 Open Access Article
Open Access ArticlePeptide–LNA oligonucleotide conjugates†
I. Kira Astakhova*a, Lykke Haastrup Hansenb, Birte Vesterb and Jesper Wengela
aNucleic Acid Center and the Biomolecular Nanoscale Engineering Center, Department of Physics, Chemistry and Pharmacy, University of Southern Denmark, Campusvej 55, DK-5230 Odense M, Denmark. E-mail: ias@sdu.dk; Fax: +45 6615 8780; Tel: +45 6550 2523
bDepartment of Biochemistry and Molecular Biology, University of Southern Denmark, Campusvej 55, DK-5230 Odense M, Denmark. E-mail: b.vester@bmb.sdu.dk; Fax: +45 6550 2467; Tel: +45 6550 2406
First published on 24th April 2013
Abstract
Although peptide–oligonucleotide conjugates (POCs) are well-known for nucleic acids delivery and therapy, reports on internal attachment of peptides to oligonucleotides are limited in number. To develop a convenient route for preparation of internally labeled POCs with improved biomedical properties, peptides were introduced into oligonucleotides via a 2′-alkyne-2′-amino-LNA scaffold. Derivatives of methionine- and leucine-enkephalins were chosen as model peptides of mixed amino acid content, which were singly and doubly incorporated into LNA/DNA strands using highly efficient copper(I)-catalyzed azide–alkyne cycloaddition (CuAAC) “click” chemistry. DNA/RNA target binding affinity and selectivity of the resulting POCs were improved in comparison to LNA/DNA mixmers and unmodified DNA controls. This clearly demonstrates that internal attachment of peptides to oligonucleotides can significantly improve biomolecular recognition by synthetic nucleic acid analogues. Circular dichroism (CD) measurements showed no distortion of the duplex structure by the incorporated peptide chains while studies in human serum indicated superior stability of the POCs compared to LNA/DNA mixmers and unmodified DNA references. Molecular modeling suggests strong interactions between positively charged regions of the peptides and the negative oligonucleotide backbones which leads to clamping of the peptides in a fixed orientation along the duplexes.
Introduction
Interactions between nucleic acids and proteins are vital for important biological processes including, for example, storage and controlled processing of genetic information,1,2 translation,3 degradation of foreign polynucleotides by cellular restriction systems4 and apoptosis.5 Recently obtained crystal structures of natural nuclear DNA of eukaryotic cells and histone peptides6 demonstrate the high importance of precise recognition between the biomolecules (Fig. 1). Furthermore, clinically important nucleic acid-based aptamers also utilize complex biomolecular recognition mechanisms when targeting proteins.7–9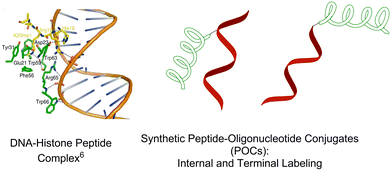 | ||
| Fig. 1 Crystal structure of the histone peptide–nuclear DNA complex; schematic illustration of peptide–oligonucleotide conjugates (internal and terminal attachment). | ||
Synthetic peptide–oligonucleotide conjugates (POCs) are artificial tools of choice for detailed studies of the abovementioned protein– and peptide–nucleic acid interactions, as well as being promising bioconjugates for nucleic acid delivery and therapy.10 Two main strategies are currently available to prepare synthetic POCs.10,11 The first is internal incorporation of a peptide residue via a chemically modified nucleotide scaffold of choice,12,13 and the second is attachment of a peptide at terminal positions of an oligonucleotide (Fig. 1).10,11,14,15 Internal labelling promotes active interaction between peptide and oligonucleotide chains, especially when they contain internally attached cationic peptides. Thereby the peptides have a high propensity for interacting with the nucleobases and the anionic backbones of oligonucleotides, and hence, to improve target binding and enzymatic stability of the POCs.16–18
Among various DNA linking strategies developed to date, the copper(I)-catalyzed azide–alkyne cycloaddition (CuAAC; or “click”) reaction is one of the most selective and versatile.19–21 As recently optimized by Finn et al., the CuAAC reaction allows preparation of various oligonucleotide conjugates in high yields and of remarkable purity.22–24 CuAAC click chemistry has been applied for preparation of several synthetic POCs.14,25 This was carried out using azido-functionalized derivatives of membrane penetrating and nuclear localization signalling peptides.14 Alternatively, azido-modified PNA and alkyne-modified amino acids were conjugated by the CuAAC reaction.25 In both cases the desired POCs were obtained in high purity and yields.14,25
Locked nucleic acids (LNA) display excellent biomedical properties such as improved target binding affinity and enzymatic stability.26–29 Moreover, incorporation of LNA has recently been shown to improve the therapeutic potential of oligonucleotides in siRNA and aptamer approaches.30,31 Internal attachment of cationic amino acids to the 2′-amino group of 2′-amino-LNA by peptide coupling reactions resulted in increased binding affinity of the conjugates to complementary DNA/RNA targets.32 Notably, the observed increase in thermal denaturation (Tm) values of the resulting conjugates was proportional to the total positive charge of the attached lysine residues. This was thought to be caused, at least in part, by decreased electrostatic repulsion between the negatively charged phosphate backbones and positively charged amino acid residues.32
Herein, we describe novel POCs in which peptide chains are internally incorporated into oligonucleotides using a 2′-alkyne-2′-amino-LNA scaffold (Fig. 2). The attachment of the selected methionine- and leucine-enkephalin peptides (Met- and Leu-enkephalins, respectively)33 to 21-mer LNA/DNA mixmer strands was performed using highly efficient so-called postsynthetic CuAAC click chemistry (performed after the oligonucleotide is synthesized). There are several advantages to this design. First, internally attached enkephalin peptides having lysine extensions at the N-termini have high potential to interact with oligonucleotide strands.16–18,32 Second, using highly efficient click chemistry, peptide chains can be incorporated into any position of oligonucleotide strands. Hence, the distance and orientation of the peptide residues could be estimated from well-known nucleic acid structure parameters. Third, a great diversity of other peptides, such as cell-penetrating34 and for targeting of specific cells,35 can be covalently tethered to the 2′-alkyne-2′-amino-LNA, opening up for various applications of this scaffold.
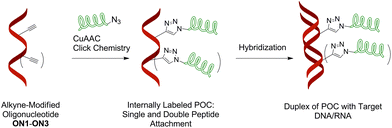 | ||
| Fig. 2 General concept of CuAAC preparation of peptide–oligonucleotide conjugates (POCs) applied in this work. | ||
In this paper we report the influence of internally attached enkephalin peptides on the structure and properties of synthetic oligonucleotides. We have prepared and studied single and double insertions of the peptides into LNA/DNA strands. We demonstrate that our novel synthesis strategy is rapid and efficiently provides series of POCs in high yields. Moreover, this approach allows us for the first time to evaluate target binding affinity, selectivity and enzymatic stability in human serum of an oligonucleotide containing covalently attached lysine-enkephalin hybrid peptides at internal positions.
Results and discussion
Strategy and design of enkephalin peptides “clicked” to LNA/DNA oligonucleotides
The general concept of POCs of this study is shown in Fig. 2. First, we synthesized 21-mer oligonucleotides containing single and double internal insertions of 2′-alkyne-LNA36 monomer M1. Similar M1-labelled oligonucleotides were already applied for preparation and studies of fluorescent LNA/DNA probes (ON1–ON3, Table 1).36,37 By combining the bicyclic LNA skeleton and a 2′-alkyne group, the monomer M1 is a very promising scaffold for preparation of POCs, providing a possibility of post-synthetic click chemistry along with precise positioning of the attached modifications within nucleic acid complexes.37 Next, ON1–ON3 were modified with enkephalin peptides by CuAAC conjugation with two azide-functionalized peptide derivatives 3–4 (Scheme 1). The number of nucleotides between two peptide modifications affects the distance, orientation and interactions between the peptides and the oligonucleotide scaffolds. Therefore we varied the number of nucleotides between the peptide residues by double insertion of monomers M1–M3 (Table 1). Peptides of choice were short Met- and Leu-enkephalin derivatives containing three additional lysine residues at the N-termini (Scheme 1). The resulting peptides are amphiphilic and therefore display high solubility in both organic and polar aqueous media. This is of importance for efficient CuAAC reactions with oligonucleotides usually performed in mixtures of organic solvents and aqueous buffers, as well as for the purification of the products. Furthermore, Met- and Leu-enkephalins are targets of clinically important opioid receptors involved in pain and pleasure signalling.38,39 Therefore the selected peptides can be potentially used as ligands for cell-specific delivery of oligonucleotides.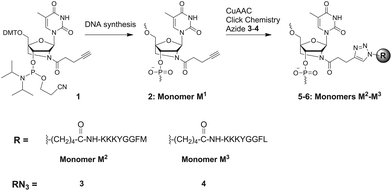 | ||
| Scheme 1 Chemical structures of modified monomers M1–M3, phosphoramidite 1 and peptido-azides 3–4 used in this study. Sequences of natural enkephalins: YGGFM (Met-enkephalin); YGGFL (Leu-enkephalin). | ||
| ON# | Sequence, 5′ → 3′ | Tm/ΔTm/°C | |||
|---|---|---|---|---|---|
| [Na+] = 110 mM | [Na+] = 20 mM | ||||
| Duplex with complementary | |||||
| DNA | RNA | DNA | RNA | ||
a Thermal denaturation temperatures Tm (°C) (change in Tm relative to corresponding reference duplex DNA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DNA/RNA, ΔTm (°C)). Tm values measured as the maximum of the first derivatives of the melting curves (A260vs. temperature) using 0.5 μM concentration of complementary strands. Reported Tm values are averages of at least two measurements with resulting Tm ± 0.5 °C. Medium and low salt phosphate buffers ([Na+] = 110 mM and 20 mM, respectively, 0.1 mM EDTA, pH 7.0). DNA/RNA, ΔTm (°C)). Tm values measured as the maximum of the first derivatives of the melting curves (A260vs. temperature) using 0.5 μM concentration of complementary strands. Reported Tm values are averages of at least two measurements with resulting Tm ± 0.5 °C. Medium and low salt phosphate buffers ([Na+] = 110 mM and 20 mM, respectively, 0.1 mM EDTA, pH 7.0). | |||||
| DNAref | TGC ACT CTA TGT CTG TAT CAT | 59.0 | 60.5 | 39.0 | 43.0 |
| ON1 | TGC ACT CTA TGM1 CTG TAT CAT | 62.0/+3.0 | 65.0/+4.5 | 41.0/+2.0 | 46.0/+3.0 |
| ON2 | TGC ACT CTA M1GT CM1G TAT CAT | 63.5/+4.5 | 69.0/+8.5 | 44.0/+5.0 | 51.5/+8.5 |
| ON3 | TGC ACM1 CTA TGT CTG TAM1 CAT | 63.0/+4.0 | 68.0/+7.5 | 43.0/+4.0 | 50.0/+7.0 |
| POC1 | TGC ACT CTA TGM2 CTG TAT CAT | 60.0/+1.0 | 63.5/+3.0 | 43.0/+4.0 | 47.0/+4.0 |
| POC2 | TGC ACT CTA M2GT CM2G TAT CAT | 65.0/+6.0 | 69.0/+8.5 | 47.0/+8.0 | 52.0/+9.0 |
| POC3 | TGC ACM2 CTA TGT CTG TAM2 CAT | 63.0/+4.0 | 70.0/+9.0 | 39.0/±0.0 | 42.0/−1.0 |
| POC4 | TGC ACT CTA TGM3 CTG TAT CAT | 62.0/+2.0 | 64.5/+4.0 | 47.0/+8.0 | 46.0/+3.0 |
| POC5 | TGC ACT CTA M3GT CM3G TAT CAT | 64.0/+5.0 | 68.5/+8.0 | 48.0/+9.0 | 52.0/+9.0 |
| POC6 | TGC ACM3 CTA TGT CTG TAM3 CAT | 58.0/−1.0 | 67.0/+6.5 | 43.0/+4.0 | 50.0/+7.0 |
Post-synthetic CuAAC click chemistry for the preparation of POCs
Phosphoramidite 1 was prepared by following the recently described method37 and was used in solid-phase oligonucleotide synthesis of conjugates ON1–ON3 (characterized by MALDI-MS spectra and IE HPLC profiles) (Table 1, ESI Tables S1–S2, Fig. S1–S2†). Next, post-synthetic click chemistry of ON1–ON3 with peptido-azide derivatives 3–4, obtained from commercial suppliers, was performed. DMSO, 0.2 M carbonate buffer (pH 8.5), aminoguanidine hydrochloride, peptido-azide 3–4,40 CuSO4–tri(benzyltriazolylmethyl)amine (TBTA)41 (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) and sodium ascorbate were added to a deaerated aqueous solution of the starting oligonucleotide. The aminoguanidine hydrochloride was added in order to prevent aggregation and cross-linking of the peptide chains under the CuAAC conditions.22,40 After 12–24 h at room temperature the product conjugates were purified on Sephadex (NAP-10) resin according to the protocol of the manufacturer followed by precipitation from cold acetone. The identity and purity of the POCs were confirmed by MALDI-MS spectra and IE HPLC profiles, respectively (Fig. S1–S2, ESI; Table S2†). The purity of POCs was ≥95% as estimated by IE HPLC, which was sufficient for their application in the studies described below without additional purification.
1) and sodium ascorbate were added to a deaerated aqueous solution of the starting oligonucleotide. The aminoguanidine hydrochloride was added in order to prevent aggregation and cross-linking of the peptide chains under the CuAAC conditions.22,40 After 12–24 h at room temperature the product conjugates were purified on Sephadex (NAP-10) resin according to the protocol of the manufacturer followed by precipitation from cold acetone. The identity and purity of the POCs were confirmed by MALDI-MS spectra and IE HPLC profiles, respectively (Fig. S1–S2, ESI; Table S2†). The purity of POCs was ≥95% as estimated by IE HPLC, which was sufficient for their application in the studies described below without additional purification.Yields of the products POC1–POC6 were estimated using absorbance at 260 nm by a comparison of the amount of a POC product to the corresponding starting oligonucleotide and were 79%–88%. Notably, the yields were only slightly decreased for double incorporation of the peptide residues compared to single incorporation, and were similar for the double labelling in a 3 nucleotides and 11 nucleotides distance (ESI†).
Alternatively, the CuAAC reactions performed under microwave conditions in an argon atmosphere at 60 °C for 15 min resulted in similar yields of the POCs as following the procedure described above, although the purity of the products was lowered down to 87–93% (data not shown). This might be due to side reactions, e.g. cross-linking of the peptide residues, promoted by the microwave conditions and/or elevated temperature.
Hybridization properties and duplex structure
Strong and selective binding to complementary nucleic acids is a highly desired property for synthetic oligonucleotides which ensures their utility, e.g. for targeting of nucleic acids in vitro and in vivo.42 Internally attached modifications, especially bulky and charged ones might significantly affect affinity of probes to targets.42 In order to evaluate the effects of the novel monomers M2 and M3 on the hybridization properties of LNA/DNA oligonucleotides, we determined thermal denaturation temperatures (Tm) of the duplexes formed by POC1–POC6 with complementary DNA/RNA, and we compared the resulting Tm values to those of the unmodified (DNAref) and corresponding M1-functionalized duplexes (Table 1). The thermal denaturation experiments were performed in medium and low salt phosphate buffers ([Na+] = 110 mM and 20 mM, respectively, 0.1 mM EDTA, pH 7.0) using 0.5 μM concentration of the two complementary strands (Table 1). All the Tm curves recorded for modified oligonucleotides displayed S-shaped monophasic transitions similar to those of the unmodified reference duplexes (Fig. S3, ESI†). Generally duplexes of the prepared POCs displayed higher Tm values when bound to the RNA target than to the DNA target. This is in good agreement with previous data on stability of nucleic acid complexes containing derivatives of 2′-amino-LNA.27,29 Single insertion of monomer M2 and M3 did not affect Tm values of the resulting duplexes compared to insertion of M1 but improved binding affinities (ΔTm + 2–4 °C) were observed relative to the unmodified reference duplexes. In the case of double modification, conjugates containing monomers M2 or M3 spaced by 3 nucleotides showed generally higher Tm values than conjugates having an 11 nucleotide distance between the attached peptides. This implies a stabilizing interaction between the attached moieties arising in the former complexes (Table 1, data for POC2 and POC5 compared to POC3 and POC6, respectively). The superior duplex stability obtained with the double-labeled POCs relative to unmodified and M1-containing references could furthermore result from the increased flexibility of the former probes, which might be beneficial for a more favourable entropy term for complex formation.43The Tm values of duplexes formed by the POCs were strongly decreased in a low salt medium although a significant stabilizing effect was observed when comparing the POCs with the unmodified references under similar conditions. It is noteworthy that proximate and distant incorporation of peptides resulted in much higher difference of Tm values measured in the low salt buffer than in the medium salt buffer. This was especially significant for M2-labeled POC2 and POC3 which displayed up to 10 °C higher Tm values with a 3 nucleotide linker than with an 11 nucleotide linker. Altogether, the observed stabilization of the duplexes formed by POCs in both medium and low salt media suggests successful orientation of the attached peptides along the double-stranded nucleic acid complexes.
Next, we investigated the sensitivity of the selected POCs and corresponding M1-labeled precursors toward single-nucleotide mismatches at different sites along the target DNA/RNA (Table 2 and Tables S3–S4, ESI†). Single incorporation of monomer M2 or M3 resulted in a decrease in Tm values in the presence of a single-nucleotide mismatch in all the investigated positions along the target DNA/RNA. The decrease in binding affinity was of similar magnitude for POC1, POC4 and the LNA reference ON1 (ΔTm ∼ 12 °C and ∼14 °C for DNA and RNA targets, respectively, in comparison to fully complementary complexes; Table S3, ESI†). In contrast, double insertion of the peptides resulted in superior mismatch sensitivity of the POCs relative to M1-labeled oligonucleotides (Table 2 compared to Table S4, ESI†).
| POC: target duplexa |
| Tm/°C | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| DNA target | RNA target | |||||||||
| A | C | T | G | A | C | U | G | |||
| a Sequences of DNA target variants are displayed; cc = complementary complex; n.t. = no clear transition detected. Medium salt phosphate buffer ([Na+] = 110 mM, 0.1 mM EDTA, pH 7.0). Tm values measured using 0.5 μM concentration of complementary strands. | ||||||||||
| 5′-TGC ACT CTA M2GT CM2G TAT CAT | 54.0 | 65.0cc | 58.0 | 55.0 | 50.0 | 69.0cc | 52.0 | 58.0 | ||
3′-ACG TGA GAT ACA GA ATA GTA ATA GTA | ||||||||||
| 5′-TGC ACT CTA M2GT CM2G TAT CAT | 65.0cc | 57.0 | 58.0 | 55.0 | 69.0cc | 53.5 | 61.0 | 55.0 | ||
3′-ACG TGA GAT ACA G C ATA GTA C ATA GTA | ||||||||||
| 5′-TGC ACT CTA M2GT CM2G TAT CAT | 65.0cc | 55.0 | 57.0 | 57.0 | 69.0cc | 61.0 | 60.5 | 63.5 | ||
3′-ACG TGA GAT  CA GAC ATA GTA CA GAC ATA GTA | ||||||||||
| 5′-TGC ACT CTA M2GT CM2G TAT CAT | 57.0 | 56.0 | 65.0cc | 52.0 | 61.0 | 69.0cc | 61.0 | 63.0 | ||
3′-ACG TGA GA ACA GAC ATA GTA ACA GAC ATA GTA | ||||||||||
| 5′-TGC ACM2 CTA TGT CTG TAM2 CAT | 53.0 | 56.0 | 53.0 | 63.0cc | 50.0 | 50.0 | 49.0 | 70.0cc | ||
3′-ACG TGA GAT ACA GAC ATA 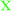 TA TA | ||||||||||
| 5′-TGC ACM2 CTA TGT CTG TAM2 CAT | 63.0cc | 52.0 | 55.0 | 53.0 | 70.0cc | 52.0 | 50.0 | 49.0 | ||
3′-ACG TGA GAT ACA GAC AT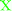 GTA GTA | ||||||||||
| 5′-TGC ACM2 CTA TGT CTG TAM2 CAT | 63.0cc | 54.0 | 54.0 | 55.0 | 70.0cc | 55.0 | 48.0 | 50.0 | ||
3′-ACG TGA GAT AC GAC ATA GTA GAC ATA GTA | ||||||||||
| 5′-TGC ACM2 CTA TGT CTG TAM2 CAT | 49.0 | 51.0 | 48.0 | 63.0cc | n.t. | 48.0 | 49.0 | 70.0cc | ||
3′-ACG TGA  AT ACA GAC ATA GTA AT ACA GAC ATA GTA | ||||||||||
Generally, a mismatch opposite to M2 within POC2 and POC3 was discriminated in a similar manner as if being located 1 nucleotide next to the modification. This is opposed to the typical behaviour of LNA/DNA conjugates, usually displaying better discrimination of a mismatch located opposite to an LNA nucleotide.29 Among the double-labeled conjugates, POC3 showed superior mismatch discrimination in binding both DNA and RNA targets (Table 2). The Tm decrease for POC3 in the presence of a single-nucleotide mismatch was 8–13 °C and 5–10 °C for DNA and RNA targets, respectively, when compared to M1-labeled reference ON3. It is noteworthy that Tm values for all the POCs investigated were less dependent on the type of mismatched nucleotide when compared to the references ON1–ON3. The observed sensitivity of the double-labeled POCs to single-nucleotide mismatches points to the strong impact of the attached peptides on the probes’ binding properties.42 Importantly, a low sensitivity of the prepared POCs to the nature of mismatched nucleobase and its position along the target strand exclude the possibility of intercalation or placing of the attached peptide moieties within the groove of the duplexes, since both these modes involve active interaction between the modification and the nucleic acid complex.29
To investigate the structure of the duplexes we measured UV-Vis and CD spectra. As can be seen from Fig. 3, all the duplexes involving the POCs displayed exclusively characteristic oligonucleotide absorption bands in the UV region at λmax ∼ 259–262 nm. Next, CD curves indicated intermediate A/B duplex geometry of the duplexes similar to the control LNA/DNA duplexes.44 Compared to the unmodified reference duplexes, LNA-induced perturbation of duplex geometry resulted in simultaneously shifted CD maxima and changed peak intensities. Thus, A/B-type duplexes formed by the POCs prepared herein displayed strong CD signals around 210 nm and 260 nm of negative and positive magnitude, respectively, accompanied by a week negative signal at ∼240 nm (Fig. 3b). However, having very low molar extinction coefficients ε ∼ 200–1500 cm−1 M−1, the attached peptides did not provide any additional signals in the UV-Vis or CD spectra. Furthermore, the similarity of CD curves observed for the POCs and reference LNA/DNA oligonucleotides excludes structural perturbation of the complexes by the modification with peptides, and we can assume that the attached peptides successfully allow the formation of a stable A/B-type duplex structure typical for LNA/DNA conjugates. Finally, no difference was observed between UV-Vis and CD spectra of POCs having monomers M2 and M3, most likely owing to the structural similarity of the Met- and Leu-enkephalins, and to the low optical impact of the peptides (Fig. S4, ESI†).
 | ||
| Fig. 3 Representative UV-Vis (a) and CD spectra (b) of POCs and reference oligonucleotides/duplexes. The spectra were recorded in a medium salt buffer at 19 °C using 2.0 μM concentration of complementary strands. | ||
Stability of POCs in human serum
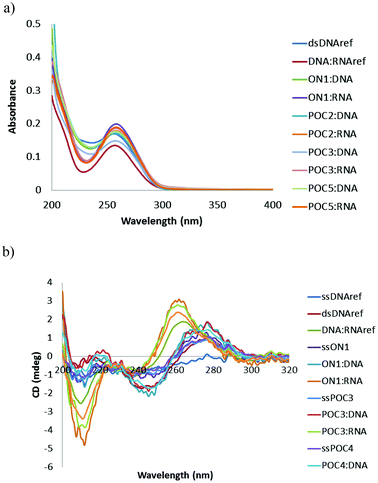
Stability of synthetic oligonucleotides in diverse biological media, including human serum (HS), is of crucial significance for their application as diagnostic and therapeutic tools in vivo.45 Chemical modification with synthetic nucleotide analogues, especially LNA,46 is known to dramatically improve the enzymatic stability of oligonucleotides.42,46 In our study we investigated the stability of the novel POCs in a weakly diluted HS, a complex medium containing several classes of enzymes, which is very similar to the media of in vivo studies and clinical trials.47 In these experiments, unmodified DNA reference and M1-labeled analogues were used as controls.We chose POC2 and POC5 for the serum stability studies, since these conjugates displayed high target binding affinities and selectivity (Tables 1 and 2). The POCs of choice and the corresponding controls were 5′-end labeled with 32P and incubated in HS according to the protocol in the ESI.† Samples were withdrawn after 2, 5, 10, 30 min and after 1, 4, 8, and 24 h and resolved on denaturing (7 M urea) acrylamide gels (Fig. 4; Fig. S5, ESI†). As is evident from Fig. 4, the unmodified DNAref was degraded already within 30 min, even when HS was diluted to 10% (Fig. 4a and b). The LNA/DNA reference ON2 was degraded within 1 h in a 90% HS (Fig. 4c). Interestingly, the partial products of degradation of ON2 displayed substantial stability (>8 h, Fig. 4c). The autoradiogram observed for ON2 points to stability of the LNA modified conjugates after cleavage close to modified position 14. In turn, the peptide-modified analogues POC2 and POC5 demonstrated significant resistance to enzymatic cleavage in 90% HS (>8 h; Fig. 4d and Fig. S5, ESI†). The two characteristic bands observed for both POCs could be referred to the heterogeneity of the prepared conjugates (purity ≥ 95% as evaluated by IE HPLC) or following radiolabelling, as two bands were observed in the initial samples (0 min). Finally, the complete disappearance of the bands after 8 h might be due to phosphatase activity of HS removing the 32P-labels from the POCs.47
 | ||
| Fig. 4 Gel electrophoresis of 5′-32P-labeled oligonucleotides after incubation in human serum; (a and b) unmodified reference DNA in 90% and 10% serum, respectively, (c) ON2 (in 90%, serum), and (d) POC2 (in 90% serum). | ||
Notably, the resistance of the investigated POC2 and POC5 to degradation up to 8 h was comparable and even superior to previously reported stabilities of other modified oligonucleotides in complex biological media.47 For example, J. Kurreck et al. investigated the stability of LNA/DNA antisense mixmers in diluted HS, which digested unmodified control 18-mer DNA in 3–4 h.47 The authors observed stability up to 15 h for the LNA/DNA chimeras, when the probes had four sequential incorporations of LNA nucleotides at both the 3′- and 5′-termini. Notably, in a recent work by N. K. Andersen et al., a dramatic degradation of internally-labeled oligonucleotides having phenyl-triazole modifications was reported within 15 min after treatment with diluted fetal calf serum.47 The increased stability of the POCs prepared herein is due to the peptide residues attached to single-stranded oligonucleotides,18 combined with the stabilization achieved due to the presence of the 2′-amino-LNA nucleotides.47
Additional digestion experiments with HS pre-treated with 1 mM diethyl 4-nitrophenyl phosphate (paraoxon-ethyl), selectively inhibiting the proteolytic activity of the serum, explained the mechanism of the observed digestion.48 Thus, experiments with HS pre-treated with paraoxon-ethyl revealed similar degradation patterns for POC2 and POC5 as in the case of the initial assay, confirming the nuclease attack as a main digestion mechanism (Fig. S5c and d, ESI†).
Molecular modelling
To further rationalize the obtained experimental data, molecular models of the single-stranded POCs and their duplexes were built (Fig. 5 and Fig. S6, ESI†). The initial structures were built and modified with LNA monomers in the B-type duplex conformation using the MacroModel suite of programs.49,50 Having minimized the structures using the AMBER force field as implemented by MacroModel,51 the models were imported into the Chem3D Pro 12.0 program, where the peptide residues, created using ChemDraw Ultra 12.0, were attached to the 2′-amino-LNA scaffolds. The resulting structures were minimized and afterwards subjected to MM2 dynamics (ESI†).52 The obtained complexes were further analyzed in Swiss-Pdb Viewer 4.1.0, where a structure of the dsDNA binding human exonuclease 1 was imported (Fig. 5c and ESI†).53 Finally, graphical images were processed in the PyMOL Molecular Graphics System. Models of the resulting single-stranded POCs and duplexes, as well as of the lack of interaction with human exonuclease 1, are shown in Fig. 4. As one can see, modeling indicated successful positioning of the peptides along single-stranded oligonucleotides and duplexes (Fig. 5a and b). The modelling does not support intercalation and/or positioning of the attached residues within the grooves, as previously reported for 2′-amino-modified analogues of 2′-amino-LNA.29 The minor influence of the attached peptide modifications on the duplex structure agrees with the observed low sensitivity of the POCs to a single-nucleotide mismatch in DNA/RNA targets (Table 2). Remarkably, but in good agreement with the CD spectra described above, we did not observe any difference in duplex parameters between conjugates possessing monomers M2 and M3 (Fig. S6, ESI†). We can also conclude that a small variation at the C-termini of the peptide residue within monomers M2 and M3 has only a minor effect on the duplex geometries, hydration and electrostatic properties, resulting in similar melting temperatures and CD profiles of the duplexes, containing internally attached Met- and Leu-enkephalins.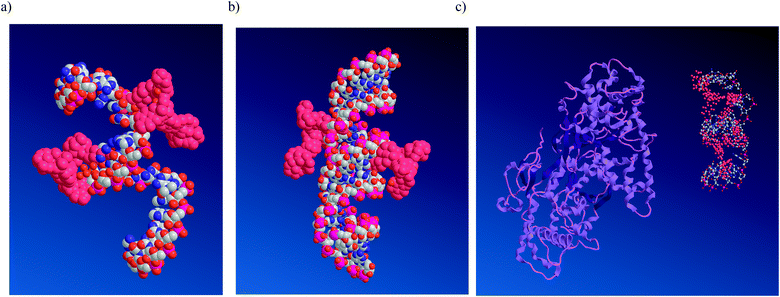 | ||
Fig. 5 Representative energy-minimized structures: (a) ssPOC5, (b) POC5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DNA, and (c) ssPOC5 and human exonuclease 1 (no interaction due to shielding by the peptide residues). Nucleic acids are shown as white, red, pink and blue balls corresponding to carbon, oxygen, phosphorus and nitrogen atoms, respectively; hydrogen atoms are not shown; peptide residues are indicated in crimson; human exonuclease 1 is shown in purple (c). DNA, and (c) ssPOC5 and human exonuclease 1 (no interaction due to shielding by the peptide residues). Nucleic acids are shown as white, red, pink and blue balls corresponding to carbon, oxygen, phosphorus and nitrogen atoms, respectively; hydrogen atoms are not shown; peptide residues are indicated in crimson; human exonuclease 1 is shown in purple (c). | ||
Finally, an effective association of the peptide residues with single-stranded oligonucleotide chains was observed in the model of ssPOC5 (Fig. 5a). The proposed association is most likely caused by electrostatic interactions and hydrogen bonding between the peptide residues and nucleic acid phosphate backbone, as well as by hydrogen bonding between the peptides and nucleobases.18 This type of structure is in good agreement with the high enzymatic stability of the POCs, as the internally attached peptides provide shielding of the oligonucleotide strand, which prevents interaction of the POCs with the enzymes present in HS, e.g. human exonuclease 1 (Fig. 5c).
Experimental methods
Reagents
Phosphoramidite 1 was prepared as described.37 Peptide strands derivatized with terminal azide groups 3–4 and TBTA41 for click chemistry were obtained from Caslo ApS, Denmark, and Lumiprobe LLC, respectively. Other reagents and solvents were used as received. Stock solutions for click chemistry were prepared as described.23,24 Human serum (HS), paraoxon-ethyl and Hank's buffered salt solution (HBSS) were purchased from Sigma-Aldrich; enzymatic activity of the serum was confirmed by following a published protocol.48Synthesis of oligonucleotides ON1–ON3
Oligonucleotide synthesis was carried out on a PerSpective Biosystems Expedite 8909 instrument in the 1 μmol scale using the manufacturer's standard protocols. For the incorporation of monomer M1 a hand-coupling procedure was applied (25 min coupling). The coupling efficiencies of standard DNA phosphoramidites and reagent 1 based on the absorbance of the dimethoxytrityl cation released after each coupling varied between 95% and 100%. Cleavage from a solid support and removal of nucleobase protecting groups were performed using 32% aq. ammonia and methylamine 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v, for 4 h at rt. The resulting oligonucleotides were purified by DMT-ON RP-HPLC using the Waters System 600 equipped with an Xterra MS C18-column (5 μm, 150 mm × 7.8 mm). Elution was performed starting with an isocratic hold of A-buffer for 5 min followed by a linear gradient to 70% B-buffer over 40 min at a flow rate of 1.0 mL min−1 (A-buffer: 0.05 M triethyl ammonium acetate, pH 7.4; B-buffer: 25% buffer A, 75% CH3CN). RP-purification was followed by detritylation (80% aq. AcOH, 30 min), precipitation (acetone, −18 °C, 12 h) and washing with acetone two times. The identity and purity of the oligonucleotides were then verified by MALDI-TOF mass spectrometry and IE HPLC, respectively (ESI†). MALDI-TOF mass-spectrometry analysis was performed using a MALDI-LIFT system on the Ultraflex II TOF/TOF instrument from Bruker and using a HPA-matrix (10 mg 3-hydroxypicolinic acid, 50 mM ammonium citrate in 70% aqueous acetonitrile). IE HPLC was performed using a Merck Hitachi LaChrom instrument equipped with a Dionex DNAPac Pa-100 column (250 mm × 4 mm). Elution was performed at 60 °C starting with an isocratic hold of A- and C-buffers for 2 min followed by a linear gradient to 60% B-buffer over 28 min at a flow rate of 1.0 mL min−1 (A-buffer: MQ water; B-buffer: 1 M NaClO4, C-buffer: 25 mM Tris-Cl, pH 8.0). Unmodified DNA/RNA strands were obtained from commercial suppliers and used without further purification.
1, v/v, for 4 h at rt. The resulting oligonucleotides were purified by DMT-ON RP-HPLC using the Waters System 600 equipped with an Xterra MS C18-column (5 μm, 150 mm × 7.8 mm). Elution was performed starting with an isocratic hold of A-buffer for 5 min followed by a linear gradient to 70% B-buffer over 40 min at a flow rate of 1.0 mL min−1 (A-buffer: 0.05 M triethyl ammonium acetate, pH 7.4; B-buffer: 25% buffer A, 75% CH3CN). RP-purification was followed by detritylation (80% aq. AcOH, 30 min), precipitation (acetone, −18 °C, 12 h) and washing with acetone two times. The identity and purity of the oligonucleotides were then verified by MALDI-TOF mass spectrometry and IE HPLC, respectively (ESI†). MALDI-TOF mass-spectrometry analysis was performed using a MALDI-LIFT system on the Ultraflex II TOF/TOF instrument from Bruker and using a HPA-matrix (10 mg 3-hydroxypicolinic acid, 50 mM ammonium citrate in 70% aqueous acetonitrile). IE HPLC was performed using a Merck Hitachi LaChrom instrument equipped with a Dionex DNAPac Pa-100 column (250 mm × 4 mm). Elution was performed at 60 °C starting with an isocratic hold of A- and C-buffers for 2 min followed by a linear gradient to 60% B-buffer over 28 min at a flow rate of 1.0 mL min−1 (A-buffer: MQ water; B-buffer: 1 M NaClO4, C-buffer: 25 mM Tris-Cl, pH 8.0). Unmodified DNA/RNA strands were obtained from commercial suppliers and used without further purification.Post-synthetic click chemistry
Concentrations of oligonucleotides were calculated using the following extinction coefficients (OD260/μmol): G, 10.5; A, 13.9; T, M1, 7.9; C, 6.6; M2, 8.1; M3, 9.2. Extinction coefficients at 260 nm of monomers M2–M3 were determined by summarizing the extinction coefficient of monomer M1 and the corresponding azide 3–4. The latter values were measured at 19 °C in 5% DMSO–water, v/v.UV-visible absorbance and thermal denaturation studies
UV-visible absorbance spectra and thermal denaturation experiments were performed on a Beckman Coulter DU800 UV/VIS spectrophotometer equipped with a Beckman Coulter High Performance Temperature Controller in a medium salt phosphate buffer (100 mM NaCl, 2 mM NaH2PO4, 8 mM Na2HPO4, 0.1 mM EDTA, pH 7.0), and a low salt phosphate buffer (10 mM NaH2PO4, 10 mM Na2HPO4, 0.1 mM EDTA, pH 7.0). Concentrations of oligonucleotides and POCs were calculated using the extinction coefficients listed above. Complementary strands (0.5 μM of each strand) were thoroughly mixed, denaturated by heating for 10 min at 85 °C and subsequently cooled to the starting temperature of the experiment (15 °C). Thermal denaturation temperatures (Tm values, °C) were determined as the maximum of the first derivative of the thermal denaturation curve (A260vs. temperature). Reported Tm values are an average of at least two measurements with the resulting Tm within ±0.5 °C.CD measurements
CD spectra were recorded on a JASCO J-815 CD spectrometer equipped with a CDF 4265/15 temperature controller. Samples for CD measurements were prepared as described in the UV-visible absorbance and thermal denaturation studies section except that a concentration of 2.0 μM of both strands was used. Quartz optical cells with a path-length of 0.5 cm were used.Serum stability assay
The oligonucleotides were 32P-labeled at the 5′-end with T4 kinase by following standard procedures. 0.1 pmol radioactive oligonucleotides mixed with 3.9 pmol unlabelled oligonucleotide were added to human serum (16 μL, 90% or 10% in HBSS buffer, pH = 7.4) in a pcr tube in a total volume of 20 μL. Pre-treatment of the human serum with 1 mM solution of paraoxon-ethyl was performed as decribed.48 The samples were incubated in an Eppendorf Mastercycler personal at 37 °C. The aliquots were withdrawn after 2 min, 5 min, 10 min, 30 min, and 1 h, 4 h, 8 h and 24 h time intervals and quenched with 1 vol of ice-cold 95% formamide with excess EDTA. The incubated samples were then resolved on 13% denaturing polyacrylamide gels (7 M urea; 1× TBE) and visualized by autoradiography on a Typhoon Trio Variable Mode Imager (Amersham Biosciences).Molecular modelling
The initial oligonucleotides were built and modified with LNA monomers in the B-type duplex conformation in the MacroModel suite of programs.49 The minimization was performed using the AMBER force field as implemented by MacroModel.50 Peptide residues were incorporated into the resulting low-energy structures using ChemBioDraw Ultra 12.0 and Chem3D Pro 12.0. The structures were further minimized by MM2 as implemented in Chem3D Pro 12.0 and subjected to MM2 dynamics (step interval: 2; frame interval: 10; heating speed: 1 K; target temperature: 300 K).51 Resulting structures were further analyzed using Swiss-Pdb Viewer 4.1.0. The structure of the human exonuclease 1 used in the modeling of interactions with POC2 was adopted from Pdb databank and used without modification.53 Resulting structures were additionally processed in the PyMOL Molecular Graphics System.Conclusions
In summary, we have successfully developed a new strategy for internal CuAAC labeling of LNA/DNA oligonucleotides with peptides and applied it for the preparation of novel POCs possessing internal peptide groups. This method gives high yields and allows incorporation of peptides into desired positions of oligonucleotides via a chemically and enzymatically stable 1,2,3-triazole linker.54 Our system using 2′-amino-LNA as a new scaffold for CuAAC attachment of peptides will be suitable for the incorporation of other groups (such as cell-penetrating peptides,55 carbohydrates,56 and small molecules57) into synthetic oligonucleotides. This can significantly benefit the preparation strategy of modified oligonucleotides as advanced tools for nanobiotechnology, supramolecular chemistry and chemical biology.Moreover, attachment of peptides to 2′-amino-LNA by CuAAC chemistry allowed us for the first time to study in detail target binding affinity and selectivity, spectral behaviour and enzymatic stability of an oligonucleotide containing clinically and biologically important enkephalins in the internal positions. Of particular note is the hybrid nature of the prepared peptides, combining encephalin residues with cationic lysines, providing high propensity to interaction with nucleic acids. The performed studies indicated high affinity and selectivity of the prepared POCs to complementary DNA and RNA targets. POCs having double incorporation of peptides separated by three nucleotides showed up to 8 h stability in weakly diluted human serum (90%), whereas LNA/DNA and unmodified DNA controls were degraded in 1 h and 30 min, respectively. The experimental data of the POCs were in good agreement with molecular modeling. The oligonucleotide chain, which is typically rapidly degraded by complex enzyme composition of human serum, appears to be protected from digestion by shielding peptide chains located outside the helix. The results presented demonstrate that the properties of oligonucleotides can be strongly influenced by internally conjugated peptides. The novel internally labeled POCs described herein have a high potential for applications both in vitro and in vivo.
Acknowledgements
The Sapere Aude programme of the Danish Council for Independent Research is acknowledged for funding. The VILLUM FOUNDATION is thanked for funding the Biomolecular Nanoscale Engineering Center.Notes and references
- M. Parisien, K. F. Freed and T. R. Sosnick, PLoS One, 2012, 7, e32647 CAS.
- R. Sukackaite, S. Grazulis, G. Tamulaitis and V. Siksnys, Nucleic Acids Res., 2012, 40, 7552 CrossRef CAS; M. AlQuraishi and H. H. McAdams, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 14819 CrossRef.
- M. Cox and D. R. Nelson, Lehninger principles of biochemistry, 4th edn, W.H. Freeman, New York, 2005 Search PubMed.
- M. Simons and M. D. Szczelkun, Nucleic Acids Res., 2011, 39, 7656 CrossRef CAS.
- S. Abe, M. Kurata, S. Suzuki, K. Yamamoto, K. Aisaki, J. Kanno and M. Kitagawa, PLoS One, 2012, 7, e40129 CAS.
- D. Kim, B. J. Blus, V. Chandra, P. Huang, F. Rastinejad and S. Khorasanizadeh, Nat. Struct. Mol. Biol., 2010, 17, 1027 CAS.
- H. Kaur and L.-Y. Lanry Yung, PLoS One, 2012, 7, e31196 CAS.
- V. G. B. Ruigrok, M. Levisson, J. Hekelaar, H. Smidt, B. W. Dijkstra and J. van der Oost, Int. J. Mol. Sci., 2012, 13, 10537 CrossRef CAS.
- L. Meng, L. Yang, X. Zhao, L. Zhang, H. Zhu, C. Liu and W. Tan, PLoS One, 2012, 7, e33434 CAS.
- B. A. R. Williams and J. C. Chaput, Curr. Protoc. Nucleic Acid Chem., 2010, 42 Search PubMed , 4.41.1; D. A. Stetsenko and M. J. Gait, Methods Mol. Biol., 2004, 288, 205 Search PubMed; V. Marchán, E. Pedroso and A. Grandas, Chem.–Eur. J., 2004, 10, 5369 CrossRef.
- J. D. Carter and T. H. LaBean, J. Nucleic Acids, 2011, 2011, 926595 CrossRef.
- F. Diezmann, H. Eberhard and O. Seitz, Biopolymers, 2010, 94, 397 CrossRef CAS.
- M. Frieden, A. Aviñó, G. Tarrasón, M. Escorihuela, J. Piulats and R. Eritja, Chem. Biodiversity, 2004, 1, 930 Search PubMed; D. Gottschling, H. Selinger, G. Tarrasón, J. Piulats and R. Eritja, Bioconjugate Chem., 1998, 9, 831 CrossRef CAS.
- M. Wenska, M. Alvira, P. Steunenberg, Å. Stenberg, M. Murtola and R. Strömberg, Nucleic Acids Res., 2011, 39, 9047 CrossRef CAS.
- T. P. Wang, N. C. Ko, Y. C. Su, E. C. Wang, S. Severance, C. C. Hwang, Y. T. Shih, M. H. Wu and Y. H. Chen, Bioconjugate Chem., 2012, 23, 2417 CrossRef CAS.
- W. Gong, T. Zhou, J. Mo, S. Perrett, J. Wang and Y. Feng, J. Biol. Chem., 2012, 287, 8531 CrossRef CAS; A. E. Rapoport and E. N. Trifonov, Gene, 2011, 488, 41 CrossRef.
- C. M. Santiveri, B. C. Lechtenberg, M. D. Allen, A. Sathyamurthy, A. M. Jaulent, S. M. Freund and M. Bycroft, J. Mol. Biol., 2008, 382, 1107 CrossRef CAS.
- J. G. Harrison and S. Balasubramanian, Nucleic Acids Res. Suppl., 1998, 26, 3136 CrossRef CAS , and references therein.
- C. Spiteri and J. E. Moses, Angew. Chem., Int. Ed., 2010, 49, 31 CrossRef CAS , and references therein.
- C. W. Tornøe, C. Christensen and M. Meldal, J. Org. Chem., 2002, 67, 3057 CrossRef.
- V. V. Rostovtsev, L. G. Green, V. V. Fokin and K. B. Sharpless, Angew. Chem., Int. Ed., 2002, 41, 2596 CrossRef CAS.
- V. Hong, S. I. Presolski, C. Ma and M. G. Finn, Angew. Chem., Int. Ed., 2009, 48, 9879 CrossRef CAS.
- A. V. Ustinov, I. A. Stepanova, V. V. Dubnyakova, T. S. Zatsepin, E. V. Nozhevnikova and V. A. Korshun, Rus. J. Bioorg. Chem., 2010, 36, 401 CrossRef CAS , and references therein.
- A. H. El-Sagheer and T. Brown, Chem. Soc. Rev., 2010, 39, 1388 RSC , and references therein.
- K. Gogoi, M. V. Mane, S. S. Kunte and V. A. Kumar, Nucleic Acids Res., 2007, 35, e139 CrossRef.
- A. A. Koshkin, S. K. Singh, P. Nielsen, V. K. Rajwanshi, R. Kumar, M. Meldgaard, C. E. Olsen and J. Wengel, Tetrahedron, 1998, 54, 3607 CrossRef CAS; S. Obika, D. Nanbu, Y. Hari, J. Andoh, K. Morio, T. Doi and T. Imanishi, Tetrahedron Lett., 1998, 39, 5401 CrossRef.
- S. K. Singh, R. Kumar and J. Wengel, J. Org. Chem., 1998, 63, 6078 CrossRef CAS.
- M. D. Sørensen, L. Kvaernø, T. Bryld, A. E. Håkansson, L. Keinicke, B. Verbeure, P. Herdewijn and J. Wengel, Nucleic Acids Res. Suppl., 2001, 1, 25 CrossRef.
- P. J. Hrdlicka;, B. R. Babu, M. D. Sørensen, N. Harrit and J. Wengel, J. Am. Chem. Soc., 2005, 127, 13293 CrossRef CAS; M. Kalek, A. S. Madsen and J. Wengel, J. Am. Chem. Soc., 2007, 129, 9392 CrossRef; I. V. Astakhova, V. A. Korshun, K. Jahn, J. Kjems and J. Wengel, Bioconjugate Chem., 2008, 19, 1995 CrossRef.
- Y. Gao, X.-L. Liu and X. R. Li, Int. J. Nanomed., 2011, 6, 1017 CrossRef CAS , and references therein; F. J. Hernandez, N. Kalra, J. Wengel and B. Vester, Bioorg. Med. Chem. Lett., 2009, 19, 6585 CrossRef; M. Terrazas and E. T. Kool, Nucleic Acids Res., 2009, 37, 346 CrossRef.
- K. K. Karlsen and J. Wengel, Nucleic Acid Ther., 2012, 22, 366 CAS , and references therein.
- M. W. Johannsen, L. Crispino, M. C. Wamberg, N. Kalra and J. Wengel, Org. Biomol Chem., 2011, 9, 234 Search PubMed.
- M. Noda and Y. Teranishi, Nature, 1982, 297, 431 CrossRef CAS.
- E. S. Olson, M. A. Whitney, B. Friedman, T. A. Aguilera, J. L. Crisp, F. M. Baik, T. Jiang, S. M. Baird, S. Tsimikas, R. Tsien and Q. T. Nguyen, Integr. Biol., 2012, 4, 595 RSC.
- A. P. Johnston, M. M. Kamphuis, G. K. Such, A. M. Scott, E. C. Nice, J. K. Heath and F. Caruso, ACS Nano, 2012, 6, 6667 CrossRef CAS.
- I. K. Astakhova and J. Wengel, Chem.–Eur. J., 2012, 19, 1112 CrossRef.
- A. Søndergaard Jørgensen, P. Gupta, J. Wengel and I. K. Astakhova, ChemBioChem Search PubMed , submitted.
- H. Wu, et al., Nature, 2012, 485, 327 CrossRef CAS.
- A. A. Thompson, et al., Nature, 2012, 485, 395 CrossRef CAS.
- S. I. Presolski1, V. P. Hong and M. G. Finn, Curr. Protoc. Chem. Biol., 2011, 3, 153 Search PubMed.
- Tris[(1-benzyl-1H-1,2,3-triazol-4-yl)methyl] amine (TBTA): T. R. Chan, R. Hilgraf, K. B. Sharpless and V. V. Fokin, Org. Lett., 2004, 6, 2853 CrossRef CAS.
- M. Manoharan, Biochim. Biophys. Acta, 1999, 1, 117 Search PubMed.
- M. Mandel and J. Marmur, Methods Enzymol., 1968, 12, 198 Search PubMed.
- W. Saenger, Principles of Nucleic Acid Structure, Springer-Verlag, New York, Berlin, Heidelberg, Tokyo, 1983 CrossRef CAS; F. Yuan, L. Griffin, L. Phelps, V. Buschmann, K. Weston and N. L. Greenbaum, Nucleic Acids Res., 2007, 35, 2833 CrossRef CAS.
- J. Wengel, B. Vester, L. B. Lundberg, S. Douthwaite, M. D. Sørensen, B. R. Babu, M. J. Gait, A. Arzumanov, M. Petersen and J. T. Nielsen, Nucleosides, Nucleotides Nucleic Acids, 2003, 22, 601 CAS.
- For example, L. Crouzier, C. Dubois, J. Wengel and R. N. Veedu, Bioorg. Med. Chem. Lett., 2012, 22, 4836 CrossRef CAS.
- J. Kurreck, E. Wyszko, C. Gillen and V. A. Erdmann, Nucleic Acids Res., 2002, 30, 1911 CrossRef CAS; N. K. Andersen, H. Døssing, F. Jensen, B. Vester and P. Nielsen, J. Org. Chem., 2011, 76, 6177 CrossRef.
- O. S. Gudmundsson, K. Nimkar, S. Gangwar, T. Siahaan and R. T. Borchardt, Pharm. Res., 1999, 16, 16 CrossRef CAS.
- MacroModel, version 9.1, Schrödinger, LLC, New York, NY, 2005 Search PubMed.
- S. J. Weiner, P. A. Kollman, D. T. Nguen and D. A. Case, J. Comput. Chem., 1986, 7, 230 CrossRef CAS.
- S. J. Weiner, P. A. Kollman, D. A. Case, U. C. Singh, C. Ghio, G. Alagona, S. Profeta and P. K. Weiner, J. Am. Chem. Soc., 1984, 106, 765 CrossRef CAS.
- U. Burkert and N. L. Allinger, Molecular Mechanics, ACS Monograph 177, American Chemical Society, Washington, DC, 1982 Search PubMed.
- J. Orans, E. A. McSweeney, R. R. Iyer, M. A. Hast, H. W. Hellinga, P. Modrich and L. S. Beese, Cell, 2011, 145, 212 CrossRef CAS.
- A. H. El-Sagheer and T. Brown, Acc. Chem. Res., 2012, 45, 1258 CrossRef CAS , and references therein.
- M. J. Gait, Cell. Mol. Life Sci., 2003, 60, 1 CrossRef.
- D. J. Lee, S. H. Yang, G. M. Williams and M. A. Brimble, J. Org. Chem., 2012, 77, 7564 CrossRef CAS; G. Pourceau, A. Meyer, J.-J. Vasseur and F. Morvan, J. Org. Chem., 2009, 74, 1218 CrossRef.
- A. Salic and T. J. Mitchison, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 2415 CrossRef CAS.
Footnote |
† Electronic supplementary information (ESI) available: Characterization of oligonucleotide conjugates, representative melting curves, CD spectra, autoradiograms of oligonucleotides incubated with HS, and molecular model of POC2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DNA are included in the ESI. See DOI: 10.1039/c3ob40786a DNA are included in the ESI. See DOI: 10.1039/c3ob40786a |
| This journal is © The Royal Society of Chemistry 2013 |

