stored at −20 °C without separation.
Human serum (10 μL) from a healthy volunteer was denatured by heating for 5 minutes in 8 M urea/0.2 M Tris–HCl buffer. Then the sample was treated with 25 mM dithiothreitol (DTT) at 57 °C for 30 minutes and alkylated with 30 mM iodoacetamide (IAA) at room temperature for 45 minutes in the dark. The denaturant and other low-molecular-weight molecules were removed by ultrafiltration. One μL PNGase F solution was added and the enzymatic deglycosylation reaction was carried out at 37 °C for 24 h. The released N-glycans were obtained by repeated ultrafiltration with pure water and lyophilized before MALDI analysis.
Mass spectrometric measurements
One μL of sample and one μL of matrix solution were spotted on the MALDI plate for MS analysis. The MALDI-TOF measurements were performed on a 5800 Proteomics Analyzer (Applied Biosystems, Framingham, MA, USA). Mass spectra were acquired in reflection mode with a Nd:YAG laser at 355 nm, a repetition rate of 400 Hz, and an acceleration voltage of 20 kV. MS/MS spectra were interpreted manually with the assistance of GlycoWorkbench software. External mass calibration was performed by using standard peptides from myoglobin digests.MSI acquisition and data processing
Mass spectrometry imaging was conducted on a 5800 Proteomics Analyzer (Applied Biosystems, Framingham, MA, USA) using 4800 imaging software. Mass conditions were set as described above. Mass spectrometric analysis was performed in the positive reflection mode and MALDI-MS spectra were recorded in the m/z range of 1000–2000. Two-dimensional ion density maps were created using the image reconstruction software (BioMap, Novartis, Basel, Switzerland).Results and discussion
Evaluation of the HYNIC matrix for highly sensitive detection of standard oligosaccharides
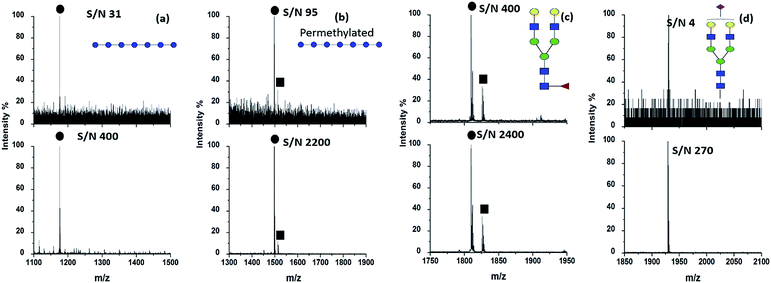
The concentration of HYNIC was optimized on the basis of the minimum amount needed to produce uniform dried crystals throughout a target spot as well as a clear mass spectrum. Our preliminary optimized work contributed to confirm that the HYNIC matrix dissolved in methanol at the concentration of 2.0 mg mL−1 containing 2% HAc can generate a high quality spectrum. A higher concentration of HYNIC generated more cluster peaks of the matrix, while a lower concentration of HYNIC did not crystallize well with the analytes. We also tried other solvents including the system of acetonitrile and water and the system of methanol and water, in which HYNIC did not crystallize on the target well after solvent evaporation. After optimization of the concentration and matrix buffer, HYNIC was used as the matrix to help ionize different kinds of oligosaccharides. Fig. 1 displays the MALDI mass spectra of four kinds of representative oligosaccharides including DP7, permethylated DP7, fucosylated oligosaccharide NA2F and sialylated oligosaccharide A1 using DHB and HYNIC as the matrix respectively. The signals of neutral oligosaccharides are often observed in the form of alkali metal adducts like [M + Na]+ or [M + K]+ because oligosaccharides have a high affinity for alkali cations.18 Therefore, [M + Na]+ is chosen for the comparison of the signal to noise (S/N) ratio between the DHB and HYNIC. The signals of sialylated oligosaccharides are obtained in negative modes. An obviously higher S/N and superior spectrum quality are obtained using the HYNIC matrix (Fig. 1, lower panel) than the DHB matrix (Fig. 1, upper panel) for all the four types of oligosaccharides. From the spectra (Fig. 1) we believed that HYNIC did not react with the reducing end of oligosaccharides because the corresponding product signals were not detected. Therefore, HYNIC acted as a matrix but not as a derivatization reagent to help improve the ionization efficiency of oligosaccharides. In addition, the molecular weight of HYNIC is 153.1 Da and the matrix peaks of HYNIC can be observed in the low mass range of the mass spectrum (data not shown), which will not interfere with the analysis of oligosaccharides. DP7 was used as a model glycan to further compare the ionization ability of HYNIC and DHB. Table S1† makes a comparison of S/N with DP7 with different loading amounts using DHB and HYNIC. With the decrease of the loading amount, DHB lost the ability to ionize DP7. By contrast, HYNIC was demonstrated as a more effective matrix. DP7 was also used as a model sample to determine the LOD. The loading amount of DP7 was decreased to the amount on a single well at which the S/N just exceeded 3. When the HYNIC matrix was used, the LOD for DP7 is 1 amol (Fig. S1b†). For comparison, the LOD provided by the traditional matrix DHB is 100 fmol (Fig. S1a†). The LOD provided by HYNIC increases by five orders of magnitude than that of DHB, and it is almost one order of magnitude lower than the LOD that was reported in a recent study.13 Dextran 1000, a more complex, branched polysaccharide composing of many glucose molecules joined into chains of different lengths19 was used to further evaluate the performance of the HYNIC matrix. Several research studies have developed a proper matrix and derivatization approach to enhance the ionization efficiency of dextran.12,20 In our study, a degree of polymerization (DP) from 3 to 15 could be detected with fine intensity and higher S/N with HYNIC (Fig. S2b†) compared to DHB (Fig. S2a†). Table S2† lists the exact m/z values of the signals detected by HYNIC and DHB in the form of sodium adducts [M + Na]+ and potassium adducts [M + K]+. Due to the structural similarity between HYNIC and nicotinic acid, we also compared the ionization effect between them. Fig. S2c† shows the MALDI mass spectrum of the same amount of Dextran 1000 obtained by nicotinic acid. Only five oligosaccharides (DP from 3 to 7) were successfully detected. Therefore HYNIC exhibits advantage over nicotinic acid by comparing the ionization effect for oligosaccharides. To summarize, the HYNIC matrix demonstrated outstanding ionization efficiency covering from a simple oligosaccharide to complex oligosaccharides.Evaluation of the HYNIC matrix for highly selective detection of oligosaccharides from peptide mixtures
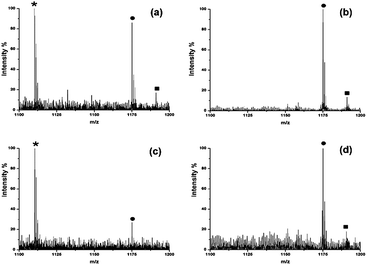
Another striking advantage of the HYNIC matrix is that it can be readily extended to selectively enhance the ionization efficiency of oligosaccharides and the suppress ionization of other components (e.g. peptides). A mixture of standard peptide (LSITGTYDLK) and DP7 was used as a model sample to evaluate its ionization selectivity toward oligosaccharides. We blend DP7 and the standard peptide in different molar ratios (with 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 1
1 and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10) to examine whether the highly abundant peptide would affect the oligosaccharide ionization efficiency. DHB was adopted as the matrix for comparison. As shown in Fig. 2, when the molar ratio of DP7 and peptide was 1
10) to examine whether the highly abundant peptide would affect the oligosaccharide ionization efficiency. DHB was adopted as the matrix for comparison. As shown in Fig. 2, when the molar ratio of DP7 and peptide was 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, both DP7 and the peptide could be detected with DHB (Fig. 2a). By contrast, HYNIC reveals the excellent ionization selectivity toward oligosaccharides (Fig. 2b). When the molar ratio of DP7
1, both DP7 and the peptide could be detected with DHB (Fig. 2a). By contrast, HYNIC reveals the excellent ionization selectivity toward oligosaccharides (Fig. 2b). When the molar ratio of DP7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) peptide decreased to 1
peptide decreased to 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10, DP7 could still be selectively detected with the HYNIC matrix (Fig. 2d), but DP7 almost could no longer be detected using DHB (Fig. 2c). To further verify the selectivity effect of HYNIC, DP7 was mixed with tryptic peptides of BSA in different mass ratios (with 1
10, DP7 could still be selectively detected with the HYNIC matrix (Fig. 2d), but DP7 almost could no longer be detected using DHB (Fig. 2c). To further verify the selectivity effect of HYNIC, DP7 was mixed with tryptic peptides of BSA in different mass ratios (with 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 1
1 and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10) and analyzed by MALDI-MS. As shown in Fig. S3a and c,† lots of peptide signals coming from BSA were detected using DHB. Although the signal of DP7 was also observed, it was suppressed by the presence of large amounts of peptides. However, when HYNIC was used, DP7 could be easily detected with a clean background in the mass spectra resisting interference of the peptides, which made it easy for later identification. The ability to selectively ionize glycans facilitated the direct identification of glycans from the deglycosylated glycopeptide mixture without the need for purifying the glycans before analysis. Ribonuclease B (RNase B) is a glycoprotein containing five different glycosylated variants with 5–9 mannose residues attached to the chitobiose core at the site Asn34.21 Without purification of the glycans, a series of five ion peaks with an interval of 162 Da was observed after detaching the glycans and using HYNIC as the matrix for analysis (Fig. 3b), corresponding to [M + Na]+ peaks from GlcNAc2Man5, GlcNAc2Man6, GlcNAc2Man7, GlcNAc2Man8, and GlcNAc2Man9. Whereas only tryptic peptides were observed when DHB was used (Fig. 3a). Therefore, we concluded that HYNIC was advantageous for the efficient oligosaccharide-specific ion embossment on the MALDI-TOF analysis.
10) and analyzed by MALDI-MS. As shown in Fig. S3a and c,† lots of peptide signals coming from BSA were detected using DHB. Although the signal of DP7 was also observed, it was suppressed by the presence of large amounts of peptides. However, when HYNIC was used, DP7 could be easily detected with a clean background in the mass spectra resisting interference of the peptides, which made it easy for later identification. The ability to selectively ionize glycans facilitated the direct identification of glycans from the deglycosylated glycopeptide mixture without the need for purifying the glycans before analysis. Ribonuclease B (RNase B) is a glycoprotein containing five different glycosylated variants with 5–9 mannose residues attached to the chitobiose core at the site Asn34.21 Without purification of the glycans, a series of five ion peaks with an interval of 162 Da was observed after detaching the glycans and using HYNIC as the matrix for analysis (Fig. 3b), corresponding to [M + Na]+ peaks from GlcNAc2Man5, GlcNAc2Man6, GlcNAc2Man7, GlcNAc2Man8, and GlcNAc2Man9. Whereas only tryptic peptides were observed when DHB was used (Fig. 3a). Therefore, we concluded that HYNIC was advantageous for the efficient oligosaccharide-specific ion embossment on the MALDI-TOF analysis.
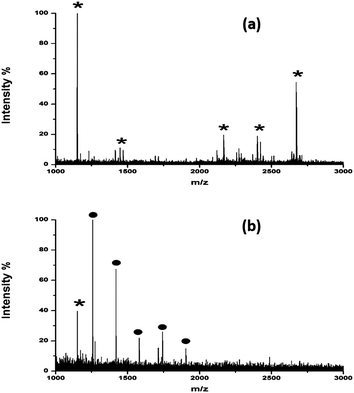 | ||
| Fig. 3 MALDI mass spectra of tryptic and PNGase F digests of RNase B (equivalent to 10 pmol amount of glycoprotein) using (a) DHB and (b) HYNIC. * denotes peptides and ● denotes glycans. | ||
Evaluation of reproducibility of the HYNIC matrix
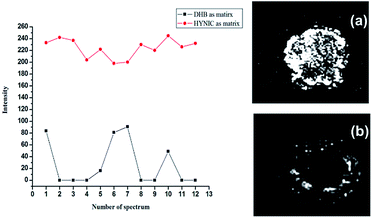
As previously mentioned, DHB suffered from the disadvantage of forming hot spots due to inhomogeneous crystallization. In most cases, the center of the target spot is devoid of crystals.22 Therefore, it is essential to upgrade crystal formation within the analyte–matrix system, aiming towards a more homogenous embedding of analytes on the target spot and finally improving the mass resolution as well as reproducibility.23–25 DP7 was chosen as a standard oligosaccharide to evaluate the performance of the HYNIC matrix. All samples were spotted on one target plate with a thin layer spotting method and analyzed in the same round of experiments to ensure reliability of the comparison. Higher signal intensity and better shot-to-shot reproducibility were obtained by comparison of the peak intensity, shown in Fig. 4 (left panel). The MALDI imaging analysis was utilized to further verify that the HYNIC matrix formed more homogeneous crystals with the analyte on the target well. As shown in Fig. 4b (right panel), the signals of DP7 using DHB were detected around the rim of the MALDI spot, which is in accordance with a previous study.22 The signals of DP7 from the HYNIC matrix located uniformly throughout the spot as shown in Fig. 4a (right panel), which is thought to be quite useful for automated data acquisition and quantitative analysis using MALDI-MS. In fact, the spotting technique can make a large difference in the crystallization form. We adopted three other spotting techniques including acetonitrile recrystallization of DHB, dried droplet method and double-layer method. As shown in Fig. S4,† acetonitrile recrystallization and dried droplet method helped to form more homogeneous crystals but no obvious improvement of signal intensity was observed (Fig. S5†). Whereas, more uniform crystals and better shot-to-shot reproducibility were obtained by HYNIC without deposition method optimization.Evaluation of the buffer tolerance ability of the HYNIC matrix
Although MALDI-TOF MS is considered to have tolerance for a low concentration of salts, analyte signals are usually severely suppressed or undetectable when the salt concentration reaches a certain amount.26 Several desalting methods including HPLC, C18Ziptip, and microextraction chip have been extensively applied to sample pretreatment prior to MALDI MS analysis.27 Nevertheless, sample loss and poor recovery of the analyte have been observed. Therefore, a matrix with high salt tolerance to enable direct MALDI MS analysis of analytes is favorable. Sodium chloride (NaCl) buffer was chosen as a representative non-volatile salt to test the salt tolerance of the HYNIC matrix, with the concentration of NaCl ranging from 25 mM to 200 mM. The HYNIC matrix demonstrated excellent buffer tolerance as it enabled the detection of the DP7 in all buffer concentrations, despite a little decrease of S/N (Fig. S6e–h†). However, for the DHB matrix, the signals of DP7 of the same amount could rarely be detected with poor S/N (Fig. S6a–d†). The high buffer tolerance ability brought the HYNIC matrix more extensive application scope, because trace concentrations of analyte dissolved in buffer can be directly analyzed without desalting. We also evaluated the salt tolerance of HYNIC by mixing the sample (DP7) with diluted cell lysis solution (140 mM urea and 40 mM sulfocarbamide) and commonly used digestion buffers (50 mM ammonium bicarbonate). As shown in Fig. S7,† more than an order of magnitude higher S/N was obtained by the HYNIC matrix than DHB. The dominant peaks are observed in the form of sodium adducts [M + Na]+, although there are some contaminated peaks which may come from the lysis solution.Evaluation of the impact of the HYNIC matrix on the fragmentation pattern in tandem mass spectrometry
To further investigate the performance of the HYNIC matrix, we performed the collision-induced dissociation (CID) for the MS/MS analysis of sodium adducts [DP7 + Na]+ from DP7 as the precursor in positive ion mode. Fragmentation of the [DP7 + Na]+ generated with DHB was used as a comparison. 10 pmol DP7 was loaded when DHB was used and 1 pmol of HYNIC was used to ensure similar intensity. Suitable laser energy was adjusted in order to get a relatively high quality spectrum. The legend for fragment ions is provided on the top in Fig. 5. The two spectra were very similar in most of the places in the spectra between the DHB and HYNIC. Fragments representing glycosidic bond dissociation (B and Y ions) are the most abundant features both in Fig. 5a and b. Differences are also observed, the most notable being the high signal intensity of cross-ring cleavage of HYNIC (Fig. 5b) relative to DHB (Fig. 5a). Corresponding m/z values and fragmentation types are summarized in Table S3.† What is more, the use of HYNIC resulted in improved quality MS/MS spectra due to a much higher signal-to-noise ratio compared to the spectra obtained using DHB. Therefore, HYNIC allowed more deposition of the sample ionized per amount on a target and provided a more informative tandem MS analysis, which is quite effective for the structural characterization of oligosaccharides.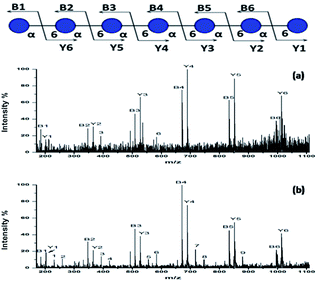 | ||
| Fig. 5 MALDI-TOF/TOF mass spectra of (a) 10 pmol of DP7 with the DHB matrix and (b) 1 pmol of DP7 with the HYNIC matrix. | ||
Real sample analysis
In order to further test the feasibility of HYNIC, we adopted glycans released and purified from human serum as a more complex sample. Human serum is a significant body liquid and serum glycan profiles can provide information which has potential in early disease diagnosis. Alterations of glycans are correlated with many kinds of diseases, such as cancer, diabetes and immune disorders. It is crucial for the analysis of limited quantities of glycans in this complex sample. Herein we used glycans obtained from healthy human serum to test whether HYNIC is still applicable to real samples. After treatment of PNGase F digestion and ultrafiltration, the sample was lyophilized and sent to be analyzed by MALDI with HYNIC directly. More than 40 glycan signals were successfully detected (Fig. S8†) and the detailed information of structures is summarized in Table S4.† Tandem mass analysis was performed to further verify the identification results. Displayed in Fig. S9,† the fragments of the precursor m/z 1663.6 were assigned to the precursor whose structure had been reported earlier.28 Therefore, HYNIC turns out to be applicable to many kinds of oligosaccharides.Conclusions
A new matrix HYNIC was found to be advantageous for oligosaccharides as it enabled high ionization efficiency in MALDI analysis. Direct profiling of the oligosaccharides without derivatization and desalting procedures streamlined the analysis process by minimizing the number of handling steps in sample preparation. HYNIC not only boasts of forming more uniform crystals with analytes on the MALDI target but also dramatically increases the detection sensitivity as low as 1 amol loading amount per target well. Besides, selectively enhancing the ionization efficiency of oligosaccharides and significantly suppressing ions of other contaminants, such as peptides, minimized the sample preparation steps. To some extent, it provides a new way to solve the inherent limitation of mass spectrometry in the direct analysis of oligosaccharides from complex mixtures. Furthermore, a high quality spectrum of tandem mass spectrometry was achieved. Abundant with B and Y ions, the MS/MS spectra of oligosaccharides obtained from the HYNIC matrix also contained the necessary signals from cross-ring fragments, facilitating the elucidation of the oligosaccharides. Compatibility with a high concentration of buffer made it possible to omit the process of desalting and avoid the loss of trace analytes. In a word, the adoption of the HYNIC matrix delivered outstanding results by quick, simple, straight and sensitive analysis of oligosaccharides in MALDI mass spectrometry measurements.Acknowledgements
The work was supported by the NST (2012CB910602, 2012AA020203, and 2015CB910401), NSF (21025519, 21335002 and 21375026), MOE (20130071110034) and Shanghai Projects (Eastern Scholar and B109).Notes and references
- R. Raman, S. Raguram, G. Venkataraman, J. C. Paulson and R. Sasisekharan, Nat. Methods, 2005, 2, 817–824 CrossRef CAS PubMed.
- J. B. Lowe, Cell, 2001, 104, 809–812 CrossRef CAS.
- A. Helenius and M. Aebi, Science, 2001, 291, 2364–2369 CrossRef CAS.
- Y. Zhang, H.-R. Yin and H.-J. Lu, Glycoconjugate J., 2012, 29, 249–258 CrossRef CAS PubMed.
- Y. Zhang, M. Kuang, L.-J. Zhang, P.-Y. Yang and H.-J. Lu, Anal. Chem., 2013, 85, 5535–5541 CrossRef CAS PubMed.
- S. Hua, H. J. An, S. Ozcan, G. S. Ro, S. Soares, R. DeVere-White and C. B. Lebrilla, Analyst, 2011, 136, 3663–3671 RSC.
- D. J. Harvey, Mass Spectrom. Rev., 1999, 18, 349–450 CrossRef CAS.
- C. A. Serrano, Y. Zhang, J. Yang and K. A. Schug, Rapid Commun. Mass Spectrom., 2011, 25, 1152–1158 CrossRef CAS PubMed.
- L.-W. Cao, Y. Zhang, L.-L. Chen, A.-J. Shen, X.-W. Zhang, S.-F. Ren, J.-X. Gu, L. Yu and X.-M. Liang, Analyst, 2014, 139, 4538–4546 RSC.
- A. Dell and H. R. Morris, Science, 2001, 291, 2351–2356 CrossRef CAS.
- M. Rohmer, B. Meyer, M. Mank, B. Stahl, U. Bahr and M. Karas, Anal. Chem., 2010, 82, 3719–3726 CrossRef CAS PubMed.
- H. Lavanant and C. Loutelier-Bourhis, Rapid Commun. Mass Spectrom., 2012, 26, 1311–1319 CrossRef CAS PubMed.
- K. Kaneshiro, Y. Fukuyama, S. Iwamoto, S. Sekiya and K. Tanaka, Anal. Chem., 2011, 83, 3663–3667 CrossRef CAS PubMed.
- M. A. Hashir, G. Stecher and G. K. Bonn, Rapid Commun. Mass Spectrom., 2008, 22, 2185–2194 CrossRef CAS PubMed.
- H.-Q. Qin, L. Zhao, R.-B. Li, R.-A. Wu and H.-F. Zou, Anal. Chem., 2011, 83, 7721–7728 CrossRef CAS PubMed.
- L.-J. Zhang, H.-J. Lu and P.-Y. Yang, Anal. Bioanal. Chem., 2010, 396, 199–203 CrossRef CAS PubMed.
- C.-L. Wu, C.-C. Wang, Y.-H. Lai, H. Lee, J.-D. Lin, Y.-T. Lee and Y.-S. Wang, Anal. Chem., 2013, 85, 3836–3841 CrossRef CAS PubMed.
- D. Asakawa, N. Smargiasso and E. De Pauw, Anal. Chem., 2012, 84, 7463–7468 CrossRef CAS PubMed.
- R. Čmelík, M. Štikarovská and J. Chmelík, J. Mass Spectrom., 2004, 39, 1467–1473 CrossRef PubMed.
- N.-Y. Hsu, W.-B. Yang, C.-H. Wong, Y.-C. Lee, R. T. Lee, Y.-S. Wang and C.-H. Chen, Rapid Commun. Mass Spectrom., 2007, 21, 2137–2146 CrossRef CAS PubMed.
- M. Thaysen-Andersen, S. Mysling and P. Højrup, Anal. Chem., 2009, 81, 3933–3943 CrossRef CAS PubMed.
- G. McCombie and R. Knochenmuss, Anal. Chem., 2004, 76, 4990–4997 CrossRef CAS PubMed.
- A. I. Gusev, W. R. Wilkinson, A. Proctor and D. M. Hercules, Anal. Chem., 1995, 67, 1034–1041 CrossRef CAS.
- M. D. Mohr, K. O. Börnsen and H. M. Widmer, Rapid Commun. Mass Spectrom., 1995, 9, 809–814 CrossRef CAS PubMed.
- J. M. Asara and J. Allison, J. Am. Soc. Mass Spectrom., 1999, 10, 35–44 CrossRef CAS.
- U. Kallweit, K. O. Börnsen, G. M. Kresbach and H. M. Widmer, Rapid Commun. Mass Spectrom., 1996, 10, 845–849 CrossRef CAS.
- S.-Y. Xu, M.-L. Ye, D.-K. Xu, X. Li, C.-S. Pan and H.-F. Zou, Anal. Chem., 2006, 78, 2593–2599 CrossRef CAS PubMed.
- D. Aldredge, H. J. An, N. Tang, K. Waddell and C. B. Lebrilla, J. Proteome Res., 2012, 11, 1958–1968 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c4an01659a |
| This journal is © The Royal Society of Chemistry 2015 |