Reverse self-assembly of lipid onions induced by gadolinium and calcium ions†
Hee-Young
Lee
a,
Kaname
Hashizaki
ab,
Kevin
Diehn
a and
Srinivasa R.
Raghavan
*a
aDepartment of Chemical and Biomolecular Engineering, University of Maryland, College Park, MD 20742-2111, USA. E-mail: sraghava@umd.edu
bSchool of Pharmacy, Nihon University, 7-7-1 Narashinodai, Funabashi, Chiba 274-8555, Japan
First published on 16th October 2012
Abstract
Self-assembly of lipids in water is well-known to result in nanostructures such as vesicles in dilute solution. In contrast, self-assembly in nonpolar organic solvents (oils) is much less established and there are hardly any known routes to forming structures such as reverse vesicles. Here, we build such structures based on our surprising recent discovery that salts can influence self-assembly of lipids in oil. We induce the self-assembly of nanoscale multilamellar vesicles (“onions”) in cyclohexane and toluene by combining the saturated phospholipid, 1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC), with salts of di- or trivalent cations like calcium (Ca2+) or gadolinium (Gd3+) in the absence of water. DMPC–Gd3+ onions can be seen by a transmission electron microscope (TEM) without additional staining. They have sizes between ∼100 and 400 nm and have 6–18 concentric bilayer shells (lamellae) that are uniformly spaced. The presence of lamellae is further confirmed by small-angle X-ray scattering (SAXS), from which we find the inter-lamellar spacing to be 6.0 nm. The formation of reverse onions is driven by the binding of these multivalent cations with the lipid headgroups, which in turn brings adjacent lipids close and causes a tighter packing of the lipid tails. Evidence for such binding is provided by the cation-induced lowering of the lipid melting temperature.
Introduction
The spontaneous assembly (self-assembly) of surfactants and lipids into nanoscale assemblies is a well-known phenomenon that is discussed in detail in textbooks of colloid science.1,2 When added into water in dilute amounts, surfactants, i.e., single-tailed amphiphiles, typically form micelles, which can be spherical or cylindrical in shape. On the other hand, lipids, i.e., twin-tailed balanced amphiphiles, typically form vesicles, which are spherical water-filled containers having a shell in which the lipids are arranged as bilayer(s).3 Unilamellar vesicles (ULVs) have a single-bilayer shell while multilamellar vesicles (MLVs, also called onions) have a shell of many concentric bilayers. Many systematic trends or patterns in aqueous self-assembly are well established. For example, adding salt to a solution of ionic surfactants causes micelles to transform in shape from spheres to cylinders.2 Combining a cationic and an anionic surfactant can lead to vesicles.4 These trends can be captured qualitatively by invoking a geometric factor termed the critical packing parameter p, which is the ratio of the average area of the amphiphile's tail portion (atail) to the average area of its head portion (ahead).1,2Self-assembly of amphiphiles can also occur in solvents other than water – in particular, it can occur in nonpolar liquids or oils. However, much less is known about self-assembly in oil and this subject is only briefly discussed in textbooks.1,2 Typically, there is a brief mention of reverse micelles, which can be formed by a few amphiphiles in dilute solutions in oil. The term “reverse” refers to the structure of the micelles, which is the reverse of “normal” micelles in water: i.e., the amphiphiles orient their tails towards the solvent (oil) and shield their hydrophilic heads from the oil in the micellar core.2 Apart from reverse micelles, much less is known about other reverse structures, especially in dilute solutions in oil. Moreover, few systematic patterns have been established in the context of reverse self-assembly, such as the effects of ionic additives or cosurfactants.5–7
Recently, we made a surprising discovery that the structure of reverse assemblies could be systematically tuned by adding salt.7 We started with a dilute solution of soybean lecithin, an unsaturated phospholipid, in n-decane. The lipid formed reverse spherical micelles.5 We showed that small amounts of inorganic salts could be dissolved in these (water-free) solutions and that the salt cations critically influenced self-assembly. Specifically, if the cation was a di- or trivalent one like calcium (Ca2+) or lanthanum (La3+), it induced the micelles to grow from spheres to long cylinders.7 Thus, salt influenced the shape and size of the reverse micelles, although it did not transform the micelles into other nanostructures.
In this paper, we demonstrate that salts can induce certain lipids to form not just reverse micelles but also reverse MLVs (onions). Reverse onions have many concentric reverse bilayers surrounding an oily core. In each reverse bilayer, the lipid molecules are arranged in a tail–head–head–tail fashion, i.e., the tails are outward while the heads are in the core and shielded from the oil; this is the reverse of a bilayer in water. The main lipid studied here is 1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC), which has two saturated C14 tails, and this is combined with salts of di- or trivalent cations like calcium (Ca2+) or gadolinium (Gd3+) in oils like cyclohexane and toluene in the absence of water. We chose to work with Gd3+ since it is an interesting trace element that finds application as a contrast agent in magnetic resonance imaging (MRI).8,9 Our results show that, with increasing molar ratio of salt![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) lipid, a transition from reverse micelles to reverse onions occurs. Reverse onions of DMPC–Gd3+ in toluene are directly visualized by a transmission electron microscope (TEM) without the use of additional contrast-enhancing stains. The images clearly reveal onion-like structures with more than 15 concentric bilayers in some cases. The presence of discrete onions is further confirmed by small-angle X-ray scattering (SAXS) and dynamic light scattering (DLS). We further show that effects of the salt on reverse self-assembly correlate with cation-binding to lipid headgroups,7 which is inferred from changes in the lipid melting temperature measured by differential scanning calorimetry (DSC).
lipid, a transition from reverse micelles to reverse onions occurs. Reverse onions of DMPC–Gd3+ in toluene are directly visualized by a transmission electron microscope (TEM) without the use of additional contrast-enhancing stains. The images clearly reveal onion-like structures with more than 15 concentric bilayers in some cases. The presence of discrete onions is further confirmed by small-angle X-ray scattering (SAXS) and dynamic light scattering (DLS). We further show that effects of the salt on reverse self-assembly correlate with cation-binding to lipid headgroups,7 which is inferred from changes in the lipid melting temperature measured by differential scanning calorimetry (DSC).
Overall, this paper presents a simple, straightforward, and inexpensive method for preparing reverse vesicles in oils. The structures are stable without precipitation or aggregation for several weeks. Much like normal vesicles, reverse vesicles could find use for encapsulation and controlled delivery of solutes.10 We should note that there are only a few reports of reverse vesicles in the literature, based on polyoxyethylene ethers,11 sucrose esters,10 amino acid derivatives,12 metal-complexed surfactants,13–15 macrocycles,16 or phospholipids.6,17 Currently, reverse vesicles are not widely used, partly due to questions regarding their stability and ease of preparation. We hope that the present work will shed new light on these unusual self-assemblies and provide researchers with a well-controlled formulation for future studies.
Materials and methods
Materials
1,2-Dimyristoyl-sn-glycero-3-phosphocholine (DMPC, C14), 1,2-dilauroyl-sn-glycero-3-phosphocholine (DLPC, C12), 1,2-dipalmitoyl-sn-glycero-3-phosphocholine (DPPC, C16), and 1,2-distearoyl-sn-glycero-3-phosphocholine (DSPC, C18) were all purchased in powder form from NOF Corporation, Japan. Anhydrous (>99.99% purity) NaCl, CaCl2 and GdCl3 salts were purchased from Sigma-Aldrich. Cyclohexane (>99.9% purity) was obtained from JT Baker, while toluene (99.5% purity) was purchased from EMD Chemicals. Deuterated cyclohexane (99.5%D) was purchased from Cambridge Isotopes.Sample preparation
Mixed solutions containing lipid and salt in a given nonpolar solvent were prepared as follows. The salt was dissolved in methanol to form a 100 mM stock solution. The lipid was dissolved in chloroform. Desired compositions of the samples were achieved by mixing the above two solutions. The solvents were removed by drying the samples under a fume hood for 24 h and then in a lyophilizer connected to a vacuum pump for at least 48 h. The final sample was prepared by adding the nonpolar solvent, followed by stirring and heating at 70 °C until the solution became homogeneous and transparent. The above procedure ensured the removal of any residual water from the sample, and thereby facilitated reproducible sample preparation. The samples were equilibrated for 2–3 days at room temperature and stored in a desiccator prior to conducting experiments.Dynamic light scattering (DLS)
A Photocor-FC light scattering instrument with a 5 mW laser light source at 633 nm was used at 25 °C, with the scattering angle being 90°. The autocorrelation function was measured using a logarithmic correlator. The average hydrodynamic radius as well as its distribution were extracted from the autocorrelation function using the Dynals software package supplied by Photocor.TEM
TEM was conducted on a Jeol JEM 2100 microscope at 80 keV. A carbon-coated copper grid was dipped into the solution for approximately 10 s, and this grid was then dried in a fume hood for 24 h before imaging was conducted.Small angle X-ray scattering (SAXS)
SAXS measurements were performed at 25 °C using a three-pin-hole-type camera (Bruker AXS, NanoSTAR) and a rotating-anode X-ray generator equipped with a copper target. The incident X-rays of CuKα radiation (1.54 Å) were monochromated by a cross-coupled Göbel mirror and passed through the sample placed in a 2 mm capillary made of soda glass. A two-dimensional position-sensitive proportional counter collected the scattered X-rays. The distance between the sample and the counter was 1060 mm, allowing the value of the scattering vector q to range from 0.15 to 3.0 nm−1. The data shown are for the normalized intensity I (arbitrary units) versus q = (4π/λ)sin(θ), where λ is the wavelength of the X-rays and 2θ is the scattering angle.Small angle neutron scattering (SANS)
SANS data were collected at 25 °C on the NG-7 and NG-3 (30 m) beamlines at NIST in Gaithersburg, MD. Neutrons with a wavelength of 6 Å were pre-selected. Three sample–detector distances (1, 4, 13 m) were used to obtain data over a range of scattering vectors from 0.004 to 0.4 Å−1. The scattering spectra were corrected and placed on an absolute scale using calibration standards provided by NIST. The data shown are for the absolute intensity I versus the scattering vector q = (4π/λ)sin(θ/2), where λ is the wavelength of incident neutrons and θ is the scattering angle.Rheology
Dynamic rheological experiments were performed on an AR2000 stress-controlled rheometer (TA Instruments). A cone-and-plate geometry of 20 mm diameter and a cone-angle of 2° was used. Samples were studied at 25 °C. A solvent trap was used to minimize cyclohexane evaporation. Dynamic frequency spectra were conducted in the linear viscoelastic regime of the samples, as determined from dynamic strain sweep measurements.Differential scanning calorimetry (DSC)
The phase transition temperature of the lipid was measured on a DSC-8230 calorimeter (Rigaku) at a heating rate of 2 °C min−1 and over a temperature range of 0–70 °C. Samples were studied under a N2 atmosphere and the N2 flow rate was 50 mL min−1. The solution (15 mg) was placed in a sample pan made of stainless steel with a diameter around 4 mm, following which the pan was sealed. The pan was rated to withstand a pressure of 50 atm. An empty sample pan was used as the reference.Results and discussion
Phase behavior
We first show results for mixtures of DMPC and anhydrous gadolinium chloride (GdCl3) in water-free cyclohexane. DMPC does not dissolve in cyclohexane at room temperature and forms a solid precipitate due to its melting temperature being around 35 °C (see Fig. 5). In addition, GdCl3 also does not dissolve in cyclohexane. However, when mixed at certain molar ratios, both components dissolve in cyclohexane at room temperature and give rise to homogeneous samples. Fig. 1 shows photographs of mixtures of 18.5 mM DMPC with varying [Gd3+]. For 4 mM Gd3+ or lower, precipitation occurs (Fig. 1a). At slightly higher [Gd3+], the sample becomes homogeneous and transparent, and its viscosity begins to increase. Samples with about 7–8 mM Gd3+ are transparent gels, and they hold their weight in the inverted vial (Fig. 1b). Dynamic rheology data (ESI, Fig. S1†) confirm the gel-like nature of this sample: i.e., the elastic G′ and viscous G′′ moduli are nearly independent of frequency ω, with G′ exceeding G′′. This result is quite similar to that obtained for mixtures of unsaturated lecithin with di- and trivalent ions.7 As in that case, we attribute the gel-like behavior to the entanglement of long cylindrical reverse micelles.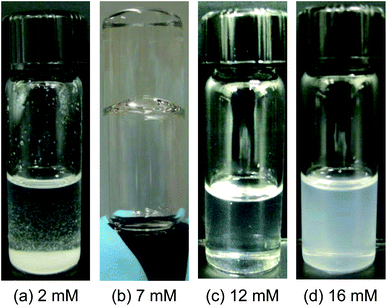 | ||
| Fig. 1 Photographs of mixtures of DMPC (18.5 mM) with varying concentrations of Gd3+ in cyclohexane. (a) At 2 mM Gd3+, the sample precipitates out. (b) At 7 mM Gd3+, the sample is viscous and gel-like, indicating long cylindrical reverse micelles. (c) At 12 mM Gd3+, the sample is of low viscosity and has a slight turbidity, indicating the onset of reverse vesicles. (d) At 16 mM Gd3+, the sample is still of low viscosity, but is much more turbid, indicating a mixture of reverse vesicles and lamellar structures. | ||
Beyond about 9 mM, further increase in [Gd3+] causes the viscosity of the samples to rapidly decrease. By about 12 mM Gd3+, the viscosity became almost identical to that of the solvent and the sample shows a slight bluish tinge (Fig. 1c), indicating the onset of larger structures that strongly scatter light. This bluish tinge intensifies as the [Gd3+] is increased further. At a Gd3+ of 16 mM, the sample is quite turbid (Fig. 1d), indicating a higher concentration and/or sizes of the scattering objects. In the latter case, a slight precipitation was observed with time. However, samples in the range of 12–14 mM remained homogeneous and unchanged for several weeks in a desiccator. As shown below, these contain reverse vesicles. Analysis of the 12 mM Gd3+ sample by DLS gave an average hydrodynamic diameter dh of 86 nm. The corresponding values of dh were 136 nm for the 14 mM Gd3+ sample and 432 nm for the 16 mM Gd3+ sample; both had relatively high polydispersities (see ESI, Fig. S3†). It should be noted that DLS gives a reliable size only if the structures are discrete, non-interacting and well-separated; this is not always the case for our samples, as shown by the TEM images in Fig. 2.
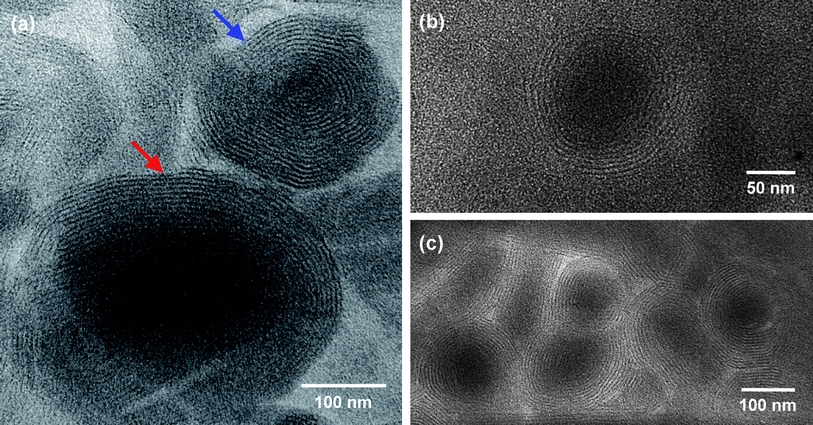 | ||
| Fig. 2 TEM images of structures present in a sample of 18.5 mM DMPC + 12 mM Gd3+ in toluene. Reverse onions are visible in these images due to the presence of Gd3+ in the bilayers (no additional staining has been done). In (a), the onion highlighted with a blue arrow has numerous bilayer shells and a negligible core region, while the onion highlighted by a red arrow has fewer shells and a distinct core region. | ||
The above pattern of phase behavior was also observed with other trivalent cations (e.g., La3+) or divalent cations (e.g., Mg2+ and Ca2+). Fig. S2 (ESI†) illustrates the behavior for mixtures of DMPC (20 mM) with anhydrous CaCl2. The results for increasing [Ca2+] are similar to those for [Gd3+] in Fig. 1: from precipitate to transparent gels to low-viscosity bluish solutions to more turbid solutions. Again, in the range of 12–14 mM Ca2+, we see evidence from visual observation (and correspondingly, DLS) for reverse vesicles. In contrast to the above results with di- and trivalent cations, monovalent cations such as Na+ did not induce reverse vesicles. In fact, mixtures of DMPC and NaCl formed a precipitate at all concentrations tested. We also examined other nonpolar solvents in addition to cyclohexane – in particular, toluene, n-decane, and benzene. The same pattern as in Fig. 1 was repeated for DMPC–Gd3+ mixtures in each of these solvents.
In addition to DMPC, we also examined other saturated phospholipids, including those with shorter tails, i.e., DLPC (C12), as well as those with longer tails, i.e., DPPC (C16) and DSPC (C18). In the case of DLPC, mixtures with di- and trivalent cations formed transparent gels at certain molar ratios, but the reverse vesicle region with non-viscous, bluish/turbid solutions was not observed. DPPC and DSPC have higher melting temperatures than DMPC, and in those cases, the addition of multivalent cations was not sufficient to solubilize the lipids at room temperature in cyclohexane. Reverse vesicle regions seem to occur in those systems at higher temperatures, but that is not discussed here and is a topic of future study. Since DMPC conveniently provides access to reverse vesicles at room temperature, we focused the rest of our studies on this lipid.
Nanostructure from TEM
TEM micrographs of DMPC–Gd3+ samples in the reverse vesicle region were obtained for two solvents, cyclohexane and toluene. Similar results were obtained for the two sets of samples, but since the contrast in the images was better for the toluene samples, those are shown here in Fig. 2 and 3 while the images of the cyclohexane samples are provided in the ESI, Fig. S4 and S5.† In all these TEM experiments, no staining was done to increase contrast. The Gd3+ (bound to the lipid headgroups; see below) acts as an intrinsic contrast agent, allowing the structures to be seen clearly.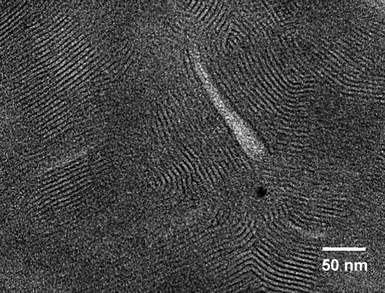 | ||
| Fig. 3 TEM image of structures present in a sample of 18.5 mM DMPC + 15 mM Gd3+ in toluene. Multilamellar stacks are visible in this image due to the presence of Gd3+ in the bilayers (no additional staining has been done). | ||
Fig. 2 shows images of a sample of 18.5 mM DMPC + 12 mM Gd3+ in toluene. This was a slightly bluish, non-viscous sample, comparable to the cyclohexane sample in Fig. 1c, and the average hydrodynamic diameter dh from DLS was 150 nm. The images in Fig. 2 show a number of spherical structures with multiple concentric bilayer shells that seem regularly spaced. These are reverse multilamellar vesicles (reverse onions). In Fig. 2a, two discrete onions are visible. The first (blue arrow) has a diameter of about 200 nm and about 16–18 concentric bilayers. The bilayers extend all the way to the center of the onion and there is hardly any core region; in effect, this structure resembles a real onion! The second onion (red arrow) is much larger (>300 nm in diameter) and it has fewer bilayers (10–12) as well as a distinct core region. This onion looks to be partially disrupted, which we suspect is an artifact caused by collapse of the bilayers when the sample is dried on the TEM grid. Fig. 2b shows a close-up of a discrete onion that looks intact: it has a diameter of ∼100 nm and a shell of 6–8 bilayers. Fig. 2c shows several onions that seem to be merging into one another: again, such a rearrangement is likely an artifact of the drying process on the TEM grid. Considering the core regions of the onions in Fig. 2c, they seem to have sizes between 100 and 200 nm and they each have 6–12 bilayers in their shell.
Overall, the TEM images in Fig. 2 clearly show the presence of discrete reverse onions in the DMPC–Gd3+–toluene sample. The size from TEM is in rough agreement with the size from DLS, especially given the fact that significant structural rearrangement is unavoidable during TEM sample preparation. Similar images are shown in the ESI, Fig. S4† for a DMPC–Gd3+–cyclohexane sample (composition corresponding to Fig. 1c). Here again, discrete onions are discernible despite the low contrast. The onions have a diameter of ∼100 to 150 nm and 6–8 bilayer shells. To the best of our knowledge, clear images of reverse onions such as those shown in Fig. 2 have not been published before. Moreover, it is unusual and possibly unprecedented to find onions (in oil or in water)18 that have as many concentric bilayers as the top structure in Fig. 2a.
TEM images were also acquired on a toluene sample containing 18.5 mM DMPC and 15 mM Gd3+. This sample was highly turbid and similar to the one in Fig. 1d. In this case, the TEM images show a series of regularly spaced multilamellar stacks, but there are no discrete reverse vesicles. Similar images showing multilamellar stacks were also obtained for the corresponding cyclohexane sample (ESI, Fig. S5†). It is not clear if the lamellae were formed by rupture of MLVs during TEM sample preparation. Alternately, these lamellae could indeed be the true structure in the sample (evidence from SAXS suggests the latter; see below). Similar TEM images have been published for lamellar phases in water and have been termed the “fingerprint pattern”.19 Additional images of the same sample showing the fingerprint pattern are provided in the ESI, Fig. S6.† Overall, the evidence from TEM is that at low [Gd3+] there are discrete onions, whereas at higher [Gd3+], the onions fuse to form extended lamellar stacks.
Nanostructure from SAXS and SANS
To further probe the structure in these samples, we made use of SAXS and SANS. SAXS spectra (intensity I vs. scattering vector q) for samples in cyclohexane with 18.5 mM DMPC and with three different concentrations (7, 12 and 16 mM) of Gd3+ are shown in Fig. 4. The presence of heavy metal atoms like Gd is beneficial in SAXS because it increases the contrast between the structures and the solvent. We find a q−1 decay of the intensity at low q for the 7 mM Gd3+ sample (Fig. 4a), which corresponds to cylindrical structures. This is consistent with the gel-like nature of this sample, which can be attributed to the entanglement of these cylindrical (wormlike) chains. In the case of the sample with 12 mM Gd3+, the data show a q−2 decay of the intensity at low q (Fig. 4a), and such a plot is indicative of discrete bilayered structures.6,20 This correlates with the presence of reverse vesicles in this sample, as shown by TEM. Finally, the sample with 16 mM Gd3+ shows a sharp peak at a q value of 1.05 nm−1 (Fig. 4b). This type of plot is most likely indicative of lamellar structures,20 which is also in good agreement with the TEM image of this sample (Fig. 3). The appearance of the peak indicates that the bilayers are relatively uniformly spaced, which is consistent with the image. From the peak position q0, the inter-bilayer spacing d = 2π/q0 is determined to be 6.0 nm.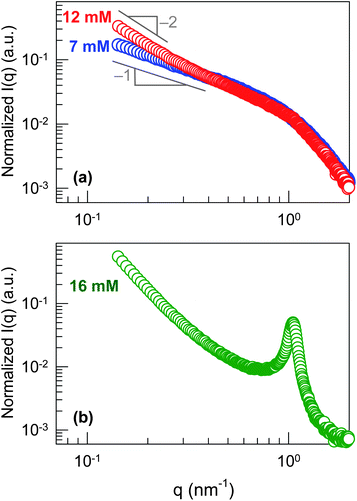 | ||
| Fig. 4 SAXS data at 25 °C from mixtures of DMPC and Gd3+ in cyclohexane. Each of the samples contains 18.5 mM DMPC while the Gd3+ concentrations were: (a) 7 mM, 12 mM and (b) 16 mM. | ||
A similar set of data was also acquired using SANS. The SANS experiments were done for mixtures in deuterated cyclohexane of DMPC and Ca2+ ions. Plots of intensity I vs. scattering vector q for 20 mM DMPC along with 9, 11 and 13 mM of Ca2+ are shown in the ESI, Fig. S7.† The data show an increase in I at low q as [Ca2+] increases. The sample with 9 mM Ca2+ shows a q−1 decay, indicating cylindrical structures. The sample with 13 mM Ca2+ shows a q−2 decay at low q, indicating bilayer structures. Again, this correlates with the visual observations from ESI, Fig. S2† since the former sample is a transparent gel whereas the latter is a bluish, non-viscous solution. Thus, the same trends are seen for DMPC–Ca2+ using SANS as for DMPC–Gd3+ using SAXS. We conclude that both di- and trivalent cations drive a transition from cylinders to vesicles to lamellae.
Thermal response
We believe that cations direct the self-assembly of DMPC by binding to the lipid headgroups. One way to infer such binding is by measuring the melting temperature Tm of the lipid in the presence of the cations. We therefore used DSC to measure Tm of DMPC at 18.5 mM in cyclohexane with different [Gd3+]. Below Tm, the two C14 tails in DMPC will be in a frozen, ordered state (“gel phase”), whereas above Tm, the tails will be liquid-like and flexible (“liquid-crystalline phase”).2 Experimental values of Tm for DMPC in water are around 23 °C.21 Here, DMPC is in a nonpolar organic liquid and thus the Tm is likely to be different. Indeed, the sample of DMPC alone in cyclohexane (Fig. 5a) shows an endothermic peak at 35.0 °C. We attribute the low solubility at room temperature of DMPC in cyclohexane to this high Tm, i.e., to the fact that the chains will be ordered and well-packed.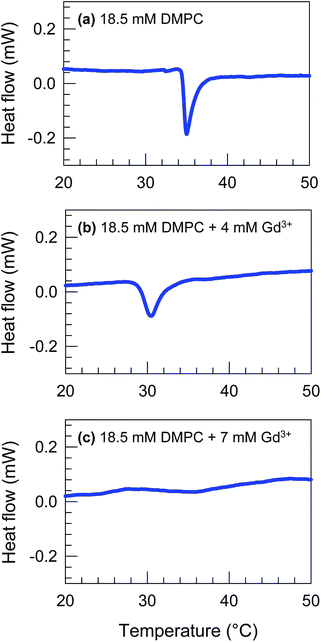 | ||
| Fig. 5 DSC scans for samples of 18.5 mM DMPC + (a) 0 mM, (b) 4 mM and (c) 7 mM Gd3+ in cyclohexane. Upon addition of Gd3+, the lipid melting peak shifts from 35.0 °C in (a) to 30.4 °C in (b) and the peak is absent in (c). | ||
Next, we note the DSC curves for the cases when Gd3+ is added to the DMPC–cyclohexane sample. For 4 mM Gd3+ (Fig. 5b), the endothermic peak shifts down to 30.4 °C. When the Gd3+ concentration is raised to 7 mM (Fig. 5c), the peak vanishes. This shows that the addition of Gd3+ reduces and then eliminates the melting peak due to DMPC. The lack of a peak at 7 mM Gd3+ implies that the melting temperature is too low to be measured (note that cyclohexane freezes around 6.7 °C, which poses a necessary limitation in the DSC experiment). From these data, we conclude that there is a strong interaction between Gd3+ and DMPC, one effect of which is to liquefy the tails of DMPC. These data help to explain why mixtures of DMPC–Gd3+ in cyclohexane are soluble and homogeneous even though the individual components are each insoluble in the same solvent. By extension, the same effects are expected for other di- and trivalent cations (e.g., Ca2+), but not for univalent cations (e.g., Na+).
Mechanism
We now discuss how cations like Gd3+ and Ca2+ induce DMPC to assemble into reverse vesicles. The basic idea is similar to that discussed in our previous work on mixtures of lecithin with such cations.7 The difference is that, while lecithin showed a transition from spherical to cylindrical micelles, it did not give rise to bilayers or vesicles. So why do we obtain vesicles with DMPC, but not with lecithin?The first point to reiterate here is that the cations that modulate lipid self-assembly in oil are the same ions that have been shown to bind to phospholipids in water. Interactions between cations and phospholipids in water have been well-studied, both by experiments22–25 and by simulations.26,27 It is known that the binding of monovalent Na+ is weak whereas di- and trivalent cations like Ca2+ and Gd3+ bind strongly to the phosphate part of the phosphocholine headgroup.22,26 Also, these multivalent cations can bind simultaneously to more than one lipid, and this has the effect of forcing the lipid tails to pack more tightly (sometimes called a “condensing effect”).28–30 In turn, simulations have shown that the lipid tails adopt a more extended configuration and that the average cross-sectional area of the tails atail decreases with cation-binding.31,32 Cation-binding may also expand ahead, but this is possibly a minor effect.
We believe that the decrease in atail is a key to explaining the differences between lecithin and DMPC. In the case of lecithin, one of its two tails has two cis-double bonds, which leads to kinks in the tail and thereby a larger overall tail area. Because of the constraints imposed by the double bonds, this tail will be able to undergo only a limited extent of straightening or compression. In comparison, both tails of DMPC are saturated, and as shown by DSC, both tails are in a flexible state in DMPC–cation mixtures. As a result, the DMPC tails may be able to pack tighter when cations bind to the lipid, which means atail will decrease more. This eventually leads to a molecular geometry where atail and ahead are comparable, and in turn, the critical packing parameter p = atail/ahead ≈ 1. Such molecules, which correspond to an overall cylindrical shape, will preferentially pack into reverse bilayers, as shown in Fig. 6.2,6 Note that the p = 1 state is reached only when the cation![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DMPC molar ratio is relatively high. At lower cation
DMPC molar ratio is relatively high. At lower cation![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DMPC ratios, the molecular geometry resembles a truncated cone because the atail is still relatively large compared to ahead (Fig. 6), and this favors the formation of cylindrical reverse micelles.2,6,7 Note also that DMPC by itself is not soluble in the solvents considered here at room temperature, but by analogy with lecithin, it would be expected to have a larger p and form spherical reverse micelles at high temperatures.
DMPC ratios, the molecular geometry resembles a truncated cone because the atail is still relatively large compared to ahead (Fig. 6), and this favors the formation of cylindrical reverse micelles.2,6,7 Note also that DMPC by itself is not soluble in the solvents considered here at room temperature, but by analogy with lecithin, it would be expected to have a larger p and form spherical reverse micelles at high temperatures.
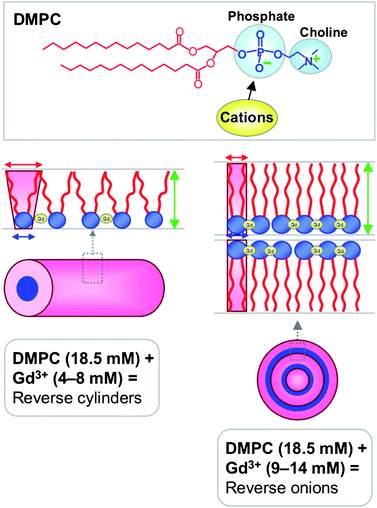 | ||
Fig. 6 Mechanism by which cations such as Gd3+ modulate the reverse self-assembly of saturated lipids such as DMPC. The top panel shows the structure of DMPC – the phosphate portion of the phosphocholine headgroup is expected to be the binding site for cations. The bottom left panel shows that at moderate Gd3+![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DMPC ratios, the net geometry resembles a truncated cone, causing assembly into cylindrical reverse micelles. In contrast, the bottom right panel depicts the scenario at higher Gd3+ DMPC ratios, the net geometry resembles a truncated cone, causing assembly into cylindrical reverse micelles. In contrast, the bottom right panel depicts the scenario at higher Gd3+![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DMPC ratios. In this case, the bound ions force the lipid molecules to be closer to each other, and as a result, the lipid tails straighten and slightly elongate. As a result, the cross-sectional area of the tail region atail (red arrow) is reduced and it becomes comparable to the cross-sectional area of the head region ahead (blue arrow). The net geometry is cylinder-like, which leads to the formation of reverse bilayers. DMPC ratios. In this case, the bound ions force the lipid molecules to be closer to each other, and as a result, the lipid tails straighten and slightly elongate. As a result, the cross-sectional area of the tail region atail (red arrow) is reduced and it becomes comparable to the cross-sectional area of the head region ahead (blue arrow). The net geometry is cylinder-like, which leads to the formation of reverse bilayers. | ||
Finally, we should point out that the geometry-based arguments above can only substantiate the presence of bilayered structures, but cannot distinguish between reverse ULVs, reverse onions (MLVs), and lamellar stacks. Stated differently, the lamellar phase is the equilibrium corresponding to p = 1.1,2 Discrete vesicles may have a more stable configuration than lamellar stacks at low volume fractions simply because a lamellar stack is exposed on its edges to the solvent whereas a vesicle is a closed structure that avoids such exposure.2 But it is difficult to pinpoint if or when the transition will occur from discrete vesicles to bulk lamellae. It is also very intriguing that the reverse onions in Fig. 2 have as many as 16–18 concentric bilayers. This would imply that the innermost bilayer in the onion has significant curvature compared to the outermost one. It is not clear why a series of small ULVs are not formed instead of one such large onion. These aspects are open questions for future studies.
Conclusions
This study has demonstrated that di- and trivalent cations such as Ca2+ or Gd3+ can systematically modulate the self-assembly of saturated phospholipids like DMPC in organic solvents and in the absence of water. The cations are expected to bind to the headgroups of the lipid, and in turn modulate the packing of the lipid tails. At low concentrations, the cations induce reverse cylindrical micelles, which entangle to produce gel-like samples. At higher concentrations, the cations induce reverse onions with an overall size of 100–400 nm and with 6–18 concentric bilayer shells. At even higher concentrations of the cations, lamellar stacks are formed. Reverse onions and reverse lamellae are very unusual structures, and our study shows how these can be readily assembled in nonpolar solvents. The approach of using cations to modulate lipid self-assembly is powerful and simple, and it is capable of being extended and generalized to include other cations with more complex chemistries as well as other types of lipids. Future studies will determine if reverse vesicles are potentially useful as controlled release vehicles or as contrast agents for imaging.Acknowledgements
We acknowledge the Maryland NanoCenter for facilitating the TEM work and NIST for facilitating the SANS experiments. We also acknowledge helpful discussions with Prof. Dganit Danino from Technion-IIT, and the assistance of Hyuntaek Oh, graduate student at UMD, during manuscript preparation.References
- J. N. Israelachvili, Intermolecular and Surface Forces, Academic Press, New York, 1992 Search PubMed.
- D. F. Evans and H. Wennerstrom, The Colloidal Domain: Where Physics, Chemistry, Biology, and Technology Meet, Wiley-VCH, New York, 2001 Search PubMed.
- D. D. Lasic, Liposomes: From Physics to Applications, Elsevier, Amsterdam, 1993 Search PubMed.
- E. W. Kaler, A. K. Murthy, B. E. Rodriguez and J. A. N. Zasadzinski, Science, 1989, 245, 1371–1374 CAS.
- S. H. Tung, Y. E. Huang and S. R. Raghavan, J. Am. Chem. Soc., 2006, 128, 5751–5756 CrossRef CAS.
- S. H. Tung, H. Y. Lee and S. R. Raghavan, J. Am. Chem. Soc., 2008, 130, 8813–8817 CrossRef CAS.
- H. Y. Lee, K. K. Diehn, S. W. Ko, S. H. Tung and S. R. Raghavan, Langmuir, 2010, 26, 13831–13838 CrossRef CAS.
- P. Caravan, J. J. Ellison, T. J. McMurry and R. B. Lauffer, Chem. Rev., 1999, 99, 2293–2352 CrossRef CAS.
- W. J. M. Mulder, G. J. Strijkers, G. A. F. van Tilborg, A. W. Griffioen and K. Nicolay, NMR Biomed., 2006, 19, 142–164 CrossRef CAS.
- H. Mollee, J. De Vrind and T. De Vringer, J. Pharm. Sci., 2000, 89, 930–939 Search PubMed.
- H. Kunieda, K. Nakamura and D. F. Evans, J. Am. Chem. Soc., 1991, 113, 1051–1052 CrossRef CAS.
- C. Boettcher, B. Schade and J. H. Fuhrhop, Langmuir, 2001, 17, 873–877 CrossRef CAS.
- D. Dominguez-Gutierrez, M. Surtchev, E. Eiser and C. J. Elsevier, Nano Lett., 2006, 6, 145–147 CrossRef CAS.
- Y. Yan, B. Li, W. Li, H. L. Li and L. X. Wu, Soft Matter, 2009, 5, 4047–4053 RSC.
- W. Li, B. Li, Y. L. Wang, J. Zhang, S. Wang and L. X. Wu, Chem. Commun., 2010, 46, 6548–6550 RSC.
- X. N. Xu, L. Wang and Z. T. Li, Chem. Commun., 2009, 6634–6636 RSC.
- H. Kunieda, K. Nakamura, M. R. Infante and C. Solans, Adv. Mater., 1992, 4, 291–293 CAS.
- M. Gradzielski, J. Phys.: Condens. Matter, 2003, 15, R655–R697 CrossRef CAS.
- A. Sein, J. F. L. Vanbreemen and J. Engberts, Langmuir, 1995, 11, 3565–3571 CrossRef CAS.
- J. S. Pedersen, Adv. Colloid Interface Sci., 1997, 70, 171–210 CrossRef CAS.
- T. A. Harroun, M. Koslowsky, M. P. Nieh, C. F. de Lannoy, V. A. Raghunathan and J. Katsaras, Langmuir, 2005, 21, 5356–5361 CrossRef CAS.
- H. Akutsu and J. Seelig, Biochemistry, 1981, 20, 7366–7373 CrossRef CAS.
- C. Altenbach and J. Seelig, Biochemistry, 1984, 23, 3913–3920 CrossRef CAS.
- J. Marra and J. Israelachvili, Biochemistry, 1985, 24, 4608–4618 CrossRef CAS.
- Y. X. Huang, R. C. Tan, Y. L. Li, Y. Q. Yang, L. Yu and Q. C. He, J. Colloid Interface Sci., 2001, 236, 28–34 Search PubMed.
- R. A. Bockmann and H. Grubmuller, Angew. Chem., Int. Ed., 2004, 43, 1021–1024 CrossRef.
- J. J. Perez, A. Cordomi and O. Edholm, J. Phys. Chem. B, 2008, 112, 1397–1408 CrossRef CAS.
- C. G. Sinn, M. Antonietti and R. Dimova, Colloids Surf., A, 2006, 282, 410–419 CrossRef.
- A. Yaghmur, P. Laggner, B. Sartori and M. Rappolt, PLoS One, 2008, 3, e2072 CrossRef.
- A. Yaghmur, B. Sartori and M. Rappolt, Phys. Chem. Chem. Phys., 2011, 13, 3115–3125 RSC.
- G. Pabst, A. Hodzic, J. Strancar, S. Danner, M. Rappolt and P. Laggner, Biophys. J., 2007, 93, 2688–2696 CrossRef CAS.
- U. R. Pedersen, C. Leidy, P. Westh and G. H. Peters, Biochim. Biophys. Acta, Biomembr., 2006, 1758, 573–582 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c2sm26565f |
| This journal is © The Royal Society of Chemistry 2013 |
