From natural product to marketed drug: the tiacumicin odyssey
William
Erb
*a and
Jieping
Zhu
*b
aLaboratoire de Chimie Organique, ESPCI, 10 rue Vauquelin, 75231, Paris Cedex 05, France. E-mail: w.erb@exchem.fr
bInstitut of Chemical Sciences and Engineering, École Polytechniques Fédérale de Lausanne, EPFL-SB-ISIC-LSPN, CH-1015 Lausanne, Switzerland. E-mail: jieping.zhu@epfl.ch
First published on 30th October 2012
Abstract
Covering: 1975 to 2012
The first members of the tiacumicin family of antibiotics, encompassing more than 40 compounds, were isolated in 1975. Structurally, the core aglycon is an 18-membered macrolactone having two conjugated diene units, one isolated double bond, 5 stereogenic centers and most often, at least one glycosidic linkage. Tiacumicin B, a RNA synthesis inhibitor, is a narrow-spectrum antibiotic against clostridia. For the treatment of Clostridium difficile infection (CDI), it has the same cure rate as vancomycin but with lower relapse rate and was approved by the FDA in May 2011. The aim of this review is to present an overview of the chemistry and biology of tiacumicins since their discovery.
 William Erb | William Erb studied chemistry at the University of Paris-Sud XI. He received his PhD in organic chemistry under the guidance of Pr. Jieping Zhu (Institut de Chimie des Substances Naturelles) in 2010. He then joined the group of Pr. Varinder Aggarwal at the University of Bristol, working on organocatalysis and its application to the synthesis of natural products. He is currently Attaché Temporaire d'Enseignement et de Recherche in the group of Janine Cossy in Paris, working on total synthesis and metal-catalyzed reactions. |
 Jieping Zhu | Jieping Zhu received his B. Sc from Hangzhou Normal University and his M.Sc. degree from Lanzhou University (P. R. China) under the guidance of Professor Li Yulin. He got his Ph.D. degree from University Paris XI, France under the supervision of Professor H.-P. Husson and Pr. J. C. Quirion. After 18 months post-doctoral stay with Professor Sir D. H. R. Barton at Texas A & M University in USA, he joined in the “Institut de Chimie des Substances Naturelles”, CNRS, France as Chargé de Recherche and was promoted to Director of Research 2nd class in 2000 and then 1st class in 2006. He moved to Ecole Polytechnique Fédérale de Lausanne (Swiss Federal Institute of Technology Lausanne), Switzerland in September 2010 as a full professor. |
1 Introduction
Microbial diversity represents an almost infinite pool for the discovery of novel compounds. There are more than 23![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 known microbial secondary metabolites, close to 60% of them produced by bacteria, and their usefulness in drug development is well established.1–4
000 known microbial secondary metabolites, close to 60% of them produced by bacteria, and their usefulness in drug development is well established.1–4
Actinomycetes are among the most morphologically diverse prokaryotes and are widely distributed all around the Earth.5,6 They arouse the attention of the scientific community due to the great diversity and biological activities associated with the corresponding metabolites: antimicrobial, antifungal, immunosuppressive, antitumor, etc. Compounds such as erythromycin, streptomycin, amphotericin B and rapamycin, all sold as drugs, came from such strains,7,8 and they are still considered as a promising source of new antibiotics.9–12
In 1975, Parenti and co-workers identified a new substance, lipiarmycin, which exhibited a strong activity against gram-positive bacteria from actinoplanaceae strains (a sub-class of Actinomycetes). A few years later, related compounds were isolated from parent strains and were named clostomicin and tiacumicin. This field remained broadly unexplored until the late 90's when Optimer Pharmaceuticals began the commercial development of one tiacumicin for the treatment of Clostridium difficile infection (CDI). C. difficile is an important nosocomial pathogen frequently diagnosed in infectious hospital-acquired diarrhoeas whose cost is estimated from 433–797 million dollars annually in the USA.13 The research in this field turned out to be highly rewarding since tiacumicin B has recently been approved by the FDA for the treatment of C. difficile infection. The aim of this review is to present an overview of the chemistry and biology of tiacumicin compounds and their application to the treatment of C. difficile associated infection.14–18
2 Isolation and characterization
2.1 Lipiarmycin from Actinoplanes deccanensis
In the 1970s, Parenti and co-workers reported the isolation of a novel antibiotic from a new strain, isolated from a soil sample collected in India, named Actinoplanes deccanensis ATCC 21983.19–21 Growth of the soil extract on agar led to the formation of sporangia that liberated spores by rupture of the wall (Fig. 1). The major compound isolated from this strain was named lipiarmycin, from leap year, because the strain was isolated on February 29th 1972. | ||
| Fig. 1 Sporangium obtained on soil extract-agar. Magnification ×800. | ||
Initial analysis of lipiarmycin revealed the presence of two chlorine atoms, at least one phenolic hydroxyl group, three carbonyls (one saturated and two conjugated to double bonds), a probable sugar moiety, and an aromatic nucleus, which was determined to be homodichloro-orsellinic acid by degradation studies.
A few years later, scientists from Gruppo Lepetit reported the isolation of 2-O-methyl-4-O-homodichloroorsellinate-β-rhamnoside upon acid methanolysis of lipiarmycin.22 This sugar was also found in other natural antibiotics with either an α- or β-glycosidic linkage.23–25 They also identified the second sugar in lipiarmycin as 5-methyl-β-rhamnose. Note that the absolute configurations of both sugars have not been established and were shown artificially in their L form.
Finally, in 1987, Nasini and co-workers reported that the lipiarmycin known at that time was a mixture of two products, lipiarmycin A3 (1) and A4 (2), in a 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio, separable by flash chromatography.26 Chemical degradation and extensive NMR studies allowed them to elucidate the structure of the two lipiarmycins (Fig. 2). These molecules feature a 18-membered macrolactone incorporating four stereogenic centers (unkown configuration), two conjugated dienes and one tri-substituted double bond. The macrolactone is glycosylated by 2-O-methyl-β-D-rhamnose esterified in the 4th position either by homodichloro-orsellinic acid (lipiarmycin A3) or dichloro-orsellinic acid (lipiarmycin A4). The second sugar link to the macrolactone is 4-O-isobutyrate-5-methyl-β-rhamnose, whose absolute configuration was not determined but shown as D form.
1 ratio, separable by flash chromatography.26 Chemical degradation and extensive NMR studies allowed them to elucidate the structure of the two lipiarmycins (Fig. 2). These molecules feature a 18-membered macrolactone incorporating four stereogenic centers (unkown configuration), two conjugated dienes and one tri-substituted double bond. The macrolactone is glycosylated by 2-O-methyl-β-D-rhamnose esterified in the 4th position either by homodichloro-orsellinic acid (lipiarmycin A3) or dichloro-orsellinic acid (lipiarmycin A4). The second sugar link to the macrolactone is 4-O-isobutyrate-5-methyl-β-rhamnose, whose absolute configuration was not determined but shown as D form.
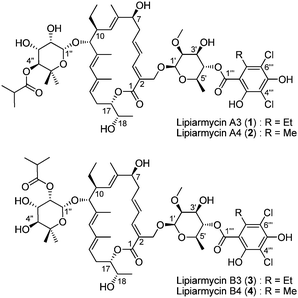 | ||
| Fig. 2 The structure of lipiarmycins. | ||
One year later, two novel lipiarmycins were isolated from the same strain:27 lipiarmycins B3 (3) and B4 (4), which differ from the corresponding A3 and A4 by the position of the isobutyric ester on the 2-O-methyl-β-D-rhamnose moiety (position 2′′ for lipiarmycin B and 4′′ for A). Lipiarmycins B3 and B4 differ through the substituent (methyl or ethyl) of the aromatic ring.
2.2 Clostomicins from Micromonospora echinospora
In 1985, Õmura and co-workers isolated a strain of Micromonospora echinospora from a soil sample collected in a rice field in Japan.28,29 Based on taxonomic studies, it appeared that it was a new strain, subsequently named Micromonospora echinospora subsp. armeniaca subsp. nov. KMR-593.Fermentation yielded five compounds: clostomicins A (5), B1, B2 (6), C (7) and D (8). NMR studies revealed that clostomicin B1 was identical to lipiarmycin A3 previously isolated and that the difference between clostomicins A and B2 resided in the substitution pattern of the 5-methyl-β-rhamnose sugar: the isobutyric ester substituent was proposed to be in the C3′′ position for clostomicin A and in the C2′′ for clostomicin B2. Therefore it was concluded that the structure of clostomicin B2 is identical to lipiarmycin B3.
The IR spectrum of clostomicins C and D showed the presence of an additional carbonyl group compared to lipiarmycin A3, which was attributed to a ketone at the C18 position. The 13C NMR spectra of clostomicin C also reveals the absence of one methyl carbon at 28.6 ppm assigned to the equatorial methyl group of the 5-methyl-β-rhamnose sugar. Therefore, the structure of clostomicins C and D was assigned as shown in Fig. 3.
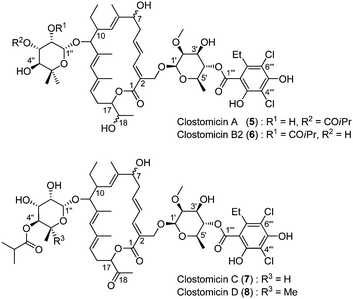 | ||
| Fig. 3 The structure of clostomicins. | ||
2.3 Tiacumicins from Dactylosporangium aurantiacum
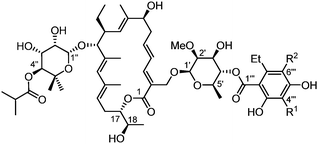
In 1986, McAlpine and co-workers reported a new strain isolated from a soil sample collected in Connecticut, which was named Dactylosporangium aurantiacum subsp. hamdenensis subsp. nov. AB718C-41 (NRRL 18085).30–32 A first fermentation experiment using 20 litres of broth yielded three compounds named tiacumicins A (9), B (10) and C (11) (10 mg, 35 mg and 24 mg, respectively). A second study using a much bigger broth (4500 litres) led to the isolation of three additional tiacumicins (D (12), E (13) and F (14)) in very low yields (7 mg, 20 mg and 13 mg, respectively) compared to tiacumicin B (3.82 g).Extensive NMR studies allowed scientists to propose the structure of tiacumicin B, which was found to be identical to lipiarmycin A3 and clostomicin B1. Tiacumicins C and F are different from tiacumicin B in the position of the isobutyric ester on the 5-methyl-β-rhamnose moiety (respectively at C2′′ and C3′′). Tiacumicin D is another isomer of tiacumicin B in which the 2-O-methyl-β-D-rhamnose is esterified in the C3′ position by homodichloro-orsellinic acid (Fig. 4). Tiacumicin E is almost identical to tiacumicin C except for the ester moiety on position C2′′, which is not an isobutyric ester but a propionate one. Finally, tiacumicin A is a simpler analog without the 2-O-methyl-β-D-rhamnose moiety and with an acetate ester on the 5-methyl-β-rhamnose sugar. Note that although described with the right configuration (2R, 3S, 4S, 5S, 6R) in the text, the 2-O-methyl-β-D-rhamnose was written as its enantiomer in the final structure of tiacumicins.31
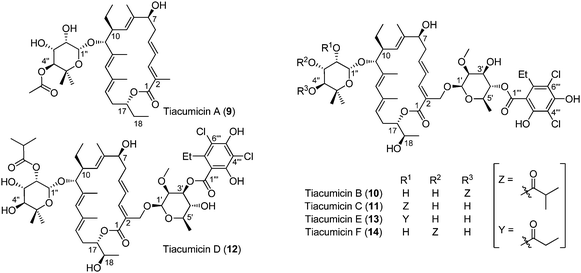 | ||
| Fig. 4 The structure of tiacumicins. | ||
The absolute configuration of the macrolactone was assigned in 2005 by X-ray crystal structure analysis of tiacumicin B. It was subsequently established that C18 has the (R) configuration for tiacumicin B and (S) configuration for lipiarmycin A4.33,34 Even though we cannot conclude definitively, it could be assumed the configuration of C18 to be (R) for other tiacumicins and (S) for lipiarmycins. The C18 configuration remained unassigned for clostomicins.
Therefore, without taking into account the C18 configuration, lipiarmycin A3, clostomicin B1 and tiacumicin B seem to be identical. So are lipiarmycin B3, clostomicin B2 and tiacumicin C or clostomicin A and tiacumicin F.
In the late 90's, with the aim of producing novel tiacumicin analogs, McAlpine and co-workers replaced the potassium chloride added to the broth with potassium bromide.35,36 Using the same Dactylosporangium aurantiacum strain as before, they isolated four new compounds (Fig. 5, 15–18) incorporating bromide on the aromatic ring, whose structure have been elucidated by mass spectroscopy and NMR studies.
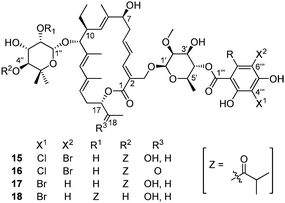 | ||
| Fig. 5 The structure of brominated tiacumicins. | ||
In 2007, scientists from Optimer Pharmaceuticals, Inc. reported seven new members of tiacumicin family (Fig. 6, 19–25), present in very low concentration in the fermentation broth, as judged by the HPLC profile of the mixture and the corresponding integrations (Fig. 6).37,38 During formulation studies, Optimer scientists also identified the new tiacumicin derivative 26. It is identical to tiacumicn B except for the presence of carbonyl function at C7.39 Although the configuration of these new compounds has not been elucidated, they are assumed to be the same as for tiacumicin B.
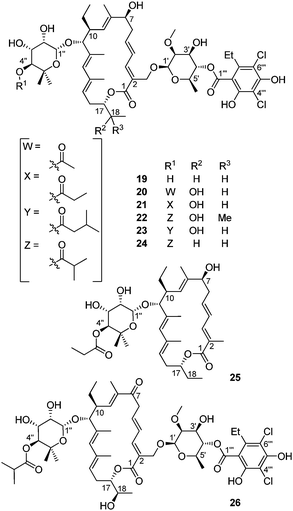 | ||
| Fig. 6 The structure of tiacumicin analogs. | ||
The most important library of tiacumicin analogs was generated by Zhang and co-workers while working on the elucidation of the biosynthesis of tiacumicin B.40–44 Indeed, from different mutants of the Dactylosporangium aurantiacum strain, they have been able to characterize 37 new analogs (Fig. 7). We can note, based on published data, that 50 is the C18 epimer of lipiarmycin A4 (2).
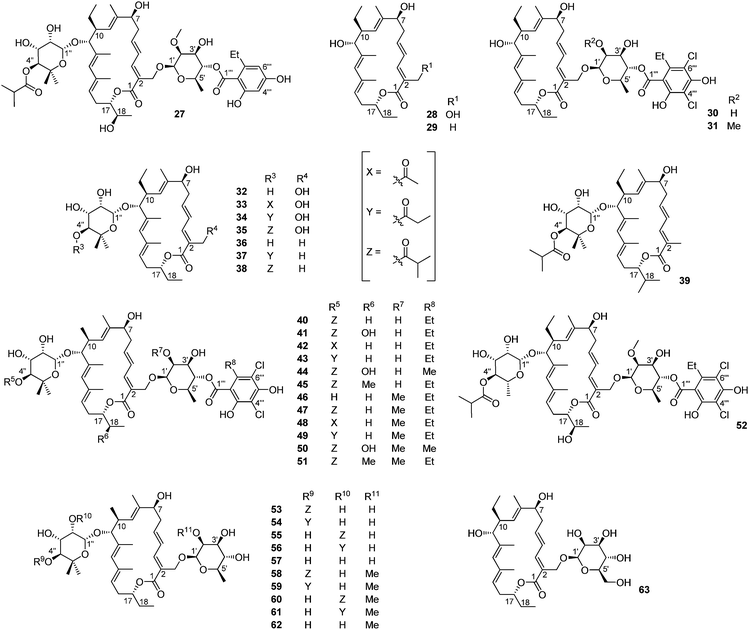 | ||
| Fig. 7 The structure of tiacumicin analogs. | ||
It is interesting to note that the yield of tiacumicin B production by D. aurantiacum has been greatly improved over the years by Optimer scientists.45 Using a growth medium mainly composed of fish powder, glucose, casamino acid, yeast extract and some inorganic salts to support microorganism growth, it is possible to obtain 100–500 mg of crude tiacumicin per litre of broth, much higher than the 18.8 mg L−1 initially reported by McAlpine. The introduction of a resin able to trap macrocycles as they are formed in the broth increased the yield of tiacumicin and facilitated the recovery of product by sieving from the broth and elution with organic solvents. Purification is mainly achieved by reversed-phase medium-pressure liquid chromatography.
2.4 Lipiarmycin from Catellatospora
In 2008, during the course of a screening programme to identify new anti-tuberculosis agents, scientists from Novartis reported the isolation of lipiarmycin A3 from Catellatospora sp. Bp3323-81, a strain from the company's screening library.463 Biosynthesis
Despite more than 30 years of history, little was known about the biosynthesis of tiacumicin until the work of Zhang and co-workers in 2011.40,41 In early studies, Parenti noted that the source of chlorine could be the meat extract used in the broth or the added sodium chloride and that omission of both chlorine sources greatly reduce the yield of lipiarmycin.19 Furthermore, McAlpine has shown that addition of potassium bromide to the broth allows the formation of brominated tiacumicin analogs.35,36 Apart from feeding experiments, the biosynthesis of tiacumicin remained undetermined.In 2011, Zhang and co-workers reported their studies on the biosynthesis of tiacumicin based on a genetic approach on Dactylosporangium aurantiacum hamdenensis NRRL 18085.47–50 Firstly, they targeted polyketide synthase (PKS) and halogenase using probes to identify genes involved in the biosynthesis of tiacumicin. They have been able to identify the complete tia-gene cluster which comprised 50 orfs (Open Reading Frame) and 110![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 633 bp (base pairing). In a first attempt, the putative functions of orfs have been deduced by comparison with protein databases and a further refinement allowed them to eliminate some orfs, which are not involved in tiacumicin biosynthesis. Thus the final gene cluster contained 31 orfs, from gene TiaG1 to TiaR2 for approximately 83 kb.
633 bp (base pairing). In a first attempt, the putative functions of orfs have been deduced by comparison with protein databases and a further refinement allowed them to eliminate some orfs, which are not involved in tiacumicin biosynthesis. Thus the final gene cluster contained 31 orfs, from gene TiaG1 to TiaR2 for approximately 83 kb.
Genes TiaA1 to TiaA4 were predicted as modular polyketide synthases, and inactivation experiments led to the conclusion that they are responsible for the tiacumicin aglycone synthesis (Fig. 8, A). The biosynthesis involved propionyl-CoA, malonyl-CoA, (2S)-methylmalonyl-CoA and (2S)-ethylmalonyl-CoA.
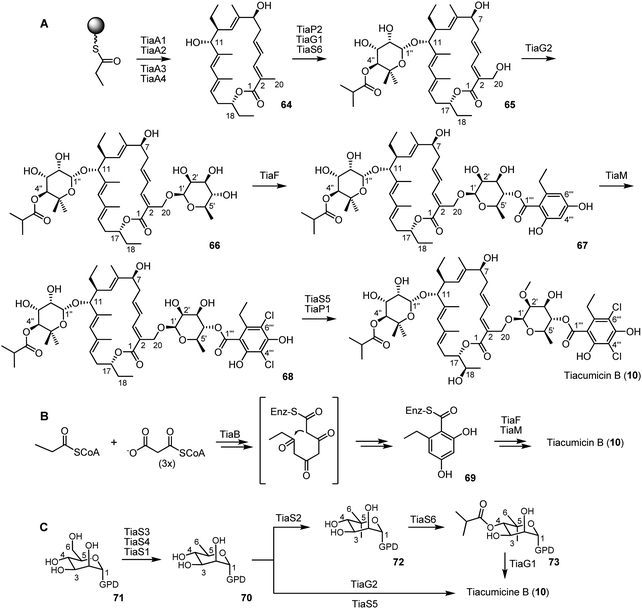 | ||
| Fig. 8 The proposed biosynthesis of tiacumicin B. | ||
Bioinformatic analysis of the TiaB gene revealed some similarity to 6-methyl-salicylic acid synthase and thus it is probably involved in the biosynthesis of the homo-orsellinic acid part 69 from a propionyl-CoA starting unit (Fig. 8, B). The aromatic moiety could then be transferred to the 2-O-methyl-D-rhamnose residue by TiaF due to similarity to acyltransferases, responsible for incorporation of aromatic parts into secondary metabolites. Finally, the gene TiaM showed some similarity to other halogenases and its selective inactivation led to the formation of tiacumicin analogs lacking chlorine atoms, therefore allowing the assignment of its function. It has also been shown that instead of chlorine (from NaCl), TiaM is able to transfer bromine atoms to the homo-orsellinic acid moiety (from NaBr). However, F− and I− are not halide donors for TiaM.
The genes TiaS1, S3 and S4 are probably involved in the biosynthesis of the D-rhamnose derivatives 70 from GDP-D-mannose 71 but the precise sequence is not currently known (Fig. 8, C). Bioinformatic analysis allowed the researchers to propose the role of TiaS2, TiaS5, TiaS6, TiaG1 and G2, which were further verified by selective inactivation. TiaG1 and TiaG2 are 5-C-methyl-D-rhamnosyl-transferase and 2-O-methyl-D-rhamnosyl-transferase, respectively, used to attach sugar moieties to the aglycone. TiaS2 and TiaS6 are a sugar C-methyltransferase and an acyltransferase, respectively, responsible for incorporation of the methyl and the isobutyryl moiety of the 4-O-isobutyrate-5-methyl-β-rhamnose. TiaS6 showed a relaxed substrate specificity for rhamnose derivatives but a great regioselectivity, being unable to methylate the C2 position of 5-C-methylrhamnose and other sugars.
The TiaS5 gene is the 2′-O-methyltransferase involved in the synthesis of the 2-O-methyl-β-D-rhamnose. Further experiments revealed that this methyltransferase was divalent metal-cation dependant: its activity is higher with Mg2+ and Mn2+, moderate with Co2+, Ni2+, Fe2+, weak with Zn2+, Cu2+ and inactive in the absence of any metal cation and in the presence of EDTA or Ca2+. It is also a pH-dependant enzyme displaying its best activity at pH 8. The two genes TiaP1 and TiaP2 encode for cytochrome P450 hydroxylase, responsible for natural product oxygenation. Inactivation experiments allowed determination of their roles as follows: TiaP1 catalyzes the hydroxylation at C18 and TiaP2 at C20.
Some others genes are involved in the precursor supply: TiaC and TiaD may be involved in isobutyryl-CoA, propionyl-CoA and acyl-CoA generation. TiaL probably catalyses the formation of (2S)-methylmalonyl-CoA from propionyl-CoA. TiaJ, TiaN and TiaK are probably involved in the ethylmalonyl-CoA pathway and TiaE, showing some similarity with thioesterases, may promote the accuracy and efficiency of the polyketide synthase. TiaR1 could be a transcription activator in bacteria, TiaR2 seems to be a negative regulator of tiacumicin biosynthesis. Genes TiaT1–T4 may constitute a system to transport the synthesized metabolites out of the cell whereas the role of TiaI is actually unknown.
In spite of this great achievement, some points remained unclear about tiacumicin biosynthesis. The right timing between genes TiaP1 and TiaS5 (hydroxylation of the C18 position and methylation of C2′ hydroxyl, respectively) is still unclear even though it seemed that TiaP1 should be the last step, directly preceded by TiaS5. Furthermore, the acylation of the C4′′ position by TiaS6, even if shown as anterior to glycosylation, could be a later step in the biosynthesis.
Fig. 9 shows the action of the most importants genes involved in the tiacumicin biosynthesis.
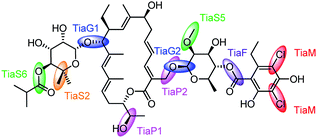 | ||
| Fig. 9 The roles of the principal genes in tiacumicin B (10) biosynthesis. | ||
4 Biological activity
4.1 Biological activity
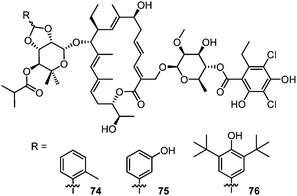
The initial report of the discovery of lipiarmycin mentioned, for the mixture of lipiarmycin A3 and A4, a fairly good activity against some Staphylococcus aureus strains and other gram-positive bacteria. A good activity against strains of cariogenic Steptococcus mutans, suggesting possible application as an antiplaque agent, was also discovered.20 A few years later, Sonenshein and co-workers reported the inhibition of bacterophage growth in Bacillus subtilis.51 During the early studies on tiacumicins, researchers from Abbott Laboratories reported a good activity of tiacumicin B against different aerobic bacteria (including S. aureus and Enterococcus faecium) but a lower activity against anaerobic bacteria.30 Tiacumicin B is also active against Staphylococcus epidermidis biofilms formed at the surface of medical devices (a major cause of nosocomial infections)52 and on some drug-resistant strains of Mycobacterium tuberculosis.46 Another study, reported by JMI Laboratories, shows a limited bacterial activity of 10 against S. aureus, CoNS, E. faecalis and E. faecium with minimal bactericidal concentrations/MIC ratios (0.5–16 μg mL−1).53In 2006 Optimer mentioned that the tiacumicins may find application in the treatment of gastrointestinal cancers, but without reporting any more detailed information.34 The first and only study for such an application of tiacumicin came from Echem Hightech Co.54 A series of tiacumicin benzylidene acetal derivatives have shown interesting activity against breast cancer cells with similar IC50 values to Tamoxifen®, a drug usually used for this type of cancer (Table 1).55 Note that the molecules drawn in this patent are tiacumicin C derivatives (with isopropyl ester on C2′′), although they are claimed to be tiacumicin B derivatives in the text (with isopropyl ester on C4′′).
|
|
||
|---|---|---|
| Compounds | IC50 for MCF7 breast cancer cells | IC50 for T-47D breast cancer cells |
| Tamoxifen® | 6.35 ± 0.45 | 5.50 ± 0.21 |
| Tiacumicin B (10) | 8.39 ± 1.00 | 5.56 ± 0.57 |
| 74 | 7.06 ± 0.83 | 3.99 ± 0.58 |
| 75 | 6.90 ± 0.36 | 4.24 ± 0.05 |
| 76 | 5.78 ± 0.81 | 4.04 ± 1.09 |
Notwithstanding these biological activities, it is in the fight against Clostridium difficile associated infection (CDI) that tiacumicin B was developed as a drug.
Clostridium difficile, a Gram-positive anaerobic bacteria, is the causative agent of between 20% and 25% of all cases of antibiotic-dependant diarrheas.56–60 Indeed, the gastrointestinal tract microbiota protects the host against most infections by pathogenic microorganisms through mechanisms known as “colonization resistance”.61 The use of broad spectrum antibiotics leads to severe perturbations of the gut microbiota, creating opportunities for the growth of bacteria usually restricted by microbial competition. C. difficile is recognized as such an opportunistic pathogen. The most encountered clinical manifestations of CDI are diarrhea (mild to moderate), pseudomembranous colitis, fever, and abdominal pain. Approximately 3% of patients will develop a fulminant colitis, with serious complications, such as colonic perforation, toxic megacolon and death.62
The treatment of these infections involves the use of metronidazole and/or vancomycin depending on the clinical presentation of the disease. However, these two broad-spectrum antibiotics have some drawbacks: a) metronidazole is easily absorbed along the gastrointestinal tract, resulting in the use of massives doses of the antibiotic and is not as efficient as vancomycin for the treatment of severe cases,63–65 b) both metronidazole and vancomycin promote the development of vancomycin-resistant Enterococci,66 c) the risk of relapse, between 15% and 35%.67,68
The fast development of tiacumicin B in CDI treatment is due to its good in vitro bioactivity and interesting characteristics:69–75 a) it is taken orally, b) tiacumicin B stays in the gastrointestinal tract and is only detectable in the blood at very low concentrations (nanomolar range), c) it shows a rate of clinical cure almost identical to vancomycin (91.7% to 90.6%) but a lower rate of recurrence of CDI (12.8% compared to 25.3%).
These results can be explained by the bactericidal action of the tiacumicins, killing C. difficile strains whereas vancomycin, as a bacteriostatic, only inhibits the development of bacteria.76 Furthermore, the narrow spectrum of tiacumicin respects the non-pathogenic species of guts,77–79 limiting the development of pathogenic microorganisms like C. difficile.80 On the other hand, vancomycin is a broad-spectrum antibiotic, facilitating the recurrence of CDI.
4.2 Mechanism of action
The synthesis of RNA from DNA, a process known as transcription, occurs in RNA polymerase (RNAP), an essential enzyme present in most organisms.81–84 The bacterial RNAP is composed of five core subunits (2α, β, β′, ω) and a σ co-factor. The two subunits β and β′ have a pincer structure, forming a channel in which the incoming DNA strand fits. The active site of the enzyme, complexing a Mg2+ ion is also in this channel. The subunit β′ is a mobile clamp, serving as a lock of the active-site, a docking site for the σ subunit and helps to position DNA template into the active site. The σ co-factor is a subunit which binds to the enzyme core to form a new complex: the holoenzyme. When formed, the holoenzyme binds to the DNA promoter, leading to the formation of a closed-complex. Initiation of transcription requires the formation of an open-complex of RNAP in which promoter DNA are melted to form the transcription bubble.The first studies directed toward the elucidation of lipiarmycin mechanism of action were conducted soon after its discovery by Lepetit's scientists.85In vivo studies on Bacillus subtilis revealed that at a low concentration, lipiarmycin could inhibit RNA synthesis and that at a higher concentration, DNA synthesis itself could be depressed. On Escherichia coli, they also noted that the inhibitory action of lipiarmycin was higher when it was added prior to the association of RNAP and DNA. If lipiarmycin is added after polymerisation has started, it does gradually reduce the extent of RNA synthesis whereas it is totally suppressed when lipiarmicin is present at the beginning. It has been proposed that lipiarmicin could bind to RNAP and thus interacts with the early steps of RNA synthesis. Similar results were obtained by Talpaert and co-workers on E. coli who proposed that lipiarmycin could interfere with initiation steps rather than elongation of RNA.86
In 1977, using B. subtilis mutants resistant to lipiarmycin, Sonenshein and co-workers showed that the core enzyme of RNAP (2α, β, β′ and ω units) is relatively resistant to the antibiotic but addition of the σ co-factor restores sensitivity.87 Mapping data revealed that mutations leading to antibiotic-resistant strains were likely to reside in the gene coding for the β subunit of the core.
Although it has been established that at least one unit of the RNAP is targeted by lipiarmycin, the mechanism of action remained unclear. The antibiotic could bind to the core enzyme and thus prevent the binding of the σ subunit or it could bind to the polymerase only after formation of the holoenzyme. In 1979, the same group reported the inhibition preference of lipiarmycin for RNAP in the holoenzyme form, an action probably not due to an interaction with the σ subunit, but with preferential interactions with the holoenzyme.88 It was still suggested that both the core and σ subunit could each provide a part of the binding site and proposed that the antibiotic could inhibit the formation of the first phosphodiester bond.
In 2006, using DNA sequencing, Leonetti and co-workers showed that mutation of the gene coding for the β′ subunit of the core confers resistance to B. subtilis.89 Other studies by Sambandamurthy and co-workers and Leonetti and co-workers also revealed that spontaneous mutations of genes coding for the RNA exit channel (β and β′ subunits) could confer resistance to lipiarmycin in Mycobacterium tuberculosis and Enterococcus faecalis.46,90
Brodolin and colleagues reported in 2011 a study aimed at establishing the mechanism of action of this antibiotic on E. coli.91 Using biochemical and genetic approaches, they proposed that lipiarmycin acts as an inhibitor of transcription, docking to both the σ70 3.2 region and the mobile-clamp domain β′ of the core, thus trapping RNAP at one of the closed (and inactive) intermediates of the enzyme. In this closed state, even if the promoter DNA remains bounded to RNAP, the single-stranded DNA template cannot fit into the active site and the transcription is inhibited.
Further studies by Ebright and colleagues, while suggesting no interaction between lipiarmycin and the σ70 subunit, still suggested that tiacumicin could interfere with the RNAP switch-region (β′), the RNA-exit channel (β and β′), or both.92
All of the studies discussed above were performed using lipiarmycin from Actinoplanes or Catellatospora strains as the antibiotic. The only study on the action of (R)-tiacumicin B (known as fidaxomicin) was reported by Artsimovitch and colleagues in 2012.93 They showed that fidaxomicin can inhibit RNAP from both C. difficile and E. coli after formation of the holoenzyme. However, the antibiotic was not able to stop RNA synthesis after formation of the open-complex of RNAP, suggesting the same mechanism of action as for lipairmycin. In addition, they showed that both lipiarmycin and fidaxomicin induce changes in the downstream DNA interactions with RNAP. However, only fidaxomicin was able to alter the DNA conformation at other regions of the RNAP–DNA complex and that its action was not dependant on the σ unit (contrary to the work of Brodolin and colleagues91 but in agreement with the work of Ebright and colleagues92). The source of theses differences remains to be determined, even if it has been suggested that the use of different promoters in these studies might be one. The source of RNAP or the structure of the antibiotics (configuration of C18 assumed to be (S) for lipiarmycin and proved to be (R) for fidaxomicin) could also play a role.
5 SAR studies
Only a few reports have dealt with the structure–activity relationship of this family of antibiotics. However, the work of Zhang directed toward the elucidation of its biosynthesis generated various analogs whose antibacterial activities have been evaluated.40,41One of the first parts of tiacumicin B which has been modified is the aromatic ring.94 The methylation of phenols afforded a di-O-methyltiacumicin B, which was 8- to 16- fold less active than tiacumicin B on various aerobic bacteria, such as S. aureus, S. epidermidis or E. faecium. In addition, the activity on C. perfringens or C. difficile strains is also lower than tiacumicin B (2- to 8- fold) but remains equal or higher than vancomycin.
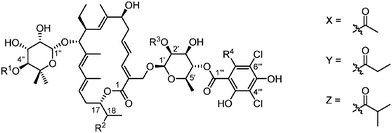
The isobutyric ester at the C4′′ position on the 5-methyl-β-rhamnose sugar is important for the bioactivity as its hydrolysis led to a known metabolite (OP-1118), characterized by a 8- to 16- fold lost in activity on various strains (Table 2 – entry 2).95
|
|
|||||||
|---|---|---|---|---|---|---|---|
| Substitution pattern | Minimum inhibitory concentration (μg mL−1) | ||||||
| Entry | R1 | R2 | R3 | R4 | C. difficile ATCC 43255 | E. faecalis ATCC 29212 | S. aureus ATCC 29213 |
| 1 | Z | (R)–OH | Me | Et | 0.5 | 2–4 | 8 |
| 2 | H | (R)–OH | Me | Et | 4 | 16–64 | >64 |
| 3 | Z | (S)–OH | Me | Et | 1 | 8 | 64 |
| 4 | Z | ![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O O |
Me | Et | 0.5 | — | — |
| 5 | Z | (S)–OH | Me | Me | 0.5 | 2 | 8 |
| 6 | Z | H | Me | Et | — | 4 | 8 |
| 7 | H | H | Me | Et | — | 32 | 128 |
| 8 | X | H | H | Et | — | 4 | 8 |
| 9 | Y | H | H | Et | — | 2 | 4 |
| 10 | Z | H | H | Et | — | 1 | 2 |
| 11 | Z | Me | H | Et | — | 1 | 1 |
| 12 | Z | (R)–OH | H | Et | — | 4 | 16 |
Interestingly, the configuration of the C18 hydroxyl group on the macrocycle has a great influence on the bioactivity as its epimerisation to the (S) configuration or its oxidation to the ketone leads to a dramatic increase of MIC on various bacteria (C. difficile, S. aureus or E. faecalis, resistant or not – Table 2, entries 3 and 4).54,96,97 However, lipiarmycin A4 with the (S) configuration at C18 and a methyl instead of an ethyl group on aromatic ring displayed almost equal activity to (S)-tiacumicin B (entry 5).
Removal of the C18 hydroxyl group was made possible with Zhang's mutants.40,41 The compound thus obtained showed a slight decrease of activity (2- to 4-fold), which still remained good (entry 6). Further removal of the isobutyric ester dramatically decrease the activity from 4- to 16-fold, results consistent with OP-1118 (entry 7). It is interesting to note that analogues in which the C18 hydroxyl group was replaced by either a hydrogen atom or a methyl group and lacking the methoxy group at C2′ showed similar or even increased bioactivities (entries 8–11). However, a compound only demethylated at C2′ was characterized by a slightly reduced activity (entries 1 vs. 12).
The presence and the nature of halogens on the aromatic ring also impacted the activity (Table 3).35,40 The replacement of both chlorines by hydrogens led to a slight increase of minimum inhibitory concentration against some Gram-positive bacteria (entry 2). The replacement of the C6′′′ chlorine atom with bromine (entry 3) or just having bromine at C4′′′ (entry 4) led to a variable decrease of activity against S. aureus or E. faecium whereas a good activity against C. difficile bacteria remained.
|
|
|||||||
|---|---|---|---|---|---|---|---|
| Substitution pattern | Minimum inhibitory concentration (μg mL−1) | ||||||
| Entry | R1 | R2 | S. aureus ATCC 29213 | S. aureus ATCC 6538P | E. faecalis ATCC 29212 | E. faecium ATCC 8043 | C. difficile ATCC 9689 |
| 1 | Cl | Cl | 8 | 0.78 | 2 | 1.56 | 0.06 |
| 2 | H | H | 8 | — | 4 | — | — |
| 3 | Cl | Br | — | 6.2 | — | 6.2 | 0.06 |
| 4 | Br | H | — | 6.2 | — | 12.5 | 0.12 |
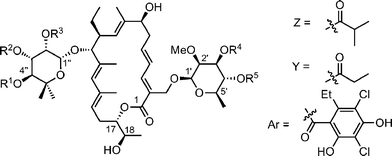
The nature and the position of subsituents on the sugar part can also impact the bioactivities and this is readily seen by comparing the bioactivities of different tiacumicins. (Table 4).30 Tiacumicin F and C differ from B by the position of the isobutyric ester on 5-methyl-rhamnose, at C3′′, C2′′ and C4′′, respectively. Tiacumicin C shows a slightly reduced activity (2- to 16-fold) against all bacterias. The trend is more pronounced for tiacumicin F (entries 2 and 3). Change of isobutyric ester at C2′′ for propanoic ester at the same position on tiacumicin E increases the activity 2- to 4-fold (entry 4) and moving the aromatic ester from C4′ to C3′ (tiacumicin D, entry 5) reduces the activity against S. epidermidis 3519.
|
|
||||||||
|---|---|---|---|---|---|---|---|---|
| Substitution pattern | Minimum inhibitory concentration (μg mL−1) | |||||||
| Entry | R1 | R2 | R3 | R4 | R5 | S. aureus ATCC 6538P | S. epidermidis 3519 | E. faecium ATCC 8043 |
| 1 | Z | H | H | H | Ar | 6.2 | 12.5 | 6.2 |
| 2 | H | Z | H | H | Ar | 12.5 | 12.5 | 6.2 |
| 3 | H | H | Z | H | Ar | 50 | 25 | 100 |
| 4 | H | H | Y | H | Ar | 25 | 12.5 | 12.5 |
| 5 | H | H | Z | Ar | H | 25 | >25 | >25 |
Zhang's investigation has shown that major modifications, such as removing one sugar, both sugars or the aromatic ring from tiacumicin B, whatever the substitution pattern was, led to a drastic reduction of the antimicrobial activity.
It is known that the antimicrobial activity of some compounds can be influenced by various parameters, such as pH, the concentration of divalent cations or bacterial density. It has been shown that, contrary to vancomycin, the inoculum density is not a significant factor affecting the activity of tiacumicin B.37,38,98 Similarly, the concentration of divalent cations, such as calcium and magnesium, didn't affect the activity of tiacumicin B. However, it has been disclosed that the minimum inhibitory concentration values for tiacumicin B increased with the increase of pH, with values at pH 8.1 being 8- to 16-fold higher than those obtained at pH 6. To explain these results, it has been proposed that at basic pH, phenolic hydroxyl groups could be deprotonated, thus forming charged species, which are expected to be less able to permeate bacterial cells.
6 Synthesis
To the best of our knowledge no total synthesis of the tiacumicins has ever been reported in the literature in spite of the great interest in these compounds. However, syntheses of the sugar and aromatic parts of tiacumicin have been reported.6.1 Homodichloro-orsellinic acid
In the early 90's, in order to validate the proposed structure of the aromatic part of lipiarmycin, Scharf and co-workers described the first synthesis of homodichloro-orsellinic acid (Fig. 10).99 Condensation of ethyl acetoacetate 77 with (E)-ethyl pent-2-enoate under basic conditions afforded the cyclohexenone 78. Aromatization took place during bromination to give the product 79. Product 81 was accessible through a two step protocol involving removal of bromine with RANEY® nickel and subsequent chlorination using sulfuryl chloride in 85% yield. Due to the easy decarboxylation of β-resorcyclic acid derivatives in basic media, the acid 82 is finally generated by hydrolysis of ethyl ester in concentrated sulfuric acid. The homodichloro-orsellinic acid 82 was thus obtained with a good 41% overall yield. The spectroscopic data of this compound were in good agreement with those published for the aromatic part of lipiarmcycin A3.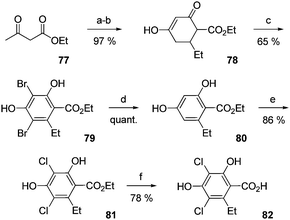 | ||
| Fig. 10 The synthesis of homodichloro-orsellinic acid. (a) (E)-Ethyl pent-2-enoate, Na, EtOH, reflux. (b) HCl aq., −10 °C to 0 °C. (c) Br2, AcOH, 40 °C. (d) Ni/Al, NaOH aq., 0 °C. (e) SO2Cl2, Et2O, reflux. (f) H2SO4 conc. | ||
6.2 2-O-Methyl-β-D-rhamnose
In 1982, Liptak and co-workers reported the first and only synthesis of 2-O-methyl-D-rhamnose 83 (Fig. 11).100,101 The methyl α-D-mannopyranoside 84 was treated with α,α-dimethoxytoluene in the presence of PTSA to give the bis-acetal 85 as a single diastereoisomer after recrystallization in a 64% yield. 86 was obtained by a selective opening of the dioxolane by hydrogenolysis followed by methylation of the C2 hydroxyl group. The 1,3-dioxane opening by lithium aluminium hydride allowed the formation of 87 in a 67% yield. The C6 position was deoxygenated by a tosylation/reduction sequence, affording 88 which was deprotected to give the desired sugar. The 2-O-methyl-β-D-rhamnose 83 was thus obtained with a 15% overall yield.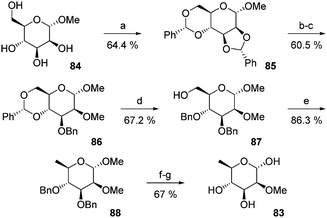 | ||
| Fig. 11 The synthesis of 2-O-methyl-β-D-rhamnose. (a) α,α-Dimethoxytoluene, PTSA, DMF, 65–75 °C. (b) LiAlH4, AlCl3, Et2O, DCM. (c) MeI, Ag2O, DMF. (d) LiAlH4, AlCl3, Et2O, DCM. (e) TsCl. (f) LiAlH4, benzene, Et2O, reflux. (g) H2, Pd/C, EtOH, AcOH. (h) H2SO4 (1 M), 100 °C. | ||
Note that Zdorovenko and co-workers have previously prepared this sugar by cultivating a strain of Bacillus faecalis alcaligenes.102
6.3 5-Methyl-β-rhamnose
The sugar 89, with the absolute stereochemistry corresponding to tiacumicin B has never been reported in the literature (Fig. 12). However, its enantiomer 90 has been prepared by Klemer and Waldmann, and Walton (drawn as the pyranose form) during studies directed toward the preparation of noviose derivatives.103,104 Note that the methylated analog on the C3 position showing the right stereochemistry was known as D-(−)-noviose (91).105–108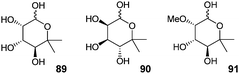 | ||
| Fig. 12 5-Methyl-rhamnose and analogs. | ||
7 Clinical application
(R)-Tiacumicin B was developed by Optimer Pharmaceuticals Inc. under the generic name fidaxomicin and the trade name Dificid®.Due to its physicochemical properties (high molecular weight 1058.04 g mol−1, large number of hydrogen bond donors and acceptors, 7 and 18, respectively), Lipinski's rules predict a poor absorption from the gastrointestinal tract.109 Indeed, both fidaxomicin and its major metabolite OP-1118 (lacking the isobutyric ester at C4′′ and which is also bactericidal110) displayed good pharmacokinetic profile with minimal systemic absorption (plasma concentration typically in the nanomolar range) and high faecal drug levels.71,73
Interactions of fidaxomicin and OP-1118 with other biological function and drugs were also assessed. Minimal inhibition of cytochrome P450 has been observed at a concentration up to 10 μg mL−1. Both fidaxomicin and OP-1118 are substrates for efflux pumps, which could be inhibited by cyclosporine, an immunosuppressant drug. Indeed, when cyclosporine is co-administrated with Dificid®, plasma concentration of fidaxomicin and OP-1118 are increased, but still remain in the ng mL−1 range.
Even if guidelines recommend discontinuation of all antibiotic therapy in the case of C. difficile infection, patients frequently require another antibiotic treatment to manage concurrent infections. The effect of other antibiotics, such as carbapenem, cephalosporin, fluoroquinoline, penicilin or streptogramin, on the efficacy of fidaxomicin has been recently evaluated by Mullane and co-workers.111 They have shown that the concomitant use of antibiotics were associated with a slightly lower cure rate (90.0% vs. 92.3%) and an extended time to resolution of diarrheas. In addition, more recurrences were observed in patients taking other antibiotics (16.8% vs. 11.9%). But even in the case of concomitant antibiotic treatment of C. difficile infection, fidaxomicin remained a better choice than vancomycin.
Fidaxomicin is also characterised by a low frequency of spontaneous resistance development by C. difficile strains (from <1.4 × 10−9 to 12.8 × 10−9). Most known mutants resistant to tiacumicin have been grown in the laboratory for research purposes. Nevertheless, a subject with recurrence of CDI has developed such resistance during clinical trials. However, no cross-resistance of fidaxomicin with other antibacterial drugs has been noted and a synergistic effect has even been revealed with the rifamycin class of compounds, ampicillin and metronidazole.
Note that both fidaxomicin and its major metabolite OP-1118 show a long post-antibiotic effect (PAE).112 Indeed on two C. difficile strains, the PAE of fidaxomicin was about 10 h and 5 h on a clinical isolate (3 h for OP-1180), whereas vancomycin shows a PAE of 0 and 1.5 h, respectively, for strains and clinical isolate. It has been proposed that this unusually long suppression of bacterial growth by fidaxomicin may be due to the specific binding of the drug to RNA polymerase or to the non-specific binding to other bacterial cell components. By these means, fidaxomicin could remain inside the cell to exert its antibacterial activity and only a slow dissociation and diffusion of the drug out of the bacteria could allow the microorganism to grow again.
In addition, it has been recently shown that fidaxomicin inhibits sporulation by C. difficile, a mechanism which may contribute to the better cure rate of this drug compared to vancomycin.113
Taking into account of all these properties and the results of phase III clinical trials in Canada and the USA (later extended to Europe), which confirm the efficacy and safety of fidaxomicin,114–117 the US Food & Drug Administration (FDA) approved its use in the treatment of CDI as a good alternative to vancomycin and metronidazole.118 However, the phase III study did not include any pediatric patients, and additional studies are therefore needed to ascertain the efficacy and safety of fidaxomicin in children.119 Note that Optimer Pharmaceuticals is currently working on new indications of fidaxomicin, such as vancomycin-resistant enterococcal infection and meticillin-resistant Staphylococcus aureus prophylaxis.120
8 Summary and outlook
The tiacumicin family encompass more than 40 molecules with the same macrolactone framework. They are isolated from the bacterial culture of different strains of the Actinomycetes group. A recent study indicated the direct involvement of 17 genes and corresponding enzymes with an unusual tailoring dihalogenase in its biosynthesis. These compounds and their derivatives displayed interesting bioactivity against pathogenic bacteria and promising anti-cancer activity.(R)-Tiacumicin B is a RNA synthesis inhibitor which acts by docking to two domains of the closed RNA polymerase enzyme, preventing its opening and thus activation. For the treatment of Clostridium difficile infection, it has the same cure rate as vancomycin but with a lower relapse rate. It was approved by the FDA in May 2011 as a marketed drug for the treatment of CDI.
It is rather surprising that in spite of its interesting structure and biological activities, no total synthesis of this macrolide has been reported in the literature. Such a synthesis could afford an interesting alternative to the actual source of supply, allowing the generation of new derivatives with new or improved activities.
9 Acknowledgements
W.E. would like to thank G. Coulthard for critically reviewing this document and for making valuable suggestions.10 References
- D. J. Newman and G. M. Cragg, J. Nat. Prod., 2007, 70, 461–477 CrossRef CAS.
- M. S. Butler and A. D. Buss, Biochem. Pharmacol., 2006, 71, 919–929 CrossRef CAS.
- F. Peláez, Biochem. Pharmacol., 2006, 71, 981–990 CrossRef.
- J. Clardy, M. A. Fischbach and C. T. Walsh, Nat. Biotechnol., 2006, 24, 1541–1550 CrossRef CAS.
- T. M. Embley and E. Stackebrandt, Annu. Rev. Microbiol., 1994, 48, 257–289 CrossRef CAS.
- W. B. Whitman, D. C. Coleman and W. J. Wiebe, Proc. Natl. Acad. Sci. U. S. A., 1998, 95, 6578–6583 CrossRef CAS.
- T. R. P. Kekuda, K. S. Shobha and R. Onkarappa, J. Pharm. Res., 2010, 3, 250–256 CAS.
- R. Solanki, M. Khanna and R. Lal, Indian J. Microbiol., 2008, 48, 410–431 CrossRef CAS.
- R. H. Baltz, Microbe, 2007, 2, 125–131 Search PubMed.
- R. H. Baltz, Curr. Opin. Pharmacol., 2008, 8, 557–563 CrossRef CAS.
- S. Dharmaraj, World J. Microbiol. Biotechnol., 2010, 26, 2123–2139 CrossRef CAS.
- O. Genilloud, I. González, O. Salazar, J. Martín, J. R. Tormo and F. Vicente, J. Ind. Microbiol. Biotechnol., 2011, 38, 375–389 CrossRef CAS.
- S. S. Ghantoji, K. Sail, D. R. Lairson, H. L. Dupont and K. W. Garey, J. Hosp. Infect., 2010, 74, 309–318 CrossRef CAS.
- W. Erb and J. Zhu, L'Act. Chim., 2012, 360–361, 83–89 CAS.
- M. Gerber and G. Ackermann, Expert Opin. Invest. Drugs, 2008, 17, 547–553 CrossRef CAS.
- M. Miller, Expert Opin. Invest. Drugs, 2010, 11, 1569–1578 CAS.
- J. S. Hardesty and P. Juang, Pharmacotherapy, 2011, 31, 877–886 CrossRef CAS.
- J. W. Lancaster and S. J. Matthews, Clin. Ther., 2012, 34, 1–13 CrossRef CAS.
- F. Parenti, H. Pagani and G. Beretta, J. Antibiot., 1975, 28, 247–252 CrossRef CAS.
- C. Coronelli, R. J. White, G. C. Lancini and F. Parenti, J. Antibiot., 1975, 28, 253–259 CrossRef CAS.
- C. Coronelli, F. Parenti, R. White and H. Pagani, GB 1458512, 1973, Gruppo Lepetit.
- E. Martinelli, L. Faniuolo, G. Tuan, G. G. Gallo and B. Cavalleri, J. Antibiot., 1983, 36, 1312–1322 CrossRef CAS.
- K. Schmidt-Bäse, M. Noltemeyer, E. Egert, E. Eigelt and A. Zeeck, Acta. Cryst., 1993, C49, 250–253 Search PubMed.
- A. Isogai, S. Sakuda, S. Matsumoto, M. Ogura, K. Furihata, H. Seto and A. Suzuki, Agric. Biol. Chem., 1984, 48, 1379–1381 CrossRef CAS.
- J. B. McAlpine, J. W. Corcoran and R. S. Egan, J. Antibiot., 1971, 24, 51–56 CrossRef CAS.
- A. Arnone, G. Nasini and B. Cavalleri, J. Chem. Soc., Perkin Trans. 1, 1987, 1353–1359 RSC.
- B. Cavalleri, A. Arnone, E. Di Modugno, G. Nasini and B. P. Goldstein, J. Antibiot., 1988, 41, 308–315 CrossRef CAS.
- S. Õmura, N. Imamura, R. Oiwa, H. Kuga, R. Iwata, R. Masuma and Y. Iwai, J. Antibiot., 1985, 39, 1407–1412 CrossRef.
- Y. Takahashi, I. Yuzuru and S. Õmura, J. Antibiot., 1985, 39, 1413–1418 CrossRef.
- R. J. Theriault, J. P. Karwowski, M. Jackson, R. L. Girolami, G. N. Sunga, C. M. Vojtko and L. J. Coen, J. Antibiot., 1987, 40, 567–574 CrossRef CAS.
- J. E. Hochlowski, S. J. Swanson, L. M. Ranfranz, D. N. Whittern, A. M. Buko and J. B. McAlpine, J. Antibiot., 1987, 40, 575–588 CrossRef CAS.
- J. B. McAlpine, M. Jackson and J. Karwowski, US 4918174, 1990, Abbott Laboratories.
- Y.-K. Shue, F. K. Babakhani, F. W. Okumu, P. S. Sears, S. L. Miller-Shangle and R. B. Walsh, WO 2005112990, 2005, Optimer Pharmaceuticals, Inc..
- Y.-K. Shue, C.-K. Hwang, Y.-H. Chiu, A. Romero, F. Babakhani, P. Sears and F. Okumu, WO 2006085838 A1, 2006, Optimer Pharmaceuticals, Inc..
- J. E. Hochlowski, M. Jackson, R. R. Rasmussen, A. M. Buko, J. J. Clement, D. N. Whittern and J. B. McAlpine, J. Antibiot., 1997, 50, 201–205 CrossRef CAS.
- J. E. Hochlowski, M. Jackson, J. B. McAlpine and R. R. Rasmussen, US 5767096, 1998, Abbott Laboratories.
- P. Sears, Y.-K. Shue, S. L. Miller-Shangle, R. B. Walsh, F. Babakhani, Y.-H. Chiu, A. Romero, S. Gorbach and T. J. Louie, WO 2007048059 A2, 2007, Optimer Pharmaceuticals, Inc..
- P. Sears, S. L. Miller-Shangle, R. B. Walsh, Y.-K. Shue, F. Babakhani, T. J. Louie, Y.-H. Chiu, A. Romero and S. Gorbach, US 20070105791, 2007, Optimer Pharmaceuticals, Inc..
- S. Sanghvi, M. Roach, J. F. Zhou, E. M. Mittleberg and P. He, US 20080176927 A1, 2008, Optimer Pharmaceuticals, Inc..
- Y. Xiao, S. Li, S. Niu, L. Ma, G. Zhang, H. Zhang, G. Zhang, J. Ju and C. Zhang, J. Am. Chem. Soc., 2011, 133, 1092–1105 CrossRef CAS.
- S. Nui, T. Hu, S. Li, Y. Xiao, L. Ma, G. Zhang, H. Zhang, X. Yang, J. Ju and C. Zhang, ChemBioChem, 2011, 12, 1740–1748 CrossRef.
- C. Zhang, Y. Xiao, S. Li, S. Niu, G. Zhang, H. Zhang, T. Hu and J. Ju, Patent CN 2010–10592416, 2011.
- C. Zhang, S. Li, Y. Xiao, S. Niu and J. Ju, Patent CN 2010–10526416, 2011.
- C. Zhang, Y. Xiao, L. Sumei, S. Niu, G. Zhang, H. Zhang, T. Hu and J. Ju, CN 102115757, 2011, Faming Zhuanli Shenqing.
- Y.-K. Shue, C.-J. F. Du, M.-H. Chiou, M.-C. Wu, Y.-T. Chen, F. W. Okumu and J. J. Duffield, WO 2004014295 A2, 2004, Optimer Pharmaceuticals, Inc..
- M. Kurabachew, S. H. J. Lu, P. Krastel, E. K. Schmitt, B. L. Suresh, A. Goh, J. E. Knox, N. L. Ma, J. Jiricek, D. Beer, M. Cynamon, F. Petersen, V. Dartois, T. Keller, T. Dick and V. K. Sambandamurthy, J. Antimicrob. Chemother., 2008, 62, 713–719 CrossRef CAS.
- T. Weber, K. Welzel, S. Pelzer, A. Vente and W. Wohlleben, J. Biotechnol., 2003, 106, 221–232 CrossRef CAS.
- C. T. Walsh and M. A. Fischbach, J. Am. Chem. Soc., 2010, 132, 2469–2493 CrossRef CAS.
- C. Olano, C. Méndez and J. A. Salas, Microb. Biotechnol., 2011, 4, 144–164 CrossRef CAS.
- A. L. Lane and B. S. Moore, Nat. Prod. Rep., 2011, 28, 411–428 RSC.
- M. S. Osburne and A. L. Sonenshein, J. Virol., 1980, 33, 945–953 CAS.
- P. Villain-Guillot, M. Gualtieri, L. Bastide and J.-P. Leoneti, Antimicrob. Agents Chemother., 2007, 51, 3117–3121 CrossRef CAS.
- D. J. Biedenbach, J. E. Ross, S. D. Putnam and R. N. Jones, Antimicrob. Agents Chemother., 2010, 54, 2273–2275 CrossRef CAS.
- Y.-K. Shue, C.-K. Hwang, Y.-H. Chiu, A. Romero, F. Babakhani, P. Sears and F. Okumu, WO 2006085838 A1, 2006, Optimer Pharmaceuticals, Inc.; M.-C. Wu, C.-C. Huang, Y.-C. Lu and W.-J. Fan, US 20090110718 A1, 2009, Echem Hightech Co. Ltd.
- M. Clemons, S. Danson and A. Howell, Cancer Treat. Rev., 2002, 28, 165–180 CrossRef CAS.
- J. G. Bartlett, Clin. Infect. Dis., 2008, 46(Suppl 1), S4–S11 CrossRef.
- Clostridium difficile, ed. K. Aktories and T. C. Wilkins, Springer, 2000 Search PubMed.
- M. Kachrimanidou and N. Malisiovas, Crit. Rev. Microbiol., 2011, 37, 178–187 CrossRef CAS.
- G. P. Carter, J. I Rood and D. Lyras, Trends Microbiol., 2012, 20, 21 CrossRef CAS.
- F. C. Lessa, C. V. Gould and L. C. McDonald, Clin. Infect. Dis., 2012, 55(Suppl 2), S65–S70 CrossRef CAS.
- E. J. Vollaard and H. A. Clasener, Antimicrob. Agents Chemother., 1994, 38, 409–414 CrossRef CAS.
- M. S. Rubin, L. E. Bodenstein and K. C. Kent, Dis. Colon Rectum, 1995, 38, 350–354 CrossRef CAS.
- R. P. Bolton and M. A. Culshaw, Gut, 1986, 27, 1169–1172 CrossRef CAS.
- F. A. Zar, S. R. Bakkanagari, K. M. L. S. T. Moorthi and M. B. Davis, Clin. Infect. Dis., 2007, 45, 302–307 CrossRef CAS.
- J. G. Bartlett, Clin. Infect. Dis., 2008, 46, 1489–1492 CrossRef CAS.
- W. N. Al-Nassir, A. K. Sethi, Y. Li, M. J. Pultz, M. M. Riggs and C. J. Donskey, Antimicrob. Agents Chemother., 2008, 52, 2403–2406 CrossRef CAS.
- F. Barbut, A. Richard, K. Hamadi, V. Chomette, B. Burghoffer and J. -C. Petit, J. Clin. Microbiol., 2000, 38, 2386–2388 CAS.
- K. Z. Vardakas, K. A. Polyzos, K. Patouni, P. I. Rafailidis, G. Samonis and M. E. Falagas, Int. J. Antimicrob. Agents, 2012, 40, 1 CrossRef CAS.
- G. Ackermann, B. Löffler, D. Adler and A. C. Rodloff, Antimicrob. Agents Chemother., 2004, 48, 2280–2282 CrossRef CAS.
- D. W. Hecht, M. A. Galang, S. P. Sambol, J. R. Osmolski, S. Johnson and D. N. Gerding, Antimicrob. Agents Chemother., 2007, 51, 2716–2719 CrossRef CAS.
- Y. K. Shue, P. S. Sears, S. Shangle, R. B. Walsh, C. Lee, S. L. Gorbach, F. Okumu and R. A. Preston, Antimicrob. Agents Chemother., 2008, 52, 1391–1395 CrossRef CAS.
- J. Karlowsky, N. M. Laing and G. G. Zhanel, Antimicrob. Agents Chemother., 2008, 52, 4163–4165 CrossRef CAS.
- T. Louie, M. Miller, C. Donskey, K. Mullane and E. J. C. Goldstein, Antimicrob. Agents Chemother., 2009, 53, 223–228 CrossRef CAS.
- News and analysis in Nature Rev. Drug Discovery, 2010, 9, 260 Search PubMed.
- For a systematic review on the comparative effectiveness of C. difficile treatments, see: D. M. Drekonja, M. Butler, R. MacDonald, D. Bliss, G. A. Filice, T. S. Rector and T. J. Wilt, Ann. Intern. Med., 2011, 155, 839–847 Search PubMed.
- R. N. Swanson, D. J. Hardy, N. L. Shipkowitz, C. W. Hanson, N. C. Ramer, P. B. Fernandes and J. J. Clement, Antimicrob. Agents Chemother., 1991, 35, 1108–1111 CrossRef CAS.
- K. L. Credito and P. C. Appelbaum, Antimicrob. Agents Chemother., 2004, 48, 4430–4434 CrossRef CAS.
- S. M. Finegold, D. Molitoris, M.-L. Vaisanen, Y. Song, C. Liu and M. Bolanos, Antimicrob. Agents Chemother., 2004, 48, 4898–4902 CrossRef CAS.
- T. J. Louie, J. Emery, W. Krulicki, B. Byrne and M. Mah, Antimicrob. Agents Chemother., 2009, 53, 261–263 CrossRef CAS.
- G. W. Tannock, K. Munro, C. Taylor, B. Lawley, W. Young, B. Byrne, J. Emery and T. Louie, Microbiology, 2010, 156, 3354–3359 CrossRef CAS.
- K. S. Murakami, S. Masuda and S. A. Darst, Science, 2002, 296, 1280–1284 CrossRef CAS.
- K. S. Murakami, S. Masuda, E. A. Campbell, O. Muzzin and S. A. Darst, Science, 2002, 296, 1285–1290 CrossRef CAS.
- D. G. Vassylyev, M. N. Vassylyeva, A. Perederina, T. H. Tahirov and I. Artsimovitch, Nature, 2007, 448, 157–162 CrossRef CAS.
- P. Villain-Guillot, L. Bastide, M. Gualtieru and J.-P. Leonetti, Drug Discovery Today, 2007, 12, 200–208 CrossRef CAS and references cited therein.
- S. Sergio, G. Pirali, R. White and F. Parenti, J. Antibiot., 1975, 28, 543–549 CrossRef CAS.
- M. Talpaert, F. Campagnari and L. Clerici, Biochem. Biophys. Res. Commun., 1975, 63, 328–334 CrossRef CAS.
- A. L. Sonenshein, H. B. Alexander, D. M. Rothstein and S. H. Fisher, J. Bacteriol., 1977, 132, 73–79 CAS.
- A. L. Sonenshein and H. B. Alexander, J. Mol. Biol., 1979, 127, 55–72 CrossRef CAS.
- M. Gualtieri, P. Villain-Guillot, J. Latouche, J.-P. Leonetti and L. Bastide, Antimicrob. Agents Chemother., 2006, 50, 401–402 CrossRef CAS.
- M. Gualtieri, A. Tupin, K. Brodolin and J.-P. Leonetti, Int. J. Antimicrob. Agents, 2009, 34, 605–606 CrossRef CAS.
- A. Tupin, M. Gualtieri, J.-P. Leonetti and K. Brodolin, EMBO J., 2010, 29, 2527–2537 CrossRef CAS.
- A. Srivastava, M. Talaue, S. Liu, D. Degen, R. Y. Ebright, E. Sineva, A. Chakraborty, S. Y. Druzhinin, S. Chatterjee, J. Mukhopadhyay, Y. W. Ebright, A. Zozula, J. Shen, S. Sengupta, R. R. Niedfeldt, C. Xin, T. Kaneko, H. Irschik, R. Jansen, S. Donadio, N. Connell and R. H. Ebright, Curr. Opin. Microbiol., 2011, 14, 532–543 CrossRef CAS and references therein.
- I. Artsimovitch, J. Seddon and P. Sears, Clin. Infect. Dis., 2012, 55(Suppl 2), S127–S131 CrossRef CAS.
- J. B. McAlpine and J. E. Hochlowski, WO 9635702 A1, 1996, Abbott Laboratories.
- Y. Ichikawa, Y.-H. Chiu, Y.-K. Shue and F. K. Babakhani, WO 2009070779 A1, 2009, Optimer Pharmaceuticals, Inc..
- Y.-K. Shue, C.-K. Hwang, Y.-H. Chiu, A. Romero, F. Babakhani, P. Sears and F. Okumu, US 20080269145 A1, 2008, Optimer Pharmaceuticals, Inc..
- Y.-K. Shue, C.-K. Hwang, Y.-H. Chiu, A. Romero, F. Babakhani, P. Sears and F. Okumu, US 20100008825 A1, 2010, Optimer Pharmaceuticals, Inc..
- F. Babakhani, J. Seddon, N. Robert, Y.-K. Shue and P. Sears, Antimicrob. Agents Chemother., 2010, 54, 2674–2676 CrossRef CAS.
- M. Alexy and H.-D. Scharf, Liebigs Ann. Chem., 1991, 1363–1364 CrossRef CAS.
- A. Lipták, Carbohydr. Res., 1982, 107, 300–302 CrossRef.
- A. Lipták, I. Czégény, J. Harangi and P. Nánási, Carbohydr. Res., 1970, 73, 327–331 CrossRef.
- G. M. Zdorovenko and E. Y. Rashba, SU 1977–2498635, 1977.
- A. Klemer and M. Waldmann, Liebigs Ann. Chem., 1986, 2, 221–225 CrossRef.
- E. Walton, US 2938900, 1960, Merck & Co..
- W. M. Pankau and W. Kreiser, Helv. Chim. Acta, 1998, 81, 1997–2004 CrossRef CAS.
- W. M. Pankau and W. Kreiser, Tetrahedron Lett., 1998, 39, 2089–2090 CrossRef CAS.
- X. M. Yu, G. Shen and B. S. J. Blagg, J. Org. Chem., 2004, 69, 7375–7378 CrossRef CAS.
- D. S. Reddy, G. Srinivas, B. M. Rajesh, M. Kannan, T. V. Rajale and J. Iqbal, Tetrahedron Lett., 2006, 47, 6373–6375 CrossRef CAS.
- C. A. Lipinski, F. Lombardo, B. W. Dominy and P. J. Feeney, Adv. Drug Delivery Rev., 2001, 46, 3–26 CrossRef CAS.
- F. Babakhani, A. Gomez, N. Robert and P. Sears, J. Med. Microbiol., 2011, 60, 1213–1217 CrossRef CAS.
- K. M. Mullane, M. A. Miller, K. Weiss, A. Lentnek, Y. Golan, P. S. Sears, Y.-K. Shue, T. J. Louie and S. L. Gorbach, Clin. Infect. Dis., 2011, 53, 440–447 CrossRef CAS and erratum: Clin. Infect. Dis., 2011, 53, 1312.
- F. Babakhani, A. Gomez, N. Robert and P. Sears, Antimicrob. Agents Chemother., 2011, 55, 4427–4429 CrossRef CAS.
- F. Babakhani, L. Bouillaut, A. Gomez, P. Sears, L. Nguyen and A. L. Sonenshein, Clin. Infect. Dis., 2012, 55(Suppl 2), S162–S169 CrossRef CAS.
- T. J. Louie, M. A. Miller, K. M. Mullane, K. Weiss, A. Lentnek, Y. Golan, S. Gorbach, P. Sears and Y.-K. Shue, N. Engl. J. Med., 2011, 364, 422–431 CrossRef CAS.
- O. A. Cornely, D. W. Crook, R. Esposito, A. Poirier, M. S. Somero, K. Weiss, P. Sears and S. Gorbach, Lancet Infect. Dis., 2012, 1, 281–289 CrossRef.
- D. W. Crook, A. S. Walker, Y. Kean, K. Weiss, O. A. Cornely, M. A. Miller, R. Esposito, T. J. Louie, N. E. Stoesser, B. C. Young, B. J. Angus, S. L. Gorbach and T. E. A. Peto, Clin. Infect. Dis., 2012, 55(Suppl 2), S93–S103 CrossRef CAS.
- K. Weiss, R. L. Allgren and S. Sellers, Clin. Infect. Dis., 2012, 55(Suppl 2), S110–S115 CrossRef CAS.
- http://www.fda.gov/NewsEvents/Newsroom/PressAnnouncements/ucm257024.htm .
- T. Daniels and T.-Y. So, Gastroen. Res., 2011, 4, 93–96 CAS.
- News in Drugs, 2010, 10, 37–45 Search PubMed.
| This journal is © The Royal Society of Chemistry 2013 |