InP/ZnS as a safer alternative to CdSe/ZnS core/shell quantum dots: in vitro and in vivo toxicity assessment
Virgilio
Brunetti
a,
Hicham
Chibli
b,
Roberto
Fiammengo
a,
Antonio
Galeone
a,
Maria Ada
Malvindi
a,
Giuseppe
Vecchio
a,
Roberto
Cingolani
c,
Jay L.
Nadeau
b and
Pier Paolo
Pompa
*a
aIstituto Italiano di Tecnologia (IIT), Center for Bio-Molecular Nanotechnologies@UniLe, Via Barsanti, 73010 Arnesano, Lecce, Italy. E-mail: pierpaolo.pompa@iit.it; Fax: +39-0832-295708; Tel: +39-0832-295714
bDepartment of Biomedical Engineering, McGill University, 3775 Rue University, Montreal, QC H3A 2B4, Canada
cIstituto Italiano di Tecnologia (IIT), Central Research Laboratories, Via Morego, 30, 16136 Genova, Italy
First published on 30th October 2012
Abstract
We show that water soluble InP/ZnS core/shell QDs are a safer alternative to CdSe/ZnS QDs for biological applications, by comparing their toxicity in vitro (cell culture) and in vivo (animal model Drosophila). By choosing QDs with comparable physical and chemical properties, we find that cellular uptake and localization are practically identical for these two nanomaterials. Toxicity of CdSe/ZnS QDs appears to be related to the release of poisonous Cd2+ ions and indeed we show that there is leaching of Cd2+ ions from the particle core despite the two-layer ZnS shell. Since an almost identical amount of In(III) ions is observed to leach from the core of InP/ZnS QDs, their very low toxicity as revealed in this study hints at a much lower intrinsic toxicity of indium compared to cadmium.
Introduction
Quantum dots (QDs), fluorescent semiconductor nanocrystals, are a very interesting class of nanomaterials with important imaging applications in biology and medicine.1 However, it was realized early in their development that the use of these materials poses serious concerns about toxicity and safety, especially because the most popular and well-studied QDs contain cadmium: CdSe, CdTe, and CdS. Cd is known to be highly toxic and carcinogenic for living systems. Several investigations of Cd-based QDs have quantified the toxicity of these materials in cell cultures and suggested strategies to reduce it.2 Most QDs used for biological applications are not bare core particles, but possess shell layers of other materials in order to improve optical properties and prevent QD breakdown in solution. A “shell” of one to two monolayers of ZnS is needed to improve the quantum yield of CdSe QDs to values useful for imaging. In addition to that shell, the QDs can be rendered water-soluble by highly stable silica shells or polymers.3 While a shell plus a stable organic coat significantly reduce Cd2+ leaching, it still remains questionable whether QDs could ever be approved for medical applications. A promising alternative is the use of QDs based upon Cd-free materials, such as InP. As with CdSe, InP cores must be protected by a ZnS shell to prevent core breakdown. Only a few studies on the toxicity of InP QDs have been published and most of them have been of limited breadth, focusing mostly on toxicity in cell culture. Even if InP-based QDs appear to be a safer alternative to Cd-containing QDs, there is still very little known about their toxicity in vivo, and there is so far no example for a direct comparison with Cd-containing QDs.4This work presents in vitro (cell culture) as well as in vivo (animal model) toxicity assessment of water-soluble InP/ZnS and CdSe/ZnS QDs. It provides a direct comparison of the impact of the QD core material on toxicity in cells and in animals. The only difference between the two investigated types of QDs is the chemical nature of their core (InP vs. CdSe) while all other properties known to affect cellular responses – particle size, charge, shell and surface chemistry – are kept constant. Under these identical conditions, it is confirmed that InP QDs show greatly reduced toxicity compared to CdSe QDs, making them a valuable alternative for biologically oriented applications.
Results and discussion
QD preparation and characterization
Water-soluble InP/Zns and CdSe/ZnS QDs (both with two ZnS shell layers) capped with mercaptopropionic acid5 were prepared according to published procedures.5–7 The materials were investigated via TEM (Fig. 1A and B) and found to be monodisperse and with a narrow size distribution. Comparable dimensions (11.3 ± 0.6 nm for InP/ZnS and 13.4 ± 0.7 nm for CdSe/ZnS) and narrow size distributions were confirmed by dynamic light scattering (DLS, Fig. 1E and F). The surface charge was similar for both types of nanocrystals (−26 ± 9 mV for InP/ZnS and −24 ± 5 mV for CdSe/ZnS, Fig. 1C and D). An increase in the QD dimensions to approx. 30 nm (Fig. 1G and H) was observed in both cases after 24 h incubation in cell culture medium (DMEM supplemented with 10% fetal bovine serum, FBS), which reflects the formation of a protein corona around the QDs, as already observed for other kinds of nanoparticles.8–10 | ||
| Fig. 1 (A and B) TEM images of water-dispersible CdSe/ZnS and InP/ZnS QDs; (C and D) ζ-potential analyses and (E and F) DLS plots of CdSe/ZnS and InP/ZnS QDs dispersed in water and (G and H) after 24 h incubation in DMEM supplemented with 10% FBS at 37 °C. DLS values are the average of at least 10 runs of 15 measurements. ζ-potential values are the average of at least 10 runs of 30 measurements. | ||
The release of metal ions from the QD core was quantified over a period of 96 h under simulated lysosomal conditions (37 °C, citrate buffer pH 4.5). Incubation of 40 nM QDs for 48 h, which correspond to 854 μM Cd and 835 μM In, led to leaching of approximately 3 μM core metal ions for both nanocrystal types (Fig. 2). For longer times, we observed a higher release of cadmium compared to indium, indicating a higher stability of InP/ZnS QDs towards hydrolysis, probably due to the robustness of the covalent bond in III–V semiconductors.11
 | ||
| Fig. 2 Time-dependent release of cadmium or indium ions from CdSe/ZnS and InP/ZnS QDs, respectively, in an aqueous environment mimicking lysosomal conditions (37 °C, pH 4.5). Data are reported as mean ± SD. All values were consistent among triplicates performed in the same experiment, with error bars smaller than symbols. | ||
In vitro toxicity investigations
A first set of experiments compared the toxicity of water soluble InP/ZnS and CdSe/ZnS QDs in vitro. The epithelial cell line A549 (human lung carcinoma) and the neuronal cell line SH SY5Y (human neuroblastoma) were chosen for these investigations and cultivated in the presence of increasing concentrations of CdSe/ZnS or InP/ZnS QDs (1 pM to 5 nM). Cell viability was measured after 24 and 48 h incubation using the WST-8 test (Water-Soluble Tetrazolium salt). As expected, CdSe/ZnS QDs produced a significantly larger decrease in cell viability compared to InP/ZnS QDs.5 The toxic effect of Cd-containing QDs was clearly observable after 24 h incubation at 1 nM concentration (Fig. 3A and B). In the case of prolonged exposure (48 h), a detectable reduction in cell viability was observed for CdSe/ZnS QD concentrations as low as 10 pM. The highest toxicity was found for the neuronal cell line, with a 27% viability loss after 24 h which grew up to 46% after 48 h (Fig. 3B, red bars) with 5 nM CdSe/ZnS QDs. The epithelial cell line appeared to be more resistant under these conditions, with a loss of viability of 33% after 48 h (Fig. 3A, red bars) with 5 nM CdSe/ZnS. In contrast, incubation for 48 h with 5 nM InP/ZnS QDs produced only a statistically insignificant reduction in cell viability (<10%) for both cell lines (Fig. 3A and B, blue bars).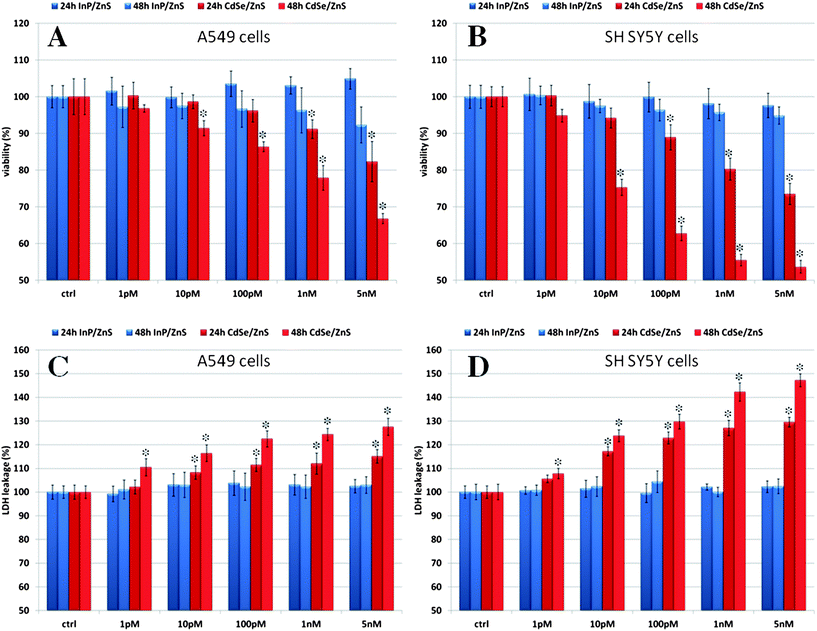 | ||
| Fig. 3 (A and B) WST-8 proliferation assay and (C and D) LDH assay on A549 and SH SY5Y cells incubated with increasing concentrations of InP/ZnS and CdSe/ZnS QDs at different times (24 and 48 h). Ctrl identifies the negative controls in the absence of QDs. Data are reported as mean ± SD from three independent experiments; *P < 0.05 compared with control (n = 8). | ||
To better understand the molecular reasons underlying the observed reduction in cell viability, we performed a series of additional assays. The LDH (lactate dehydrogenase) leakage assay was used to gain information about possible cell membrane damage. The results showed that indeed the treatment with CdSe/ZnS QDs induced significant membrane damage in both cell types, possibly at concentrations as low as 1 pM, already observable after 24 h for the neuronal cell line and after 48 h for the epithelial line (Fig. 3C and D, red bars). By comparison, no significant increase in LDH was observed upon incubation with InP/ZnS QDs at any time and tested concentration compared to the controls (Fig. 3C and D, blue bars).
QD-treated cell cultures were also investigated to detect the occurrence of oxidative stress. A positive DCF-DA (dichloro fluorescein diacetate) assay after incubation of cell cultures with as little as 1 pM Cd-containing QDs for 24 h (data not shown) indicated generation of intracellular reactive oxygen species (ROS). However, it has been reported that quantification of ROS generated by Cd-containing QDs via this assay may lead to overestimation of ROS levels, possibly due to direct interaction of photooxidized QDs with the dye.12 To confirm the induction of ROS caused by oxidative stress, we used here additionally real-time qPCR and followed the expression levels of SOD1, SOD2, CAT, and Gpx genes coding for antioxidant and detoxifying enzymes. The induction of antioxidant enzymes, such as superoxide dismutase (SOD), catalase (CAT), and glutathione peroxidase (Gpx), is observed upon treatment with low concentrations of a wide variety of chemical agents as well as a consequence of physical stress. Together with high intracellular levels of glutathione (GSH), these enzymes protect the cell from oxidative stress, which may cause lethal damage to DNA, RNA, proteins, and lipids. SOD quenches the free radical superoxide by converting it to peroxide, which can then be inactivated by reactions catalyzed by CAT or Gpx. CAT disproportionates intracellular hydrogen peroxide to water and molecular oxygen and Gpx, a selenium-containing peroxidase, catalyzes the reduction of a variety of hydroperoxides using GSH.13 The expression profiles obtained under conditions of non-photoactivation of the QDs indicated that the cells experienced oxidative stress when treated with Cd-containing QDs, most probably due to intracellular degradation of the nanocrystals with concomitant release of Cd2+ ions.14 In contrast, incubation with InP/ZnS QDs did not result in any detectable over-expression of genes correlated with oxidative stress at any tested concentration, demonstrating once again the very low in vitro toxicity of these nanocrystals under the investigated conditions (Fig. 4A and B, green bars). All results presented so far revealed a higher resistance of the epithelial cell line A549 compared to the neuronal cell line SH SY5Y towards the treatment with Cd-containing QDs. In particular, considering the expression profiles presented in Fig. 4A and B, it can be observed that the Gpx gene was over-expressed only in A549 cells. Gpx is the most important H2O2-scavenging enzyme, closely associated with the maintenance of reduced glutathione. It has been reported that A549 cells can efficiently counteract the Cd-induced generation of ROS, likely due to constitutively high levels of glutathione.15 It has also been shown that these cells are able to develop resistance to cadmium by an enhanced expression of γ-glutamylcysteine synthetase, the enzyme catalyzing the rate-determining step in cellular glutathione synthesis.15 More recently, it was found that Cd-resistant A549 subclones over-express the gene encoding for glutathione peroxidase (GPX2), possibly leading to a higher antioxidant defense in these cells. This enzyme plays a fundamental role in the protection of the airway epithelium under oxidative stress conditions.16,17
![mRNA expression level analyzed by RT-qPCR of A549 (A) and SH SY5Y (B) cells treated with 5 nM CdSe/ZnS or InP/ZnS QDs for 24 h. Data are reported as mean ± SD from three independent experiments; *P < 0.05 compared with control (n = 8). [Ca2+]i measurements for A549 (C) and SH SY5Y (D) cells treated with 10 nM InP/ZnS or CdSe/ZnS QDs for 24 h. Cells were pretreated with BAPTA/AM or EGTA in the presence or in the absence of QDs. Data are reported as mean ± SD from three independent experiments; *P < 0.05 compared with control (n = 8).](/image/article/2013/NR/c2nr33024e/c2nr33024e-f4.gif) | ||
| Fig. 4 mRNA expression level analyzed by RT-qPCR of A549 (A) and SH SY5Y (B) cells treated with 5 nM CdSe/ZnS or InP/ZnS QDs for 24 h. Data are reported as mean ± SD from three independent experiments; *P < 0.05 compared with control (n = 8). [Ca2+]i measurements for A549 (C) and SH SY5Y (D) cells treated with 10 nM InP/ZnS or CdSe/ZnS QDs for 24 h. Cells were pretreated with BAPTA/AM or EGTA in the presence or in the absence of QDs. Data are reported as mean ± SD from three independent experiments; *P < 0.05 compared with control (n = 8). | ||
The significantly higher toxicity of Cd-containing QDs shown by the neuronal cell line SH SY5Y is probably a direct consequence of the leach of Cd2+ ions from the QDs. Studies have shown that Cd2+ ions produce severe alteration of calcium homeostasis and an increase of the oxidative stress experienced by the cell, which results in apoptosis of primary neurons as well as PC12 and SH SY5Y cell lines.18 In particular, it has been shown that Cd2+ ions induce an increase of intracellular Ca2+ ([Ca2+]i), which mediates the activation of the MAPK and mTOR pathways, determining neuronal death under stress conditions. We have, therefore, measured the level of [Ca2+]i using the fluorescent dye Fluo-3/AM,18 for both cell lines after treatment with 10 nM CdSe/ZnS QDs or InP/ZnS QDs.
The results showed a significant increase in [Ca2+]i only for the treatment with Cd-containing QDs (Fig. 4C and D, red bars), while the InP-based QDs did not produce any appreciable increase (Fig. 4C and D, blue bars). The neuronal cell line gave, as expected, a higher increase of [Ca2+]i compared to the epithelial line (Fig. 4D), confirming the higher sensitivity of these cells. The detrimental increase in [Ca2+]i could be completely suppressed by pretreatment with chelating agents, such as EGTA or BAPTA-AM. Taken together, these results indicate that the observed cellular responses were indeed most likely due to the intracellular release of Cd2+ ions.
TUNEL assays were performed to reveal possible damage to cellular DNA. While the percentage of TUNEL positive nuclei remained below 1% even for treatments with 5 nM InP/ZnS QDs (Fig. 5A and B, blue bars), the same amount of CdSe/ZnS QDs produced 8% positive nuclei for A549 cells and 9% for SH SY5Y cells (Fig. 5A and B, red bars). These data hinted once again at intracellular release of Cd2+ ions, which has been shown to heavily interfere with DNA repair machinery, for instance mediating conformational changes of protein p53.19,20
 | ||
| Fig. 5 (A and B) TUNEL assay on A549 and SH SY5Y cells incubated with increasing concentrations of InP/ZnS and CdSe/ZnS QDs. Results are reported after 24 h incubation; ctrl indicates the negative control (in the absence of QDs). Data are reported as mean ± SD from three independent experiments; *P < 0.05 compared with control (n = 8). | ||
Finally, the intracellular distribution of CdSe/ZnS and InP/ZnS QDs was investigated by confocal microscopy. Representative images of the two cell lines treated with QDs are shown in Fig. 6. In both cases we found that the nanocrystals were internalized similarly by the cells according to their very similar size and surface chemistry (and protein corona), and localized predominantly in the cytoplasm and in the perinuclear region. We then quantified the cellular uptake by ICP-AES measurements. These quantitative analyses revealed that the amounts of Cd or In for a given cell line were practically indistinguishable, indicating identical uptake for the two QD types, which was again expected due to their very similar properties in the cell culture medium. In particular, 24 h incubation with 1 nM QDs gave: 1.80 ± 0.13 fmol of Cd and 1.81 ± 0.10 fmol of In for A549 cells, and 1.69 ± 0.07 fmol of Cd and 1.69 ± 0.05 fmol of In for SH SY5Y cells. However, the two investigated cell lines seem to have a different capability of internalizing nanocrystals, with the epithelial cell line A549 internalizing, at all times and concentrations tested, slightly more QDs compared to the SH SY5Y neurons.
 | ||
| Fig. 6 (Top) Bioaccumulation of QDs in A549 and SH SY5Y cells. Cells were exposed at 1, 5, and 10 nM of CdSe/ZnS and InP/ZnS QDs for 24 h. Data are reported as mean ± SD from three independent experiments; (bottom) representative confocal-microscopy images of SH SY5Y and A549 cells treated with 5 nM InP/ZnS or CdSe/ZnS QDs for 24 h. In all images, red spots represent the emission from InP/ZnS or CdSe/ZnS QDs (cells were imaged in bright field). | ||
In summary, our in vitro studies showed very clearly that, given a certain QD size and surface chemistry, CdSe/ZnS QDs produced toxic effects on A549 cells as well as on SH SY5Y cells, whereas InP/ZnS QDs were well tolerated at all investigated conditions. Furthermore, our measurements point out that, under conditions of non-photoactivation, the main toxicity cause can be ascribed to the intracellular release of Cd2+ ions, which occurred despite the ZnS shell, as shown in Fig. 2. On the other hand, the good biocompatibility shown by InP/ZnS QDs was derived from the combined lower release of metal ions from the QD core and the much reduced reactivity of indium.
In vivo toxicity investigations
Drosophila is an excellent model for toxicological21,22 and genetic studies23,24 due to its short life cycle, high genetic homology with the human genome, and limited ethical issues. Fruit flies have been recently exploited to study the toxicity of some chemicals22 and nanoparticles25–28 showing their suitability for in vivo screening of nanomaterials. Here we report the results of our studies on the toxicity of CdSe/ZnS and InP/ZnS QDs in fruit flies fed with QD-supplemented food. We focused on real-time qPCR to clarify the mechanisms underlying QD toxicity, investigating variations in expression levels of genes involved in stress response and DNA damage, such as hsp70, hsp83, p53 and Dredd genes.Flies were fed with food containing two different sub-nanomolar concentrations of CdSe/ZnS or InP/ZnS QDs (100 and 500 pM), or alternatively with food supplemented with an equal amount of nanocrystal supernatant solution and with standard food as control. As shown in Fig. 7, hsp70 and hsp83 were over-expressed in flies treated with CdSe/ZnS QDs (3 to 5 times for the 100 pM and up to 10 times for the 500 pM QD-enriched food), indicating a significant and systemic toxicity of CdSe/ZnS QDs. Hsp70 and hsp83 are proteins involved in the stress response, and are part of the heat shock protein (Hsp) family. Up-regulation of Hsps is important to counteract proteotoxic effects since these proteins play various roles, including chaperoning other proteins during synthesis, folding, assembly and degradation. The heat shock response is induced in a protective way in response to systemic toxicity, involving generation of abnormal proteins along with alteration of cellular functions. The expression of Hsps is also strictly related to increased ROS generation.29
 | ||
| Fig. 7 mRNA expression levels (analyzed by RT-qPCR) for Drosophila flies treated with two different concentrations (100 or 500 pM) of InP/ZnS or CdSe/ZnS QDs. The table lists the primers used in the RT-qPCR experiments. Data are reported as mean ± SD from three independent experiments; *P < 0.05 compared with control (n = 8). | ||
We also observed an approximately 3-fold over-expression of p53 (for 100 pM QD-enriched food), indicating the presence of genomic perturbation. p53 is a critical component of cellular mechanisms that responds to genotoxic stresses, such as DNA damage and hypoxia, maintaining genomic integrity by arresting cell-cycle progression and/or by inducing apoptosis.30,31
Finally, the expression levels of Dredd showed that cellular death induced by treatment with CdSe/ZnS QDs in Drosophila was mostly a consequence of apoptosis. The expression of Dredd was approximately 8-fold the control for 100 pM, and 25–30 times the control for 500 pM QD-enriched food. In contrast, Drosophila flies treated with InP/ZnS QDs did not present any significant increase in gene expression related to systemic stress (hsp70 and hsp83), genome damage repair (p53), or apoptosis (Dredd), compared to the controls.
A direct necrosis/apoptosis test on Drosophila larval hemocytes was also carried out (Fig. 8). These cells are primarily involved in the immune response of the larvae and their physiological function is very similar to that of nucleated blood cells in mammals. A high apoptotic rate is a sign of systemic intoxication involving all insect tissues. Combined annexin V and propidium iodide (PI) staining showed indeed a high apoptosis rate for flies treated with CdSe/ZnS QDs in comparison to the control or to flies treated with InP/ZnS QDs, in agreement with the gene expression data reported above. Yet, due to the extreme sensitivity of Drosophila larval hemocytes to genotoxic compounds, we observed a slight increase of the apoptotic rate even in the case of flies treated with InP/ZnS QDs. This might be ascribed to the local generation of some ROS species due to QD oxidation5 along with accumulation of In or Zn ions.
 | ||
| Fig. 8 (Top) Apoptosis/necrosis assay performed on circulating haemocytes. Data are reported as mean ± SD from three independent experiments; *P < 0.05 compared with control (n = 8). (Bottom) Representative confocal images of Drosophila larval haemocytes treated with CdSe/ZnS or InP/ZnS QDs. Green fluorescence shows annexin V-FITC positive haemocytes. | ||
Conclusions
Our systematic study on the toxicity of water soluble core/shell CdSe/ZnS and InP/ZnS QDs with comparable physical and chemical properties, except the nature of the particle core, very clearly shows that InP/ZnS QDs are a significantly safer alternative to CdSe/ZnS QDs. We show that this conclusion holds for in vitro (cell culture) as well as for in vivo (animal model Drosophila) applications. The CdSe/ZnS QDs were observed to induce: (1) cell membrane damage; (2) conditions of oxidative stress in the cells as indicated by the upregulation of genes coding for antioxidant and detoxifying enzymes; (3) damage of the genetic material and (4) interference with Ca2+ homeostasis. All these effects may be mainly ascribed to the presence of Cd2+ and indeed we could show leaching of ions from the particle core despite the two-layer ZnS shell. Since In(III) ions were also observed to leach in comparable amounts from InP/ZnS QDs, our study indicates that they must have a much lower intrinsic toxicity compared to Cd2+. Furthermore, the high toxicity of CdSe/ZnS QDs compared to InP/ZnS QDs does not depend on higher particle internalization and is clearly observable for cell lines which should be considered “robust” towards Cd2+-poisoning, as well as for more delicate neuronal cell lines. Finally, conditions of systemic and non-specific toxicity and strong induction of cell apoptosis are also evident upon administration of Cd-containing QDs with the diet to the animal model Drosophila, while no such effects were observed for a comparable treatment with InP/ZnS QDs. While the toxicity of Cd-containing QDs has long been recognized, this study shows the importance of having well characterized nanomaterials with comparable physical and chemical properties for meaningful comparative toxicity tests in the case of newly developed nanomaterials.Experimental
InP/ZnS synthesis
InP/ZnS QDs were synthesized according to a published procedure.5 Briefly, hexadecylamine (96 mg, 0.4 mmol), stearic acid (57 mg, 0.2 mmol), zinc undecylenate (340 mg, 0.8 mmol) and indium chloride (44 mg, 0.2 mmol) were mixed with 4 mL octadecene (ODE) under an inert atmosphere (Ar or N2) in a flask equipped with a thermometer and a condenser. The reaction mixture was heated to 270 °C and tris-(trimethylsilyl)phosphine (0.2 mmol, 2 mL of 0.1 M solution in octadecene) was rapidly added. The temperature was decreased and maintained at 240 °C for 20 min. The solution was then cooled down in a water bath at 20 °C. Zinc diethyldithiocarbamate (144 mg, 0.2 mmol) was added to the mixture at room temperature and the temperature was increased and kept at 180 °C for 10 min and 240 °C for 20 min. Timing started when the thermometer reached the desired temperature. The reaction mixture was cooled down to room temperature and 8 mL toluene were added. The mixture was centrifuged at 2500g for 5 min and the precipitate was discarded. The particles were precipitated by adding 42 mL of ethanol to the orange supernatant solution and recovered by centrifugation (20 min, 2500g). The InP/ZnS pellet was redissolved in 3 mL of toluene.QDs were transferred into an aqueous environment by addition of 800 μL butanol, 1000 μL borate buffer (pH 9, 200 mM) and 8 μL mercaptopropionic acid (MPA) (10 μmol) to 200 μL of a 5 μM QD solution. The mixture was heated at 50 °C for 15 min. The two phases were separated and the aqueous layer containing the InP/Zn–MPA was purified by four rounds of washing/filtration on a 10 kDa molecular weight cutoff filter (Vivaspin 500) with 50 mM borate buffer at pH 9. The purified QDs were dissolved in borate buffer at the desired pH and stored at 4 °C.
CdSe/ZnS synthesis
CdSe QDs were synthesized following a method adapted from the literature.32 Briefly, cadmium oxide (CdO) (0.026 g, 0.20 mmol) and oleic acid (OA, 0.895 g, 3.17 mmol) were added to 10 mL ODE. This mixture was degassed for 5 min and heated under nitrogen to 250 °C until the mixture became colorless. The selenium precursor (TOPSe) was prepared by mixing Se (0.01 g, 0.127 mmol) with trioctylphosphine (TOP, 0.415 g, 1.12 mmol) and ODE (2 mL) under N2 into a sealed vial until the solution became light yellow. This solution was rapidly injected into the CdO–ODE mixture at 260 °C. The solution was then cooled down using a water bath directly after the injection of the selenium precursor.Under a N2 atmosphere, TOP (0.415 g, 1.12 mmol), hexamethyldisilathiane ((TMS)2S) (0.084 g, 0.5 mmol), and dimethylzinc (0.3 mL, 2.0 M in toluene, 0.5 mmol) were diluted to 5 mL with ODE. 2.5 mL of this solution were injected into the CdSe QD solution at 170 °C followed by drop wise addition of the remaining 2.5 mL over 5 min. The temperature was then allowed to drop and maintained at 100 °C for 3 h for QD annealing. QDs were purified by 3 consecutive extractions with 100 mL 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 hexane–methanol in a separatory funnel. The unreacted CdO and oleic acid are soluble in the lower methanol phase which can be discarded while the QDs are only soluble in the hexane–ODE upper phase. A large excess of ethanol was finally added to the hexane phase to precipitate the QDs and the sample was centrifuged at 7000g for 1 min. The QD pellet was taken up in an appropriate volume of toluene to reach an optical density of approximately 4 at 400 nm. Aliquots of this QD solution were stored in the dark in air-tight vials.
1 hexane–methanol in a separatory funnel. The unreacted CdO and oleic acid are soluble in the lower methanol phase which can be discarded while the QDs are only soluble in the hexane–ODE upper phase. A large excess of ethanol was finally added to the hexane phase to precipitate the QDs and the sample was centrifuged at 7000g for 1 min. The QD pellet was taken up in an appropriate volume of toluene to reach an optical density of approximately 4 at 400 nm. Aliquots of this QD solution were stored in the dark in air-tight vials.
QDs were transferred into an aqueous environment by ligand exchange. To this end, 1 mL of concentrated QD solution was diluted to 5 mL with methanol. MPA (50 μL) was added and the mixture was adjusted to pH 10 using tetramethylammonium hydroxide pentahydrate (TMAH). This solution was stirred at room temperature for 12 h. At the end, QDs were separated from excess MPA by precipitation upon addition of excess ethyl acetate followed by centrifugation at 7000g for 5 min. The supernatant was discarded and the pellet was dried at room temperature for 1 h. Finally the pellet was redissolved in 2 mL of doubly distilled water and the solution was filtered through a 0.2 μM membrane filter (Millipore, Billerica, MA), and stored at room temperature, protected from light.
In vitro assays
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 U mL−1 penicillin and 10
000 U mL−1 penicillin and 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 U mL−1 streptomycin (Sigma Aldrich). The cells were maintained under standard cell culture conditions (5% CO2, 95% humidity and 37 °C in a Thermo Scientific incubator) and harvested every 3 days.
000 U mL−1 streptomycin (Sigma Aldrich). The cells were maintained under standard cell culture conditions (5% CO2, 95% humidity and 37 °C in a Thermo Scientific incubator) and harvested every 3 days.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per well and cultured for 24 and 48 h. Different amounts of QDs dispersed in cell culture medium stock solution were added into different wells obtaining final QD concentrations of 1, 10, 100, 500 pM and 1 nM. A final concentration of 5% DMSO in medium was used as the positive control for both cell lines; this treatment generated a reduction in viability to approx. 80–90% compared to the negative control (data not shown in the graphs). Eight replicates were performed for each investigated point including the controls (ctrl = untreated cells) and blanks (medium only). A 10 μL aliquot of WST-8 solution was added to each well. The 96 well microplates were incubated for 3 h in a humidified atmosphere of 5% CO2 at 37 °C. Subsequently, the orange WST-8 formazan product was measured at a wavelength of 460 nm in a Fluo Star Optima (BMG LABTECH) microplate reader. Data were collected by Control Software and elaborated with MARS Data Analysis Software (BMG LABTECH). To express the cytotoxicity, the average absorbance of the wells containing cell culture medium without cells was subtracted from the average absorbance of the solvent control, 5% DMSO or QD treated cells. The percentage of cell viability was calculated using the following equation: (absorbancetreated/absorbancecontrol) × 100.
000 cells per well and cultured for 24 and 48 h. Different amounts of QDs dispersed in cell culture medium stock solution were added into different wells obtaining final QD concentrations of 1, 10, 100, 500 pM and 1 nM. A final concentration of 5% DMSO in medium was used as the positive control for both cell lines; this treatment generated a reduction in viability to approx. 80–90% compared to the negative control (data not shown in the graphs). Eight replicates were performed for each investigated point including the controls (ctrl = untreated cells) and blanks (medium only). A 10 μL aliquot of WST-8 solution was added to each well. The 96 well microplates were incubated for 3 h in a humidified atmosphere of 5% CO2 at 37 °C. Subsequently, the orange WST-8 formazan product was measured at a wavelength of 460 nm in a Fluo Star Optima (BMG LABTECH) microplate reader. Data were collected by Control Software and elaborated with MARS Data Analysis Software (BMG LABTECH). To express the cytotoxicity, the average absorbance of the wells containing cell culture medium without cells was subtracted from the average absorbance of the solvent control, 5% DMSO or QD treated cells. The percentage of cell viability was calculated using the following equation: (absorbancetreated/absorbancecontrol) × 100.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (v/v), diluted to 10 mL with ultrapure water, and the resulting solution was directly analyzed by ICP-AES.
1 (v/v), diluted to 10 mL with ultrapure water, and the resulting solution was directly analyzed by ICP-AES.
In vivo assays
Acknowledgements
The authors gratefully acknowledge V. Fiorelli and B. Antonazzo for the expert technical assistance.References
- W. A. Hild, M. Breunig and A. Goepferich, Eur. J. Pharm. Biopharm., 2008, 68, 153–168 CrossRef CAS.
- S. J. Soenen, P. Rivera-Gil, J.-M. Montenegro, W. J. Parak, S. C. De Smedt and K. Braeckmans, Nano Today, 2011, 6, 446–465 CrossRef CAS.
- T. Pellegrino, L. Manna, S. Kudera, T. Liedl, D. Koktysh, A. L. Rogach, S. Keller, J. Rädler, G. Natile and W. J. Parak, Nano Lett., 2004, 4, 703–707 CrossRef CAS.
- J. Gao, K. Chen, R. Luong, D. M. Bouley, H. Mao, T. Qiao, S. S. Gambhir and Z. Cheng, Nano Lett., 2011, 12, 281–286 CrossRef.
- H. Chibli, L. Carlini, S. Park, N. M. Dimitrijevic and J. L. Nadeau, Nanoscale, 2011, 3, 2552–2559 RSC.
- S. J. Clarke, C. A. Hollmann, F. A. Aldaye and J. L. Nadeau, Bioconjugate Chem., 2008, 19, 562–568 CrossRef CAS.
- S. Xu, J. Ziegler and T. Nann, J. Mater. Chem., 2008, 18, 2653–2656 RSC.
- G. Maiorano, S. Sabella, B. Sorce, V. Brunetti, M. A. Malvindi, R. Cingolani and P. P. Pompa, ACS Nano, 2010, 4, 7481–7491 CrossRef CAS.
- B. Sahoo, M. Goswami, S. Nag and S. Maiti, Chem. Phys. Lett., 2007, 445, 217–220 CrossRef CAS.
- M. A. Malvindi, V. Brunetti, G. Vecchio, A. Galeone, R. Cingolani and P. P. Pompa, Nanoscale, 2012, 4, 486–495 RSC.
- D. J. Bharali, D. W. Lucey, H. Jayakumar, H. E. Pudavar and P. N. Prasad, J. Am. Chem. Soc., 2005, 127, 11364–11371 CrossRef CAS.
- D. R. Cooper, N. M. Dimitrijevic and J. L. Nadeau, Nanoscale, 2010, 2, 114–121 RSC.
- R. L. T. Matsumoto, D. H. M. Bastos, S. Mendonça, V. S. Nunes, W. Bartchewsky, M. L. Ribeiro and P. de Oliveira Carvalho, J. Agric. Food Chem., 2009, 57, 1775–1780 CrossRef CAS.
- K. G. Li, J. T. Chen, S. S. Bai, X. Wen, S. Y. Song, Q. Yu, J. Li and Y. Q. Wang, Toxicol. in Vitro, 2009, 23, 1007–1013 CrossRef CAS.
- E. L. Hatcher, Y. Chen and Y. J. Kang, Free Radical Biol. Med., 1995, 19, 805–812 CrossRef CAS.
- A. Singh, T. Rangasamy, R. K. Thimmulappa, H. Lee, W. O. Osburn, R. Brigelius-Flohé, T. W. Kensler, M. Yamamoto and S. Biswal, Am. J. Respir. Cell Mol. Biol., 2006, 35, 639–650 CrossRef CAS.
- F. Croute, B. Beau, J.-C. Murat, C. Vincent, H. Komatsu, F. Obata and J.-P. Soleilhavoup, J. Toxicol. Environ. Health, Part A, 2005, 68, 703–718 CrossRef CAS.
- B. Xu, S. Chen, Y. Luo, Z. Chen, L. Liu, H. Zhou, W. Chen, T. Shen, X. Han, L. Chen and S. Huang, PLoS One, 2011, 6, e19052 CAS.
- T. Schwerdtle, F. Ebert, C. Thuy, C. Richter, L. H. F. Mullenders and A. Hartwig, Chem. Res. Toxicol., 2010, 23, 432–442 CrossRef CAS.
- W. K. B. Khalil, E. Girgis, A. N. Emam, M. B. Mohamed and K. V. Rao, Chem. Res. Toxicol., 2011, 24, 640–650 CrossRef CAS.
- B. H. Jennings, Mater. Today, 2011, 14, 190–195 CrossRef.
- D. Bhargav, M. Pratap Singh, R. C. Murthy, N. Mathur, D. Misra, D. K. Saxena and D. K. Chowdhuri, Ecotoxicol. Environ. Saf., 2008, 69, 233–245 CrossRef CAS.
- J. Botas, Nat. Genet., 2007, 39, 589–591 CrossRef CAS.
- G. J. Lieschke and P. D. Currie, Nat. Rev. Genet., 2007, 8, 353–367 CrossRef CAS.
- M. Ahamed, R. Posgai, T. J. Gorey, M. Nielsen, S. M. Hussain and J. J. Rowe, Toxicol. Appl. Pharmacol., 2010, 242, 263–269 CrossRef CAS.
- X. Liu, D. Vinson, D. Abt, R. H. Hurt and D. M. Rand, Environ. Sci. Technol., 2009, 43, 6357–6363 CrossRef CAS.
- P. P. Pompa, G. Vecchio, A. Galeone, V. Brunetti, G. Maiorano, S. Sabella and R. Cingolani, Nanoscale, 2011, 3, 2889–2897 RSC.
- P. P. Pompa, G. Vecchio, A. Galeone, V. Brunetti, S. Sabella, G. Maiorano, A. Falqui, G. Bertoni and R. Cingolani, Nano Res., 2011, 4, 405–413 CrossRef CAS.
- M. P. Singh, M. M. K. Reddy, N. Mathur, D. K. Saxena and D. K. Chowdhuri, Toxicol. Appl. Pharmacol., 2009, 235, 226–243 CrossRef CAS.
- M. Zylicz, F. W. King and A. Wawrzynow, EMBO J., 2001, 20, 4634–4638 CrossRef CAS.
- K. Somasundaram, Front. Biosci., 2000, 5, D424–D437 CrossRef CAS.
- A. Subashini, M. K. Karl, A. Ammar, R. C. Alessandra, M. Zuze, L. C. Vicki, V. M. Nikos and S. W. Michael, Nanotechnology, 2005, 16, 2000 CrossRef.
| This journal is © The Royal Society of Chemistry 2013 |
