Ruthenium porphyrin and oxidant convert N-nitrosodialkylamines into direct-acting mutagen in the Ames assay
Keiko Inami*ab,
Kyohei Yoshimitsua,
Hideaki Seinob and
Masataka Mochizukiab
aKyoritsu University of Pharmacy, Shibakoen 1-5-30, Minato-ku, Tokyo 105-8512, Japan
bFaculty of Pharmaceutical Sciences, Tokyo University of Science, Yamazaki 2641, Noda-shi, Chiba 278-8510, Japan. E-mail: inami@rs.noda.tus.ac.jp; Fax: +81-4-7121-3641; Tel: +81-4-7121-3641
First published on 17th July 2013
Abstract
The Ames assay is used for short-term screening of mutagens/carcinogens that induce DNA damage. Most mutagens/carcinogens require enzymatic activation through oxidation by cytochrome P450 in an S-9 mix to exert their mutagenicity. Chemical models for cytochrome P450, consisting of water-soluble or water-insoluble iron porphyrin plus an oxidant, have been used to produce frameshift mutagens from aromatic amines, heterocyclic amines and polyaromatic hydrocarbons. In this study, the mutagenicity of N-nitrosodialkylamines was assayed in the presence of a chemical model, which consists of 5,10,15,20-tetrakis(2,4,6-trimethylphenyl)porphyrinatoruthenium(IV) dichloride (RuMe3) and 2,6-dichloropyridine N-oxide (Cl2pyNO). The chemical model activated symmetrical N-nitrosodialkylamines (alkyl = methyl, ethyl, propyl, butyl), and unsymmetrical N-nitroso-N-methylalkylamine (alkyl = propyl, butyl) in Salmonella typhimurium YG7108. Furthermore, the mutagenicity of N-nitrosodipropylamine (NDP) in S. typhimurium YG7108 was higher than that in S. typhimurium TA1535, suggesting that the mutagenicity derived from NDP using the chemical model was due to DNA alkylation. The results showed that the chemical model can activate N-nitrosodialkylamines to induce base substitution mutations.
Introduction
N-Nitrosodialkylamines are found in the diet, cosmetics, tobacco and other environmental sources.1–5 N-Nitrosodialkylamines form by the reaction of secondary and tertiary amines in foods and drugs with nitrite derived from nitrate in saliva, under the acidic conditions of the stomach. Under neutral conditions, N-nitrosodialkylamines also form from the reaction of amines and the activated immune system in vivo.6,7 Thus, humans are exposed endogenously and exogenously to N-nitrosamines at all times.8 Since many N-nitrosodialkylamines induce cancers in experimental animals, N-nitrosamines are suspected as one of the causative agents of human cancer.9–13 Therefore, it is important to detect the DNA damage by N-nitrosodialkylamines.14 N-Nitrosodialkylamines themselves are inactive, and their activation is required to exert their mutagenic and carcinogenic properties. A postulated pathway of activation is through α-hydroxylation. The intermediate N-alkyl-N-(1-hydroxyalkyl)nitrosamine decomposes spontaneously by heterolysis, which releases aldehyde and generates reactive electrophilic species capable of alkylating nucleophiles.15,16The Ames assay is widely used for detecting DNA-damaging compounds.17,18 The S-9 mix, which consists of cytochrome P450 and an NADPH-regenerating system, is used for activating promutagens to active forms in the assay.17,18 Since the cytochrome P450/NADPH reaction can be mimicked by iron porphyrin in the presence of iodosylbenzene,19–21 we tested a chemical model consisting of a metalloporphyrin and an oxidant, as cytochrome P450 substitutes for the S-9 mix in the Ames assay. In contrast to the S-9 mix, the chemical model has the advantage of universal activity, since it lacks substrate and species specificity due to the absence of apoproteins. The chemical model is an alternative to the use of the S-9 mix derived from experimental animals in primary screening.
We have reported the detection of the mutagenicity of aromatic amines,22 heterocyclic amines23 and polyaromatic hydrocarbons24 by a chemical model in the Ames assay. The N-nitrosodialkylamines mutagenicity was also detected in the presence of 5,10,15,20-tetrakis(pentafluorophenyl)porphyrinatoiron(III) chloride (FeF5) and tert-butyl hydroperoxide (t-BuOOH); however, an increased number of bacteria were required in the assay.25
RuMe3 and Cl2pyNO have been reported to oxidise inactive alkanes and arenes to afford alcohols and/or ketones in high yields.26–28 In this study, RuMe3 plus Cl2pyNO, used as an alternative to the S-9 mix, was tested in detecting the mutagenicity of N-nitrosodialkylamines in the Ames assay with a conventional concentration of test strain. Furthermore, the DNA-damaging pathway of N-nitrosodipropylamine (NDP) by the chemical model was also investigated by comparison with the mutagenicity in Salmonella typhimurium TA1535 and YG7108.
Materials and methods
Chemicals
Sodium ammonium hydrogenphosphate tetrahydrate was purchased from Merck (Darmstadt, Germany). Bacto agar and Bacto nutrient broth were obtained from Becton, Dickinson Microbiology Systems (Sparks, USA). F5P and t-BuOOH were purchased from Sigma-Aldrich, Inc. (St. Louis, MO) and were used as received. Other reagents used including Cl2pyNO were purchased from Wako Pure Chemical Industries (Osaka, Japan), and used as received. RuMe3 was synthesised as previously described [yield: 21.5%, λmax (log ε in dichloromethane): 407 (5.21), 517 nm (4.05)].28 The purity of RuMe3 was verified using ultraviolet spectroscopy (96.6%).Symmetrical N-nitrosodialkylamines were prepared by nitrosation of the corresponding aqueous dialkylamine with NaNO2 under acidic conditions. Unsymmetrical N-methylalkylamines were synthesised by methylation with methyl iodide, following the formation of the Schiff base of alkylamines with benzaldehyde.29 All N-nitrosodialkylamines were purified by distillation. To decompose the unknown direct-acting mutagen formed in trace amounts, N-nitrosodialkylamine was dissolved in methanol saturated with sodium hydroxide and the entire solution was stirred overnight at room temperature.30 The reaction mixture was extracted three times with CH2Cl2, and the combined organic phases were dried over Na2SO4, filtered, and then evaporated in vacuo to produce a pale yellow oil. The structures and purities of compounds were confirmed by nuclear magnetic resonance spectroscopy (JEOL JNM-LA400). Fig. 1 shows the chemicals for the model systems used in this study.
 | ||
| Fig. 1 Structure of the chemicals used. | ||
Bacterial mutation assay using the chemical model
The bacterial mutation assay was based on the Ames test,17,31 using a chemical model consisting of metalloporphyrin and an oxidant as a substitute for the metabolic activation system.22,32 Professor B. N. Ames (University of California, Berkeley, USA) provided the S. typhimurium TA1535 strain, and Dr T. Nohmi (National Institute of Health Sciences, Tokyo, Japan) provided the S. typhimurium YG7108 strain. RuMe3, Cl2pyNO and mutagen were dissolved in acetone, and t-BuOOH was dissolved in 0.1 M sodium phosphate buffer (pH 7.4). The concentrations of the reagents are shown in the figure legends. All mutagens were tested before use and found to be non-mutagenic in S. typhimurium YG7108. Each sample was assayed using duplicate plates, and the data presented are the mean revertant colonies per plate ± standard error (SE) of three independent assays. The results were considered positive if the test produced a reproducible, dose-related increase in the number of revertant colonies, or the number of colonies is double the background number of colonies.33Preincubation assay involved exposing the test strain to the reaction mixture
Aliquots of porphyrin solution (20 μL) and mutagen (20 μL) in acetone were mixed, and an oxidant solution (20 μL), 0.1 M sodium phosphate buffer (pH 7.4, 0.5 mL), and a culture of the test strain (0.1 mL) were added. The mixture was then incubated (37 °C, 120 strokes per min) for 0, 20, 30, 60, 90, 120, or 180 min, and top agar (2 mL) was added. The mixture was then poured onto a minimal-glucose agar plate. After incubation for 44 h at 37 °C, the colonies were counted. N-Nitrosodialkylamines were tested using all preincubation periods, but the highest mutagenic activity was observed at a different preincubation period for each chemical model. In this study, the optimal preincubation period for each of the assay conditions was selected for data presentation.Preincubation assay involved adding the test strain to the reaction mixture after incubation
Aliquots of porphyrin (20 μL) and mutagen (20 μL) solutions were mixed, and an oxidant solution (20 μL) was added. The mixture was then incubated for 1, 20, 30, 60, 90, or 120 min at 37 °C with shaking (120 strokes per min). Next, 0.1 M sodium phosphate buffer (pH 7.4, 0.5 mL), a culture of the test strain (0.1 mL), and top agar (2 mL) were added. The mixture was then poured onto a minimal-glucose agar plate. After incubation for 44 h at 37 °C, colonies were counted.Results
Development of the chemical model for detecting NDP mutagenicity in the Ames assay
Experimental procedures for the Ames assay using RuMe3 plus Cl2pyNO as a substitute for the S-9 mix were developed. NDP activation by RuMe3 plus Cl2pyNO was compared between procedures where the test strain was present or absent during the reaction containing NDP and the activation system (Fig. 2A). RuMe3 plus Cl2pyNO for the demonstration of NDP mutagenicity was suitable as the preincubation method where the test strain was added before the incubation periods. NDP was mutagenic using any preincubation period; however, for each mutagen, the highest mutagenic activity occurred with a different preincubation period. In this study, the optimal preincubation period for each of the assay conditions was selected for data presentation as shown in the figure legend.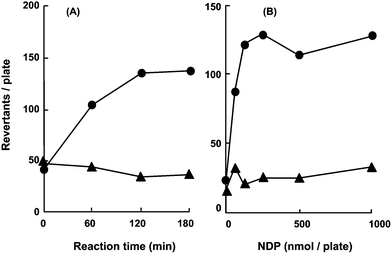 | ||
| Fig. 2 Effect of methods on NDP mutagenicity in S. typhimurium YG7108 using a chemical model. (A) NDP mutagenicity in the presence of RuMe3 plus Cl2pyNO using different methods. The preincubation assay consisted of exposing the test strain to the reaction mixture (●). The preincubation assay consisted of adding the test strain to a reaction mixture after incubation (▲). Reaction components at the indicated concentration included NDP (1000 nmol per plate), RuMe3 (25 nmol per plate) and Cl2pyNO (2000 nmol per plate). Each data point in both the assays was an average of two independent experiments because the reproducible data were obtained. All plates showed no cytotoxicity in the strains for the concentrations tested. (B) NDP mutagenicity by RuMe3 and Cl2pyNO (●) or FeF5 and t-BuOOH (▲). Preincubation assay involved exposing the test strain to the reaction mixture. NDP, 25 nmol per plate RuMe3 and 2000 nmol per plate Cl2pyNO or 25 nmol per plate FeF5 and 25 nmol per plate t-BuOOH were incubated for 120 min. Each data point in the RuMe3/Cl2pyNO and FeF5/t-BuOOH systems was an average of two independent experiments because reproducible data were obtained. All plates showed no cytotoxicity in the strains for the concentrations tested. | ||
FeF5 plus t-BuOOH and RuMe3 plus Cl2pyNO were compared for the ability to show NDP mutagenicity (Fig. 2B). FeF5 plus t-BuOOH has been reported to activate N-nitrosodialkylamines (alkyl = methyl, NDM; ethyl, NDE; NDP; butyl, NDB) by a method using an increased number of S. typhimurium YG7108.25 FeF5 plus t-BuOOH was not effective for promutagen activation in the assay with a conventional concentration of the bacterial strain, whereas RuMe3 plus Cl2pyNO converted NDP into the direct-acting mutagen (Fig. 2B). Different concentrations of the chemical models were used due to the cytotoxicity during the reaction periods. The result showed that the RuMe3 plus Cl2pyNO combination was suitable for demonstrating NDP mutagenicity by the procedures where the test strain was added before the incubation periods.
DNA damage derived from NDP with RuMe3 plus Cl2pyNO
The DNA-damaging mechanism of NDP treated with RuMe3 plus Cl2pyNO was investigated by comparison of NDP mutagenicity in S. typhimurium TA1535 versus S. typhimurium YG7108. Since S. typhimurium YG7108 is deficient in O6-methylguanine methyltransferase, the strain is highly sensitive to alkylating agents.34 The NDP in S. typhimurium TA1535 was weakly mutagenic; however, the mutagenicity of NDP in S. typhimurium YG7108 was higher than that in the parent strain S. typhimurium TA1535 (Fig. 3). | ||
| Fig. 3 Mutagenicity of NDP with RuMe3 plus Cl2pyNO in S. typhimurium YG7108 (●) and TA1535 (▲). NDP, 1000 nmol per plate; RuMe3, 25 nmol per plate; Cl2pyNO, 2000 nmol per plate. Each data point was an average of two independent experiments because reproducible data were obtained. All plates showed no cytotoxicity in the strains for the concentrations tested. | ||
Mutagenicity of N-nitrosodialkylamines under optimal conditions in the presence of the RuMe3 plus Cl2pyNO
RuMe3 plus Cl2pyNO was applied for detecting mutagenicity of other N-nitrosodialkylamines; symmetrical NDM, NDE, NDP and NDB, and unsymmetrical N-nitroso-N-methylpropylamine (NMP) and N-nitroso-N-methylbutylamine (NMB) (Fig. 4). | ||
| Fig. 4 Mutagenicity of N-nitrosodialkylamines by RuMe3/Cl2pyNO in S. typhimurium YG7108 under optimal conditions. (A) NDM mutagenicity in the presence of RuMe3 plus Cl2pyNO. NDM, 37.5 nmol per plate RuMe3 and 5000 nmol per plate Cl2pyNO were incubated for 60 min. The complete system contained mutagen, RuMe3, and Cl2pyNO (●). Control systems: NDM alone (○), without RuMe3 (□), without Cl2pyNO (△). Each data point represents the mean ± SE (n = 3). All plates showed no cytotoxicity in the strains for the concentrations tested. (B) NDE mutagenicity in the presence of RuMe3 plus Cl2pyNO. NDE, 37.5 nmol per plate RuMe3 and 2000 nmol per plate Cl2pyNO were incubated for 20 min. The complete system contained mutagen, RuMe3, and Cl2pyNO (●). Control systems: NDE alone (○), without RuMe3 (□), without Cl2pyNO (△). Each data point represents the mean ± SE (n = 3). All plates showed no cytotoxicity in the strains for the concentrations tested. (C) NDP mutagenicity in the presence of RuMe3 plus Cl2pyNO. NDP, 37.5 nmol per plate RuMe3 and 5000 nmol per plate Cl2pyNO were incubated for 90 min. The complete system contained mutagen, RuMe3, and Cl2pyNO (●). Control systems: NDP alone (○), without RuMe3 (□), and without Cl2pyNO (△). Each data point represents the mean ± SE (n = 3). All plates showed no cytotoxicity in the strains for the concentrations tested. (D) NDB mutagenicity in the presence of RuMe3 plus Cl2pyNO. NDB, 37.5 nmol per plate RuMe3 and 5000 nmol per plate Cl2pyNO were incubated for 30 min. The complete system contained mutagen, RuMe3, and Cl2pyNO (●). Control systems: NDB alone (○), without RuMe3 (□), and without Cl2pyNO (△). Each data point represents the mean ± SE (n = 3). All plates showed no cytotoxicity in the strains for the concentrations tested. (E) NMP mutagenicity in the presence of RuMe3 plus Cl2pyNO. NMP, RuMe3 37.5 nmol per plate, Cl2pyNO 5000 nmol per plate incubated for 90 min. The complete system contained mutagen, RuMe3, and Cl2pyNO (●). Control systems: NMP alone (○), without RuMe3 (□), and without Cl2pyNO (△). Each data point represents the mean ± SE (n = 3). All plates showed no cytotoxicity in the strains for the concentrations tested. (F) NMB mutagenicity in the presence of RuMe3 plus Cl2pyNO. NMB, 37.5 nmol per plate RuMe3 and 2000 nmol per plate Cl2pyNO were incubated for 60 min. The complete system contained mutagen, RuMe3, and Cl2pyNO (●). Control systems: NMB alone (○), without RuMe3 (□), and without Cl2pyNO (△). Each data point represents the mean ± SE (n = 3). All plates showed no cytotoxicity in the strains for the concentrations tested. | ||
In the entire assay, RuMe3 plus Cl2pyNO in the absence of mutagens did not show any mutagenicity, and the reaction mixture was not found to be mutagenic in the absence of RuMe3 or Cl2pyNO. This indicated that the chemical model oxidised N-nitrosodialkylamines to active forms.
Mutagenicity of N-nitrosodialkylamines by the S-9 mix
The N-nitrosodialkylamines (NDM, NDE, NDP, NDB, NMP, NMB) were mutagenic in S. typhimurium YG7108 in the presence of the S-9 mix (Fig. 5). N-Nitrosodialkylamine activation by the S-9 mix was carried out using procedures where a test strain was present in a reaction mixture containing a mutagen with the activation system for 20 min. The activity with the chemical model was 3–10 times lower than that with the S-9 mix.![Mutagenicity of N-nitrosodialkylamines [NDM (●), NDE (◆), NDP (▲), NDB (■), NMP (□) or NMB (△)] using the S-9 mix in S. typhimurium YG7108. The preincubation assay involved exposing the test strain to the reaction mixture in a 20 min incubation at 37 °C. Each data point represents the mean ± SE (n = 3).](/image/article/2013/TX/c3tx50036e/c3tx50036e-f5.gif) | ||
| Fig. 5 Mutagenicity of N-nitrosodialkylamines [NDM (●), NDE (◆), NDP (▲), NDB (■), NMP (□) or NMB (△)] using the S-9 mix in S. typhimurium YG7108. The preincubation assay involved exposing the test strain to the reaction mixture in a 20 min incubation at 37 °C. Each data point represents the mean ± SE (n = 3). | ||
Discussion
The Ames assay is used to evaluate the mutagenic potency of a DNA-damaging agent.17 Although the rat S-9 mix provides consistent data when used for activation in the Ames assay, there are difficulties in maintaining constant enzymatic activity.35 To overcome these difficulties, we tested chemical models consisting of a metalloporphyrin plus an oxidant for use in the mutation assay. Furthermore, an advantage of using chemical models is that there is less need for experimental animals.We have already reported that a chemical model, consisting of Fe porphyrin plus hydroperoxide or peracid, oxidised aromatic amines, heterocyclic amines and polyaromatic hydrocarbons to mutagenic compounds.22–24,32 RuMe3 has efficiently oxidised inactive compounds under mild conditions using Cl2pyNO as an oxidant.26–28 The Cl2pyNO in the Ames assay can be used at higher concentrations due to the low toxicity. In the present study, RuMe3 plus Cl2pyNO activated symmetrical NDM, NDE, NDP and NDB and unsymmetrical NMP and NMB with S. typhimurium YG7108. A procedure using RuMe3 plus Cl2pyNO was more effective when the test strain was also present during preincubation. Since unstable intermediates have a better chance of reacting with the test strain in the activation system, higher mutagenic activity can be detected by this procedure.32 Hence, the chemical model may activate N-nitrosodialkylamines to unstable active metabolites.
N-Nitrosodialkylamines are activated via α-hydroxylation by cytochrome P450, followed by elimination of the corresponding aldehyde. The reaction of the generated alkyldiazonium ions with DNA base nucleophiles causes DNA base substitution mutation via DNA alkylation.15,16 The NDP mutagenicity in the presence of the RuMe3 plus Cl2pyNO was examined in S. typhimurium YG7108 versus S. typhimurium TA1535 to investigate the DNA-damaging pathway of N-nitrosodialkylamines by the chemical model. NDP mutagenicity in S. typhimurium YG7108 was higher than that in S. typhimurium TA1535, indicating that NDP mutagenicity by the chemical model was due to DNA alkylation. The result suggested that the mutagenicity derived from N-nitrosodipropylamines was due to DNA alkylation using either the chemical model or the enzymatic system.
The mutagenic activation of N-nitrosodialkylamines by the chemical model and the S-9 mix was compared. Although the chemical model showed lower mutagenic activity of N-nitrosodialkylamines compared to the S-9 mix, the chemical model apparently activated them to mutagenic species in the Ames assay. The strength of mutagenicity was different in the S-9 mix and in the chemical model. The reason may be that the reactive species derived from N-nitrosodialkylamines in the S-9 mix existed for a longer period than in the chemical model because of the stabilization in non-aqueous environments owing to the apoprotein. A series of Salmonella strains expressing human P450 detected higher mutagenicity of some promutagens with NADPH-cytochrome P450 reductase in the Ames assay, since the mutagen was activated inside the bacteria.36–39 In conclusion, RuMe3 plus Cl2pyNO activated some potentially mutagenic N-nitrosodialkylamines without an enzymatic activation system in the Ames assay.
In this study, we showed that the chemical model was useful as an alternative for a metabolic activating system in the Ames assay. Then it is worth noting that the present metabolic system will be applied to other test systems: the micronuclei assay, the comet assay, and so on. The chemical model can be also used under various reaction conditions (pH, temperature, ionic strength, etc.), and then it can be easy to isolate and identify the unstable mutagenic compound from the reaction mixture. Thus using the chemical model in the oxidative activation of xenobiotics represented a useful tool for detecting unstable activated mutagens.
Abbreviations
| t-BuOOH | tert-Butyl hydroperoxide |
| Cl2pyNO | 2,6-Dichloropyridine N-oxide |
| NDB | N-Nitrosodibutylamine |
| NDE | N-Nitrosodiethylamine |
| NDM | N-Nitrosodimethylamine |
| NDP | N-Nitrosodipropylamine |
| NMB | N-Nitroso-N-methylbutylamine |
| NMP | N-Nitroso-N-methylpropylamine |
| FeF5 | 5,10,15,20-Tetrakis(pentafluorophenyl)porphyrinatoiron(III) chloride |
| RuMe3 | 5,10,15,20-Tetrakis(2,4,6-trimethylphenyl)porphyrinatoruthenium(IV) dichloride |
Acknowledgements
This work was supported in part by a Grant-in-Aid from the Ministry of Education, Culture, Sports, Science and Technology of Japan and by a Grant-in-Aid from the Science Research Promotion Fund of the Japan Private School Promotion Foundation. We thank Professor Tsunehiko Higuchi (Nagoya City University, Japan) for his helpful advice on the ruthenium porphyrin.References
- J. W. Hutchings, B. Ervens, D. Straub and P. Herckes, Environ. Sci. Technol., 2010, 44, 8128–8133 CrossRef CAS.
- A. Ghassempour, M. Abbaci, Z. Talebpour, B. Spengler and A. Römpp, J. Chromatogr., A, 2008, 1185, 43–48 CrossRef CAS.
- P. Jakszyn, A. Agudo, R. Ibáñez, R. García-Closas, G. Pera, P. Amiano and C. A. González, J. Nutr., 2004, 134, 2011–2014 CAS.
- W. Lijinsky, Mutat. Res., Genet. Toxicol. Environ. Mutagen., 1999, 443, 129–138 CrossRef CAS.
- H. Bartsch and B. Spiegelhalder, Eur. J. Cancer Prev., 1996, 5, 11–18 Search PubMed.
- I. T. Vermeer, L. Y. Henderson, E. J. Moonen, L. G. Engels, J. W. Dallinga, J. M. van Maanen and J. C. Kleinjans, Toxicol. Lett., 2004, 154, 175–182 CrossRef CAS.
- M. Miwa, D. J. Stuehr, M. A. Marletta, J. S. Wishnok and S. R. Tannenbaum, Carcinogenesis, 1987, 8, 955–958 CrossRef CAS.
- A. R. Tricker, Eur. J. Cancer Prev., 1997, 6, 226–268 CrossRef CAS.
- Y. H. Loh, P. Jakszyn, R. N. Luben, A. A. Mulligan, P. N. Mitrou and K. Khaw, Am. J. Clin. Nutr., 2011, 93, 1053–1061 CrossRef CAS.
- F. Kamangar, W. Chow, C. Abnet and S. Dawsey, Gastroenterol. Clin. North Am., 2009, 38, 27–57 CrossRef.
- R. L. Santarelli, F. Pierre and D. E. Corpet, Nutr. Cancer, 2008, 60, 131–144 CrossRef CAS.
- P. Jakszyn and C. A. González, World J. Gastroenterol., 2006, 12, 4296–4303 CAS.
- M. Dietrich, G. Block, J. M. Pogoda, P. Buffler, S. Hecht and S. Preston-Martin, Cancer, Causes Control, 2005, 16, 619–635 CrossRef.
- J. B. Guttenplan, Mutat. Res., Rev. Genet. Toxicol., 1987, 186, 81–134 CrossRef CAS.
- K. Inami, S. Ishikawa and M. Mochizuki, Genes Environ., 2009, 31, 97–104 CrossRef CAS.
- R. Preussmann and G. Eisenbrand, in Chemical Carcinogens ACS Monograph No. 182, ed. C. E. Searle, American Chemical Society, Washington, DC, 1987 Search PubMed.
- D. M. Maron and B. N. Ames, Mutat. Res., Environ. Mutagen. Relat. Subj., 1983, 113, 173–215 CrossRef CAS.
- B. N. Ames, W. E. Durston, E. Yamazaki and F. D. Lee, Proc. Natl. Acad. Sci. U. S. A., 1973, 70, 2281–2285 CrossRef CAS.
- W. Lohmann and U. Karst, Anal. Bioanal. Chem., 2008, 391, 79–96 CrossRef CAS.
- J. Bernadou and B. Meunier, Adv. Synth. Catal., 2004, 346, 171–184 CrossRef CAS.
- J. Groves, T. Nemo and R. Myers, J. Am. Chem. Soc., 1979, 101, 1032–1033 CrossRef CAS.
- K. Inami, M. Okazawa and M. Mochizuki, Toxicol. in Vitro, 2009, 23, 986–991 CrossRef CAS.
- K. Inami, M. Nagao, S. Ishikawa and M. Mochizuki, Genes Environ., 2010, 32, 7–13 CrossRef CAS.
- K. Inami, S. Ishikawa and M. Mochizuki, Toxicol. Environ. Chem., 2010, 92, 1169–1176 CrossRef CAS.
- E. Okochi, E. Namai, K. Itoh and M. Mochizuki, Biol. Pharm. Bull., 1995, 18, 49–52 CAS.
- J. Zhang and C. Che, Chem. Eur. J., 2005, 11, 3899–3914 CrossRef CAS.
- H. Ohtake, T. Higuchi and M. Hirobe, Heterocycles, 1995, 40, 867–903 CrossRef CAS.
- W. Leung, Polyhedron, 1993, 12, 2331–2334 CrossRef CAS.
- W. Hartman and L. Roll, in Organic Synthesis, John Wiley & Sons, New York, 1950, Coll. vol. 2 Search PubMed.
- K. Inami, M. Miura, N. Tsutsumi, E. Okochi, Y. Susaki, S. Ishikawa, S. Motohashi, J. Shiino, K. Takeda and M. Mochizuki, Heterocycles, 2012, 84, 1081–1088 CrossRef CAS.
- J. McCann, E. Choi, E. Yamazaki and B. N. Ames, Proc. Natl. Acad. Sci. U. S. A., 1975, 72, 5135–5139 CrossRef CAS.
- K. Inami, A. Inokawa, Y. Sugita and M. Mochizuki, J. Health Sci., 2009, 55, 109–113 CrossRef CAS.
- K. Mortelmans and E. Zeiger, Mutat. Res., Fundam. Mol. Mech. Mutagen., 2000, 455, 29–60 CrossRef CAS.
- M. Yamada, K. Matsui, T. Sofuni and T. Nohmi, Mutat. Res., Fundam. Mol. Mech. Mutagen., 1997, 381, 15–24 CrossRef CAS.
- M. Paolini and G. Cantelli-Forti, Mutat. Res., Rev. Mutat. Res., 1997, 387, 17–34 CrossRef.
- T. Shimada, E. Gillam, P. Sandhu, Z. Gou, R. Turky and F. Guengerich, Carcinogenesis, 1994, 15, 2523–2529 CrossRef CAS.
- M. Kranendonk, P. Mesquita, A. Laires, N. Vermeulen and J. Rueff, Mutagenesis, 1998, 13, 263–269 CrossRef CAS.
- Y. Yamazaki, K. Fujita, K. Nakayama, A. Suzuki, K. Nakamura, H. Yamazaki and T. Kamataki, Mutat. Res., Genet. Toxicol. Environ. Mutagen., 2004, 562, 151–162 CrossRef CAS.
- B. Emmert, J. Bünger, K. Keuch, M. Müller, S. Emmert, E. Hallier and G. A. Westphal, Toxicology, 2006, 228, 66–76 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2013 |
