Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Counting cell number in situ by quantification of dimethyl sulphide in culture headspace
Thomas W. E.
Chippendale
ab,
Patrik
Španěl
ac,
David
Smith
*a and
Alicia J.
El Haj
a
aInstitute for Science and Technology in Medicine – Keele University, Guy Hilton Research Centre, Stoke-on-Trent, ST4 7QB, UK. E-mail: d.smith@keele.ac.uk
bCobra Biologics, Stephenson Building, The Science Park, Keele ST5 5SP, UK
cJ. Heyrovský Institute of Physical Chemistry, Academy of Sciences of the Czech Republic, Dolejškova 3, Praha 8, Prague, Czech Republic
First published on 21st July 2014
Abstract
A novel, non-invasive technique is reported for determining the numbers of cells in a culture by quantifying dimethyl sulphide (DMS) in the culture headspace as produced by the cellular enzymatic reduction of dissolved dimethyl sulphoxide (DMSO). Measured DMS concentrations, as performed using selected ion flow tube mass spectrometry (SIFT-MS), in the headspace of 2D and 3D cultures of four cell lines, viz. HEK293 (kidney), MG63 (bone), hepG2 (liver) and CALU-1 (lung), linearly correlate with starting cell number. Clear differences in the rates of production of DMS by the four cell types in both the 2D and 3D situations are seen. This novel analytical technique for cell enumeration offers a significant contribution to quality assessment across cell-based research and industry, including analysis of large scale culture systems, and for routine cell biology research.
1 Introduction
Mammalian cell cultures are involved in a range of applications, including the production of viral vaccines, recombinant proteins and cell therapies, as well as for various strands of biological research. In order to determine the number of cells present in a culture, be they grown adherently or suspended by agitation in liquid media, it is usually required that all or a portion of the cells be removed from the culture vessel for assay. This can be performed by manually counting the cells using optical microscopy and by more complex methods such as MTT-type colorimetric assays1 or by automated computational image analysis systems such as Cell-IQ.2 Well-established culturing in two dimensions (2D) is now being expanded into the three dimensional (3D) systems in order to mimic physiological environs, to assess the behaviour of cells in this setting3 and to develop high-throughput low volume tissue-engineered products for clinical purposes.4 However, sampling of cultures for cell counting,1,2,5,6 colorimetrics or imaging1 can be challenging and presents a risk of contamination. Thus, we have investigated whether it is possible to monitor cell viability and proliferation by analysing in real time the concentrations of volatile metabolites (VOCs) released by the cells. Direct mass spectrometric analysis of VOCs emitted by mammalian and bacterial cells in vitro is a rapidly growing field that has recently been thoroughly reviewed.7 A major aspect of this work is to direct the search and quantification of biomarkers of disease and bacterial infection in exhaled breath. This we are pursuing with some vigour by real time quantification and monitoring of the VOCs emitted by the common respiratory pathogens Pseudomonas aeruginosa,8Staphylococcus aureus, Streptococcus pneumoniae and Haemophilus influenzae9 and Aspergillus fumigatus.10The unique cell assay described in this report is based on the enzymatic conversion of dimethyl sulphoxide (DMSO), one of the most commonly used amphipathic solvents,11 to volatile dimethyl sulphide (DMS) in cell cultures by the actions of the ubiquitous enzyme methionine sulphoxide reductase A (MsrA) that has been identified in both the mitochondria and the cytosol of animal cells.12 Thus, it is seen that the rate of DMS emission from DMSO-spiked cultures, as measured by the concentration of DMS in the culture headspace, is related to the number of cells in the culture. This new method of cell enumeration is analogous to the aforementioned MTT assays except that the analysis can be performed non-invasively on a bulk population of cells.
2 Experimental
2.1 Cell culture and sample preparation
HEK293 embryonic kidney cells were first cultured adherently in tissue culture (T-) flasks to confluence in DMEM supplemented with 10% foetal bovine serum (PBS), 2 mM L-glutamine and 1% penicillin–streptomycin. For 2D experiments, cells were removed using trypsin, counted and transferred to T75 flasks (300 mL total volume) in 15 mL of medium additionally supplemented with 0.1% v/v DMSO and 10 mM HEPES. For 3D experiments, cells were removed, counted and seeded to hydrogels of rat tail collagen I (3 g L−1; BD Biosciences). The gels were formed by neutralising the acetic acid–collagen solution using 1 M sodium hydroxide and adding 10× PBS (10%), the cell suspension (in culture medium; 10%), DMSO (0.1%) and sterile deionised water. The mixture was sealed inside clean, sterile 10 mL glass containers and allowed to set for approximately 30 minutes before a further 1 mL of culture medium containing 0.1% DMSO and 10 mM HEPES was added. In both 2D and 3D experiments, the T75 flasks and the glass vessels were sealed with septa-caps and incubated at 37 °C for 16 hours, after which the headspace above the cells was analysed for trace volatile compounds using selected ion flow tube mass spectrometry (SIFT-MS; see below). These methods are adapted from cell culture headspace analysis methods developed by our group previously.3,13,14Three other cell lines, namely MG-63, hepG2 and CALU-1, were included in this study. In the 2D experiments, following the initial culture in T-flasks, the cells were transferred to 150 mL glass bottles, sealed using septa caps and incubated overnight at 37 °C in media (DMEM with 10% FBS, 2 mM L-glutamine and 1% penicillin–streptomycin) containing 0.1% DMSO and 100 mM HEPES. This method more closely follows the previously published work wherein the headspace of cell cultures was analysed for volatile organic compounds using SIFT-MS.3,13,14 The 3D cultures were prepared in the manner described above and the headspace again analysed by SIFT-MS.
2.2 Headspace analysis by SIFT-MS
The SIFT-MS analytical technique can be used to quantify, in real time, several trace gas compounds in gaseous, humid samples; it has been described previously in several review articles.15–17 Briefly, a mixture of ions is generated by a microwave discharge through humid air, and a desired ionic species is selected from the mixture using a quadrupole mass filter and injected into the flowing helium carrier gas of the SIFT-MS instrument to act as precursor (reagent) ions for the analysis. Concurrently, the sample vapour (headspace) to be analysed is introduced into the carrier gas via a calibrated, heated capillary at a known flow rate, when the composite trace compounds of the sample undergo reactions with the injected reagent ions. The product (analyte) ions of these analytical reactions are sampled into a downstream quadrupole mass spectrometer, where a signal is generated in terms of ion counts-per-second (c s−1). The reagent ions employed in SIFT-MS are always H3O+, NO+ or O2+˙; these have been chosen because they do not react significantly with the major compounds of air (nitrogen, oxygen, argon), and so only the trace gas compounds are reactive and can be analysed. When the reaction kinetics (rate coefficients and product ions) of each trace compound are fully characterised, their concentrations can be obtained.In order to perform an analysis, the cell culture headspace it is sampled by piercing the septa that close the culture containers with a hypodermic needle attached to the sampling arm of the SIFT-MS instrument. In the present experiments, the DMS concentration in the headspace matrix was determined with NO+ reagent ions from the count rates of the m/z 62 isotopologue of the product ion (CH3)2S+ as described previously.18,19 It should be noted that due to the sample flow rate (∼30 mL min−1) and the relatively small headspace volume of the vessels used to perform the 3D experiments (8 mL), it was necessary to balance the atmospheric pressure inside the bottles by using a flow of clean cylinder air from a flexible bag coupled to the bottle via a second hypodermic needle. The finite dilution of the headspace sample due to this procedure can readily be accounted for by extrapolation of the analytical curve to the onset of sampling.20 This sampling procedure has been described in detail in previous publications.3,13,14,21,22
2.3 Colorimetric cell counting assay
Following the headspace analysis by SIFT-MS, the total cell number in the culture was assessed by the water-soluble tetrazolium salt WST-8 assay23 using Cell Counting Kit-8 (CCK-8; NBS Biologicals). Due to the relatively large volumes of the cell samples and the presence of the collagen in the 3D experiments, the protocol was modified from that recommended by the manufacturer. To the T75 cultures used for the 2D experiments (15 mL liquid volume), 150 μL of CCK-8 solution was added and the vessels were incubated for 1 hour at 37 °C. To the 10 mL vessels containing the 3D collagen scaffold cultures, 40 μL of CCK-8 solution was added, and the vessels were incubated for 4 hours. In each case, following the incubation period, 100 μL media samples were extracted from the cultures and transferred to a 96-well plate (n = 3). A Synergy 2 spectrophotometer (BioTek) was then used to measure the absorbance of the samples at 450 nm, with the absorbance at 650 nm subtracted, as recommended by the manufacturer. The absorbance reading at 450 nm correlates with the production of formazan dye by the cells that, in turn, is related to both the number of viable cells in the sample and to the metabolic activity of the cells.3 Results & discussion
3.1 Non-cytotoxicity of DMSO
This novel approach to cell number assessment obviously requires that the toxicity of DMSO be addressed if the proposed method is to be utilized. DMSO is commonly used as a cryopreservant for human cells at a concentration of 10% v/v (ref. 24) and is used clinically as a palliative treatment for interstitial cystitis.25 A study by our group revealed no change in the viability of both primary human bone marrow-derived mesenchymal stem cells (MSCs) and hepG2 hepatocyte-like cells following 16 hours incubation in medium containing 0.1% DMSO.21 In addition, a study by Da Violante and co-workers found no significant cytotoxic effects in Caco2/TC7 colon tumour cells cultured with DMSO concentrations of up to 10%.24 Whilst these data are persuasive as far as they go, more work is desirable to fully validate the non-toxicity of DMSO at the low concentrations needed for this novel cell assay method.3.2 HEK293 embryonic kidney cells
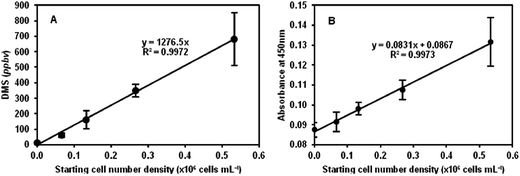
The plot in Fig. 1A shows the results obtained for 2D (sealed T75 tissue culture flasks) cell cultures for the DMS concentrations in ppbv as a function of the starting cell number density. The linearity of the plot suggests that cell number can be accurately derived using this non-invasive method. Given the gas and liquid phase volumes, the incubation period and the Henry's Law coefficient of DMS,20,26 the mean DMS production rate of the HEK293 cells can be determined from the slope of the line in this figure. Thus, the mean production rate for these 2D cultures is 8.9 × 105 DMS molecules per cell per minute (also given in Table 1). Immediately following the analyses by SIFT-MS, the cell numbers were counted by the established WST-8-based colorimetric assay as mentioned above, which additionally provides an indication of metabolic activity23 as discussed in Section 2.3. Once again, a linear relationship between the absorbance readings and cell count was obtained as shown in Fig. 1B, but in this case note the relatively large intercept on the absorbance axis indicating the “background” absorbance of the phenol red that is present in the culture medium. More importantly, as the starting cell number density doubles the relative increase in the absorbance is small, of the order of 10% only, which in some cases is barely within the error of the measurements, whereas the increase in headspace DMS concentration clearly indicates the greater sensitivity of the DMSO/DMS method of cell enumeration. Nonetheless, given the linear relationships in Fig. 1A and B, the equivalence of these two different assays is clear for cells cultures in the 2D situation, but this DMS method has the distinct advantage that it is completely non-invasive, given that the DMSO is seemingly non-toxic at the 0.1% v/v level, and the culture need not be manipulated.
| Cell type | Mean DMS production rate (×106 DMS molecules per cell per min) | ||
|---|---|---|---|
| 2D | 3D | ||
| HEK293 | Embryonic kidney | 0.89 | 0.44 |
| MG-63 | Osteoblast-like | 3.2 | 4.2 |
| hepG2 | Hepatocyte-like | 0.99 | 0.67 |
| CALU-1 | Lung epidermoid | 9.9 | 1.8 |
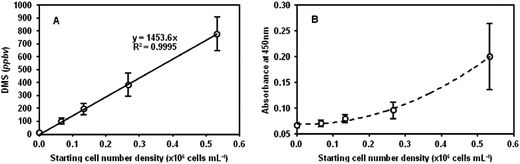
3.3 MG-63, hepG2 and CALU-1 cell lines
4 Concluding remarks
The experimental data presented in this short paper demonstrate that the measurement of headspace DMS concentration produced from DMSO by the cells can be a valuable non-invasive technique by which the number of cells in both 2D and 3D cultures can be determined. Indeed, its combination with the conventional assays may offer further insights into the behaviour of cells in the 3D situation. This new technique is applicable to cell and biochemical engineering applications as well as to tissue engineering and generic cell biological research. In fact, it has been hypothesised that the activity of the DMSO-reducing enzyme MsrA could be employed as a stem or progenitor cell marker for the isolation of cell populations.27 Given the presence of DMSO-reducing enzymes in a variety of prokaryotic and eukaryotic cells, the technique might also be applicable for monitoring the growth of microbial cell cultures in biopharmaceutical production processes. Thus, this novel approach appears to be promising for further application in a variety of biological fields.Competing financial interests
The authors have submitted a patent application relating to the work presented here (WO Application no. PCT/EP2013/054025).Acknowledgements
The work was supported by the Biodesign project and the EPSRC Centre for Innovative Manufacturing in Regenerative Medicine Analysis Frameworks.References
- Mammalian Cell Viability: Methods and Protocols, ed. M. J. Stoddart, Humana Press, 2011 Search PubMed.
- T. Toimela, H. Tahti and T. Ylikomi, ATLA, Altern. Lab. Anim., 2008, 36, 313–325 CAS.
- A. V. Rutter, T. W. E. Chippendale, Y. Yang, P. Španěl, D. Smith and J. Sulé-Suso, Analyst, 2013, 138, 91–95 RSC.
- J. L. Drury and D. J. Mooney, Biomaterials, 2003, 24, 4337–4351 CrossRef CAS.
- J. C. Y. Dunn, W. Y. Chan, V. Cristini, J. S. Kim, J. Lowengrub, S. Singh and B. M. Wu, Tissue Eng., 2006, 12, 705–716 CrossRef CAS PubMed.
- P. Thevenot, A. Nair, J. Dey, J. Yang and L. Tang, Tissue Eng., Part C, 2008, 14, 319–331 CrossRef CAS PubMed.
- K. Chingin, J. C. Liang and H. W. Chen, RSC Adv., 2014, 4, 5768–5781 RSC.
- D. Smith, P. Španěl, F. J. Gilchrist and W. Lenney, J. Breath Res., 2013, 7, 044001 CrossRef PubMed.
- T. W. E. Chippendale, F. J. Gilchrist, P. Španěl, A. Alcock, W. Lenney and D. Smith, Anal. Methods, 2014, 6, 2460–2472 RSC.
- T. W. E. Chippendale, F. J. Gilchrist, P. Španěl, A. Alcock, W. Lenney and D. Smith, Anal. Methods, 2014 Search PubMed , in press.
- N. C. Santos, J. Figueira-Coelho, J. Martins-Silva and C. Saldanha, Biochem. Pharmacol., 2003, 65, 1035–1041 CrossRef CAS.
- H. Weissbach, L. Resnick and N. Brot, Biochim. Biophys. Acta, 2005, 1703, 203–212 CAS.
- J. Sulé-Suso, A. Pysanenko, P. Španěl and D. Smith, Analyst, 2009, 134, 2419–2425 RSC.
- D. Smith, T. Wang, J. Sulé-Suso, P. Španěl and A. J. El Haj, Rapid Commun. Mass Spectrom., 2003, 17, 845–850 CrossRef CAS PubMed.
- P. Španěl and D. Smith, Mass Spectrom. Rev., 2011, 30, 236–267 CrossRef PubMed.
- D. Smith and P. Španěl, Analyst, 2011, 136, 2009–2032 RSC.
- D. Smith and P. Španěl, Mass Spectrom. Rev., 2005, 24, 661–700 CrossRef CAS PubMed.
- A. Pysanenko, P. Španěl and D. Smith, J. Breath Res., 2008, 2, 046004 CrossRef CAS PubMed.
- D. Smith, T. W. E. Chippendale and P. Španěl, Curr. Anal. Chem., 2013, 9, 550–557 CrossRef CAS.
- P. Španěl, A. M. Diskin, S. M. Abbott, T. Wang and D. Smith, Rapid Commun. Mass Spectrom., 2002, 16, 2148–2153 CrossRef PubMed.
- T. W. E. Chippendale, B. Hu, A. J. El Haj and D. Smith, Analyst, 2012, 137, 4677–4685 RSC.
- F. J. Gilchrist, H. Sims, A. Alcock, J. Belcher, A. M. Jones, D. Smith, P. Španěl, A. K. Webb and W. Lenney, Anal. Methods, 2012, 4, 3661–3665 RSC.
- H. Tominaga, M. Ishiyama, F. Ohseto, K. Sasamoto, T. Hamamoto, K. Suzuki and M. Watanabe, Anal. Commun., 1999, 36, 47–50 RSC.
- G. Da Violante, N. Zerrouk, I. Richard, G. Provot, J. C. Chaumeil and P. Arnaud, Biol. Pharm. Bull., 2002, 24, 1600–1603 Search PubMed.
- J. Parkin, C. Shea and G. R. Sant, Urology, 1997, 49, 105–107 CrossRef CAS.
- R. Sander, in NIST Chemistry WebBook, NIST Standard Reference Database Number 69, National Institute of Standards and Technology, Gaithersburg MD, p. 20899 Search PubMed.
- B. Hu and A. J. El Haj, Med. Hypotheses, 2013, 80, 663–665 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2014 |