 Open Access Article
Open Access ArticleCo(II)-detection does not follow Kco(II) gradient: channelling in Co(II)-sensing†
Carl J.
Patterson
a,
Rafael
Pernil
a,
Samantha J.
Dainty
a,
Buddhapriya
Chakrabarti
b,
Clare E.
Henry
c,
Victoria A.
Money
c,
Andrew W.
Foster
ad and
Nigel J.
Robinson
*ac
aSchool/Department of Biological and Biomedical Sciences, Biophysical Sciences Institute, Durham University, DH1 3LE, UK. E-mail: nigel.robinson@durham.ac.uk
bSchool/Department of Mathematics, Biophysical Sciences Institute, Durham University, DH1 3LE, UK
cSchool/Department of Chemistry, Biophysical Sciences Institute, Durham University, DH1 3LE, UK
dICaMB, Newcastle University, NE2 4HH, UK
First published on 31st January 2013
Abstract
The MerR-like transcriptional activator CoaR detects surplus Co(II) to regulate Co(II) efflux in a cyanobacterium. This organism also has cytosolic metal-sensors from three further families represented by Zn(II)-sensors ZiaR and Zur plus Ni(II)-sensor InrS. Here we discover by competition with Fura-2 that CoaR has KCo(II) weaker than 7 × 10−8 M, which is weaker than ZiaR, Zur and InrS (KCo(II) = 6.94 ± 1.3 × 10−10 M; 4.56 ± 0.16 × 10−10 M; and 7.69 ± 1.1 × 10−9 M respectively). KCo(II) for CoaR is also weak in the CoaR–DNA adduct. Further, Co(II) promotes DNA-dissociation by ZiaR and DNA-association by Zur in vitro in a manner analogous to Zn(II), as monitored by fluorescence anisotropy. After 48 h exposure to maximum non-inhibitory [Co(II)], CoaR responds in vivo yet the two Zn(II)-sensors do not, despite their tighter KCo(II) and despite Co(II) triggering allostery in ZiaR and Zur in vitro. These data imply that the two Zn(II) sensors fail to respond because they fail to gain access to Co(II) under these conditions in vivo. Several lines of evidence suggest that CoaR is membrane associated via a domain with sequence similarity to precorrin isomerase, an enzyme of vitamin B12 biosynthesis. Moreover, site directed mutagenesis reveals that transcriptional activation requires CoaR residues that are predicted to form hydrogen bonds to a tetrapyrrole. The Co(II)-requiring vitamin B12 biosynthetic pathway is also membrane associated suggesting putative mechanisms by which Co(II)-containing tetrapyrroles and/or Co(II) ions are channelled to CoaR.
Introduction
Cells control the buffered concentration of each metal in the cytosol through the combined actions of proteins of metal homeostasis including metal-importers, -exporters, -storage proteins, -delivery proteins and -sensors.1 In bacterial cells the metal-sensors are commonly, although not exclusively, DNA-binding, metal-binding transcriptional regulators.2 These sensors are categorised into different families and function as metal-dependent activators, de-repressors or co-repressors.3–9 The ability of the sensors to discern and respond to the correct metal(s) as opposed to all other ions is paramount, since this influences the expression of the homeostatic proteins which in turn influences which metals are available to occupy other metallo-proteins.10 Factors which dictate metal selectivity in metal-sensors have been summarised under the headings affinity, allostery and access.11The cyanobacterium Synechocystis PCC 6803 contains a Co(II)-responsive transcriptional activator of the MerR-family, CoaR.12 In elevated concentrations of cobalt CoaR activates transcription of a gene encoding a cobalt-exporting P1-type ATPase, CoaT, from a promoter with sub-optimal spacing between canonical −10 and −35 RNA polymerase binding sites.12 Variants in which this spacing is shortened confer constitutive transcriptional activity consistent with CoaR acting in a manner analogous to MerR where the inducer-bound activator under-winds the promoter region to re-align the two DNA promoter elements.13 CoaR-dependent expression of a reporter gene (lacZ) from the coaT operator–promoter is activated in response to maximum non-inhibitory concentrations of cobalt but not silver, cadmium, copper, mercury, nickel or zinc either in the host cell (Synechocystis PCC 6803) or when introduced into Escherichia coli.12 What confers this specificity and what prevents sensors for other metals, for example for Zn(II), from responding to surplus Co(II)?
Synechocystis PCC 6803 contains sensors from three other families (additional to MerR) including: a Zn(II)-responsive de-repressor of the ArsR/SmtB family, ZiaR; a Ni(II)-responsive de-repressor of the CsoR/RcnR family, InrS; and a Zn(II)-responsive co-repressor of the Fur family, Zur.8,14–19 We recently explored the hypothesis that selectivity in favour of Ni(II)-sensing by InrS is partly a function of the relative affinities of the complement of sensors in a cell.17 The Ni(II) sensor InrS was shown to have the tightest KNi(II) of the set enabling InrS to de-repress Ni(II)-efflux at a cytosolic concentration below KNi(II) of the other sensors, providing a mechanism by which the actions of InrS sustain a buffered [Ni(II)] sufficiently low to prevent the other sensors from gaining access to Ni(II).17 Here we unexpectedly discover that an analogous model cannot explain the Co(II) specificity of CoaR, prompting a search for additional mechanisms to account for this apparent anomaly.
Results
DNA and Co(II)-binding CoaR can be purified using n-dodecyl β-D-maltoside
Inability to purify sufficient CoaR for in vitro biochemical studies stalled analyses beyond an initial characterisation over a decade ago.12 Three independent algorithms now predict surface exposed, hydrophobic, potentially membrane associated regions within a domain of CoaR which can be modelled on the structure of precorrin isomerase from Pseudomonas denitrificans (Fig. S1, ESI†). Inclusion of a non-ionic detergent in buffers enabled purification of CoaR from recombinant E. coli cell extracts.17 Titration of a hexachlorofluorescein-labelled 36 bp coa operator–promoter fragment containing a 13-6-13 hyphenated inverted repeat corresponding to the CoaR binding site12 (coa-O/P) showed binding of the recombinant protein to its deduced DNA target with KDNA ∼28 nM (Fig. 1A). Moreover, addition of Co(II) to an anaerobic solution of CoaR generated Co(II)-dependent spectral features consistent with ligand to metal charge transfer (LMCT) due to formation of Co(II)-thiolate bonds (ε310 nm ≈ 2,500 M−1 cm−1), which saturate at ∼ one molar equivalent of Co(II) suggestive of two sites per CoaR dimer of KCo(II) ≤ 10−6 M (Fig. 1B). Co(II)-dependent features around 650 nm indicate d–d transitions of sufficient intensity to report upon tetrahedral coordination environments, which moreover contain at least two Cys-thiols per site based upon the intensity of the LMCTs (Fig. 1B).20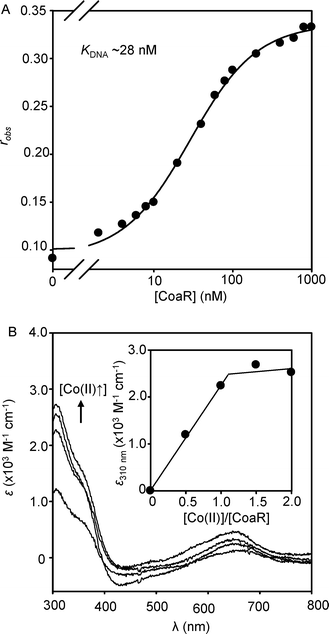 | ||
| Fig. 1 (A) Titration of coa-O/P DNA (10 nM) with recombinant CoaR monitoring fluorescence anisotropy. (B) Apo-subtracted difference spectra upon anaerobic titration of recombinant CoaR (4.7 μM) with Co(II). Inset shows the Co(II)-dependent change in absorbance at 310 nm. | ||
K Co(II) is weaker for CoaR than for ZiaR, Zur or InrS
To explore why CoaR is the cellular sensor for Co(II), by analogy to recent investigations into the specificity of Ni(II)-detection in the same cells,17KCo(II) was estimated for Zn1Zur (one atom of Zn(II) is kinetically trapped in a structural site),21 ZiaR, InrS and CoaR; representing one member from each family of cytosolic metal-sensors encoded within the Synechocystis PCC 6803 genome.17 Co(II)-dependent spectral features were described previously for InrS, and are presented here for ZiaR and Zn1Zur (Fig. S2, ESI†).17 Co(II)-titrations of ZiaR reveal LMCT and d–d-transitions requiring at least two molar equivalents of metal to reach saturation, consistent with four tetrahedral sites per dimer. Unlike the d–d feature at 585 nm, the LMCT feature at 310 nm does not show a linear [Co(II)]-dependent increase in intensity (Fig. S2A, ESI†). ZiaR contains two conserved metal binding sites; a thiol-free α5 site with predicted ligands (two His, Asp, Glu) derived from antiparallel α5 helices and an α3N site with predicted ligands (three Cys, His) derived from α3 helices and an amino terminal extension.7,15 These data (Fig. S2A, ESI†) imply that thiol-free α5 sites have the tightest KCo(II), with two to three Cys-thiols present in α3N sites of weaker KCo(II). Co(II)-titration of Zn1Zur also reveals d–d transitions at 585 nm of intensity consistent with tetrahedral coordination and LMCT features at 310 nm of intensity consistent with a single Cys-thiol, and both features saturate at ∼ one molar equivalent of Co(II) (Fig. S2B, ESI†). Co(II)-titration of InrS previously showed saturation at ∼ two molar equivalents of Co(II), a maximal stoichiometry which is consistent with the resolution of Co(II)-bound InrS via gel filtration chromatography (Fig. S3, ESI†).17 The ratiometric fluorescent metal chelator Fura-2 has been used previously to probe protein Co(II) affinities in the nM range.22 Incubation of Fura-2 with ZiaR, Zn1Zur or InrS in the presence of a sub-stoichiometric concentration of Co(II), sufficient to almost completely fill a single tightest metal binding site on each protein (Tables S2–S4, ESI†), resulted in partial Co(II)-occupancies of sensor-proteins and chelator as reported by an intermediate magnitude of quenching of Fura-2 fluorescence emission (Fig. 2). Using a determined KCo(II) for Fura-2 of 7.03 nM (Fig. S4, ESI†), KCo(II) of the tightest site per dimer for ZiaR and Zn1Zur is calculated to be 6.94 (±1.3) × 10−10 M and 4.56 (±0.16) × 10−10 M respectively (Fig. 2E; Fig. S2–S4, ESI†). KCo(II) of the tightest site per InrS tetramer is calculated to be 7.69 (±1.1) × 10−9 M (Fig. 2A and E). In contrast to the other three sensors, CoaR failed to compete with Fura-2 implying KCo(II) at least one order of magnitude weaker than Fura-2, but KCo(II) must be tighter than the half-saturation [Co(II)] (∼10−6 M) for linear Co(II)-dependent spectral features (Fig. 1B inset). Thus, unexpectedly, CoaR binds Co(II) more weakly than any of the other tested cytosolic metal sensors (Fig. 2F), raising questions about how CoaR gains access to Co(II) in vivo, in a cytosolic environment shared by the Zn(II)-sensors ZiaR and Zur.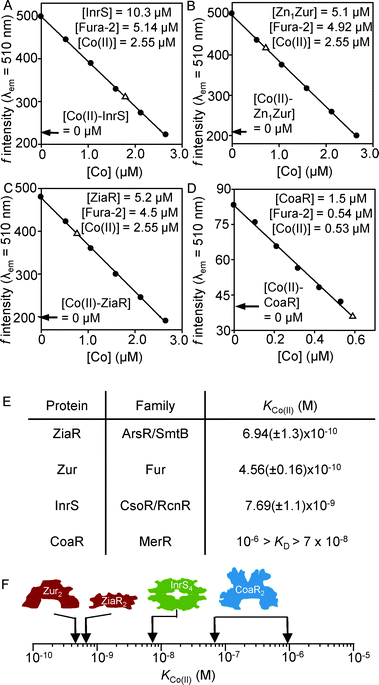 | ||
| Fig. 2 Determination of Co(II) binding constants for each sensor by competition with the Co(II) binding chelator Fura-2. Co(II)-dependent quenching of Fura-2 fluorescence emission at 510 nm (λex = 360 nm) was measured in the absence of protein (closed symbols). Open symbols show the fluorescence emission, at equilibrium, following addition of InrS (A), Zn1Zur (B), ZiaR (C) and CoaR (D) to a mixture of Fura-2 and Co(II) under anaerobic conditions. Expected fluorescence values if Fura-2 fully outcompetes the protein for Co(II) are shown (arrows). Mean KCo(II) values for ZiaR, Zn1Zur and InrS, and a KCo(II) range for CoaR, are tabulated (with standard deviations) (E) and shown in graphical form (F). Numerical parameters used to calculate mean KCo(II) values are shown in Tables S2–S4 (ESI†). | ||
Zur outcompetes CoaR for Co(II)
To test the order of affinity in Fig. 2F, the sensor with the tightest estimated KCo(II) (Zn1Zur) was competed with the weakest (CoaR). Zn1Zur does not contain tryptophan residues resulting in weak fluorescence emission when excited at 295 nm (Fig. 3A), unlike CoaR (Fig. 3B), enabling discrimination between metal-binding to the two proteins. In the presence of equimolar Zn1Zur quenching of CoaR fluorescence by Co(II) is lost, consistent with Co(II) binding to the tighter but invisible site of Zn1Zur, using λex = 295 nm (Fig. 3C). In an analogous experiment, addition of ∼ one molar equivalent of Zn(II) also gave no quenching of CoaR fluorescence, but quenching occurred upon subsequent addition of Co(II), consistent with Zn(II) preferentially binding and saturating Zn1Zur allowing Co(II) to subsequently partition to CoaR (Fig. 3D).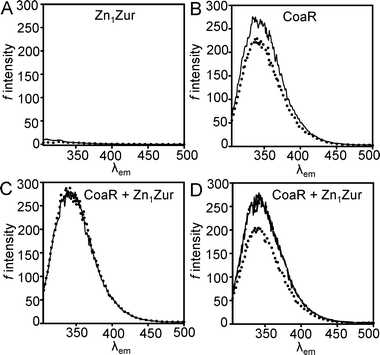 | ||
| Fig. 3 (A) Fluorescence emission spectra for Zn1Zur (5 μM) in the presence (dotted line) and absence (solid line) of one molar equivalent of Co(II). (B) Emission spectra of CoaR (5 μM) in the absence (solid line) and presence (dotted line) of 0.9 molar equivalents of Co(II). (C) Emission spectra from a solution of equimolar (5 μM) apo-CoaR and Zn1Zur (solid line) and after addition of 0.9 molar equivalents of Co(II) (dotted line). (D) Emission spectra from an experiment analogous to (C) but with 0.9 molar equivalents Zn(II) (dashed line, under solid line) followed by 1.1 molar equivalents of Co(II) (dotted line). λex = 295 nm for all experiments. | ||
K Co( II ) is relatively weak for CoaR–DNA complexes
As a MerR-like transcriptional regulator, activated Co(II)–CoaR remains associated with DNA. Therefore we explored the possibility that KCo(II) might be tighter for the DNA-adduct. Based on the estimated KDNA (Fig. 1), CoaR at 1.0 μM in the presence of surplus (1.2 μM), coa-O/P DNA (without fluorescent label), should be fully DNA-bound. Fura-2 was titrated with Co(II) in the presence or absence of this CoaR–DNA complex and in common with an analogous experiment using CoaR alone, negligible Co(II) was withheld from Fura-2 (Fig. 4A and B). In contrast, control titrations with ZiaR and Zn1Zur do show evidence of Co(II) being withheld from Fura-2 (Fig. 4C and D). Thus, DNA-binding does not enhance the KCo(II) of CoaR and its weak affinity relative to the other sensors remains anomalous.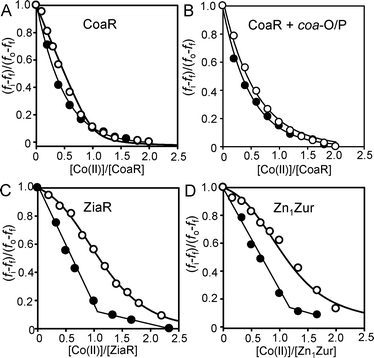 | ||
| Fig. 4 Titration of Fura-2 (1 μM) with Co(II) in the absence (closed symbols) or presence (open symbols) of CoaR (1 μM), either in the absence (A) or presence (B) of unlabelled coa-O/P DNA (1.2 μM). At these DNA and protein concentrations all of the CoaR should be DNA-bound. Titrations analogous to (A) but for ZiaR (C) and Zn1Zur (D) (1.5 μM Fura-2 and protein). All titrations in the presence of protein were performed anaerobically. | ||
Co(II) inhibits DNA-binding by ZiaR and promotes DNA-binding by Zur
Although Co(II) binds tightly to the cytosolic cyanobacterial sensors for other metals, it might be ineffective at triggering the allosteric mechanism to alter DNA-binding and hence to modulate gene expression by these other proteins. This was tested in vitro by fluorescence anisotropy (Fig. 5). Addition of Co(II) to a pre-formed complex of ZiaR and hexachlorofluorescein-labelled zia operator–promoter DNA (zia-O/P) caused a decrease in anisotropy consistent with Co(II)-dependent DNA-dissociation (Fig. 5A), analogous to previous observations with Zn(II).23 Consistent with this observation, Co(II) also inhibited the association of ZiaR with zia-O/P DNA (Fig. S5, ESI†). Addition of Co(II) to Zn1Zur caused an increase in anisotropy consistent with Co(II)-dependent DNA association by this metal-dependent co-repressor (Fig. 5B), which is similar to previous observations with Zn(II).21 Co(II) is also effective at dissociating InrS–DNA complexes (Fig. 5C). Thus, each of the other three sensors is competent to respond to Co(II) in vitro.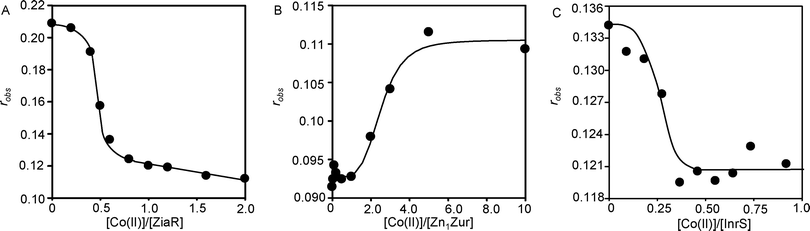 | ||
| Fig. 5 (A) Anaerobic titration of a complex of ZiaR (1 μM) and zia-O/P DNA (10 nM) with Co(II), monitoring fluorescence anisotropy. (B) Titration (in the presence of 1 mM TCEP) of znu-O/P DNA (10 nM) and Zn1Zur (100 nM) with Co(II), monitoring fluorescence anisotropy. (C) Anaerobic titration of a complex of InrS (1 μM) and nrsD-O/P DNA (10 nM) with Co(II), monitoring fluorescence anisotropy. | ||
Either α3N or α5 metal-sites of ZiaR are sufficient for allostery
Only 0.5 molar equivalents of Co(II) were required to dissociate ZiaR–DNA complexes (Fig. 5A). This was unexpected because previously it was shown that mutation of putative metal ligands at either α5 or α3N sites impaired Zn(II) sensing in vivo.15 This implied that both α5 and α3N sites were needed for the protein to respond to Zn(II) whereas the stoichiometry in Fig. 5A implies that, at least for Co(II), binding to only one of four sites (per dimer) is needed to dissociate the protein from DNA. Variants of ZiaR were generated in which metal-binding residues at each of the sites were replaced individually or in combination with non-binding alternates. Titration of these variants with Co(II) confirmed binding of ∼ one molar equivalent of metal solely to a thiol-free, and hence LMCT-free (at 310 nm), α5 site in the α3N mutants, and binding of ∼ one molar equivalent of metal solely to a thiol-containing, and hence LMCT-generating, α3N site in the α5 mutants (Fig. S6, ESI†). Double mutants missing both pairs of sites failed to dissociate from DNA upon addition of up to ten molar equivalents of Zn(II), as expected. However, mutants missing either individual pair of sites (at α5 or at α3N) remained inducer responsive (Fig. 6). Thus either site is sufficient to trigger the allosteric response and it is reasonable to consider the affinity of the tightest Co(II) site of ZiaR in comparisons with the other sensors.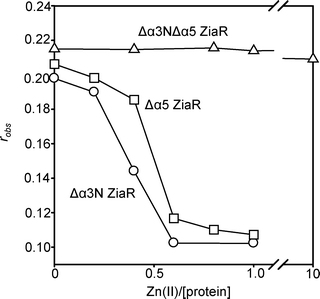 | ||
| Fig. 6 Anaerobic titration of a complex of either Δα3N ZiaR (circles), Δα5 ZiaR (squares) or Δα3NΔα5 ZiaR (triangles) (each at 1 μM) and zia-O/P DNA (10 nM) with Zn(II), monitoring fluorescence anisotropy. | ||
CoaR, but not ZiaR or Zur, responds after prolonged exposure to Co(II) in vivo
ZiaR is reported to respond to Zn(II) and not Co(II), but CoaR the opposite in vivo.12,15 In view of the observation that the two Zn(II) sensors ZiaR and Zur bind Co(II) at least two orders of magnitude more tightly than CoaR (Fig. 2) and in both cases Co(II) alters DNA-binding in a manner analogous to Zn(II) (Fig. 5), we coincidentally tested the abundance of ziaA, znuA and coaT transcripts, regulated by ZiaR, Zur and CoaR respectively, in common populations of nucleic acids isolated from cells exposed to elevated Zn(II) (16 μM) or Co(II) (2 μM) (maximum non-inhibitory concentrations) for 48 h (Fig. 7A). Transcripts encoded by the gene activated by CoaR, namely coaT, accumulated in response to Co(II) not Zn(II), transcripts de-repressed by ZiaR, ziaA, accumulated in response to Zn(II) not Co(II), while the abundance of transcripts repressed by Zn2Zur, znuA, declined in response to Zn(II) but not Co(II). Thus, the observed selectivity precisely replicates predictions based upon the literature.12,15,19 InrS is known to respond to Co(II) as well as Ni(II) and was not included in these analyses.17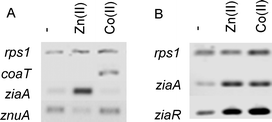 | ||
| Fig. 7 (A) Abundance of ziaA, znuA and coaT transcripts in common populations of RNA analysed by RT-PCR following 48 h exposure to Zn(II) (16 μM) or Co(II) (2 μM). (B) Abundance of ziaA and ziaR transcripts in response to the same concentrations of Zn(II) and Co(II) following 1 h exposure. rps1 is included as a control in both cases. | ||
Co(II) does initially trigger ZiaR in vivo
Failure to de-repress expression of ziaA in response to 48 h exposure to Co(II) implies that by the end of this incubation ZiaR does not have access to excess Co(II). We tested whether shorter (1 h) exposure to Co(II) might elicit a response. Fig. 7B shows accumulation of ziaA and ziaR transcripts under these conditions. Both these genes are regulated by ZiaR. Thus ZiaR is responsive to Co(II) in vivo as well as in vitro but after longer exposure (48 h) Co(II) is somehow sensed by CoaR but not by ZiaR despite the >100 fold tighter KCo(II) of the latter.A deduced tetrapyrrole binding-site in CoaR is required for cobalt-sensing
CoaR contains a domain with homology to precorrin isomerase, an enzyme which catalyses a methyl isomerisation reaction required during the biosynthesis of vitamin B12 from precursor tetrapyrrole molecules.12 Homologous proteins catalysing this reaction in organisms utilising the so-called aerobic B12 synthesis pathway are termed CobH and those from anaerobic B12 synthesis pathways are termed CbiC.24 Sequence alignment of CoaR and Pseudomonas denitrificans CobH reveals that only two out of the eleven residues (Ser17 and Ala44), that in CobH form hydrogen bonds to its reaction product hydrogenobyrinic acid (HBA), are absolutely conserved in CoaR (Ser120 and Ala192) (Fig. S7, ESI†). Using the structure of P. denitrificans CobH in which the tight-binding hydrogenobyrinic acid molecule was co-visualised,25 a dimeric model of the precorrin isomerase-like domain of CoaR was produced that included hydrogenobyrinic acid associated with modelled CoaR tetrapyrrole binding sites (Fig. S8, ESI†). By making a comparison with the analogous structure of CobH from P. denitrificans, it was possible to identify residues in CoaR in equivalent spatial locations to the residues known to form hydrogen bonds to the tetrapyrrole in CobH (Fig. 8; Fig. S8 and Table S5, ESI†).25 Three of these CoaR residues (Trp265, Leu191 and Gly 342) are incapable of forming side chain hydrogen bonds while Ala192 overlays a residue in CobH (Ala44) which forms a hydrogen bond from a backbone nitrogen atom. Site directed mutagenesis was performed on the remainder to test for effects on Co(II)-dependent transcriptional activation. Conspicuously, Tyr287 protrudes into the predicted tetrapyrrole binding site at the opposite face to a ligand in CobH, Tyr14, and this has also been mutated. Loss and gain of Co(II)-responsive expression of a reporter gene (lacZ) driven from the coaT operator–promoter correlated with Tyr287 substitutions which lose (Phe) and restore (Glu) hydrogen bonding, respectively (Fig. 8). Co(II)-responsive expression was lost or impaired by Ala substitutions at five of the remaining six deduced ligand positions (Fig. 8). The exception (Ser290) overlays with a residue (Thr140) which forms one of two hydrogen bonds to a functional group in the tetrapyrrole and hence its loss may be compensated by an alternate bond. Three of the substitutions (Ser167Ala, His267Ala and Tyr287Phe) gave no Co(II)-response (Fig. 8) and two of these variants were over-expressed and purified from E. coli, and both confirmed to form complexes with the coa-O/P DNA with similar KDNA to wild type CoaR (Fig. 9), consistent with loss of effector recognition rather than global mis-folding. In Synechocystis PCC 6803 ΔcbiE, unable to synthesise the substrate for precorrin isomerase, expression of a reporter gene from the coaT operator–promoter was enhanced and this was taken to imply that intermediates in the vitamin B12 biosynthetic pathway are inhibitory to transcriptional activation by CoaR.12 In contrast, the observations reported here suggest that a deduced binding site for such molecules in CoaR is required for Co(II)-dependent transcriptional activation.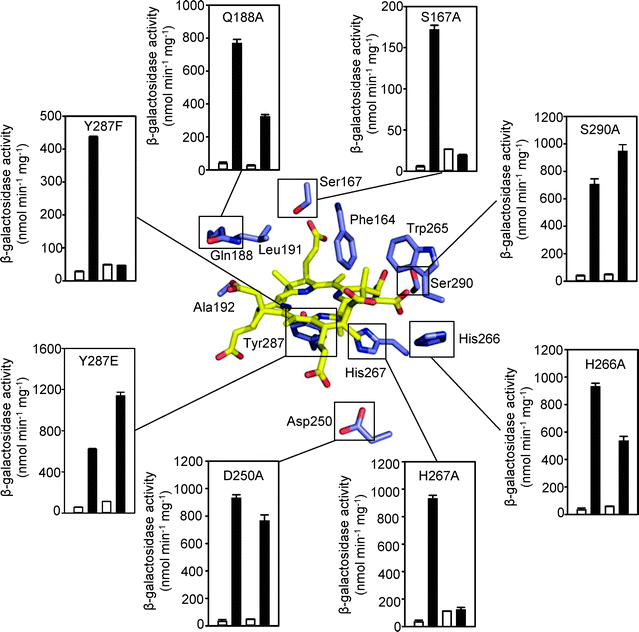 | ||
| Fig. 8 A simplified model of the predicted tetrapyrrole binding site of CoaR is shown (central panel) in complex with hydrogenobyrinic acid. Residues spatially analogous to tetrapyrrole ligands in CobH are annotated (see Fig. S8, ESI†). Outer panels show expression from the coaT promoter in E. coli cells containing pET3acoa and variants of coaR, in the absence (open bars) or presence (closed bars) of 100 μM CoCl2. In each dataset the left hand bars represent a wild-type CoaR control performed in the same assay. All data are mean activities from assays performed in triplicate shown with standard deviations. | ||
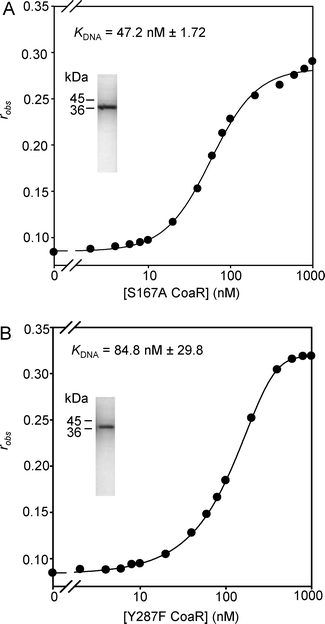 | ||
| Fig. 9 Titrations of coa-O/P (10 nM) with CoaR variants S167A (A) and Y287F (B) that are Co(II) non-responsive, monitoring fluorescence anisotropy. Purity and molecular mass of recombinant CoaR variants are shown by SDS-PAGE (insets). | ||
A carboxyl-terminal Co(II)-site in CoaR is required for cobalt-sensing
If CoaR sensed a tetrapyrrole rather than directly sensing Co(II) this could account for its response to Co(II) in vivo despite its relatively weak KCo(II). However, previously a triple mutant within a carboxyl-terminal CysHisCys motif in CoaR was shown to lose Co(II)-dependent transcriptional activation and it was suggested that this was a likely Co(II) binding site.12 Another residue (Cys121) is conserved in CoaR and acts as a metal ligand in the homologous E. coli MerR-like sensors ZntR (Zn(II)) and the CueR (Cu(I)).26 We have examined the effects of mutation of Cys121 and the first Cys residue from the carboxyl-terminal CysHisCys motif (Cys363) on the Co(II)-dependent spectral features of purified CoaR (Fig. 10 inset). Conversion of either Cys to Gly reduced the intensity of the LMCT features at 310 nm by ∼103 M−1 cm−1 consistent with loss of a single Co(II)–S bond in each mutant. Both variants showed either no, or negligible, Co(II)-responsive expression of a reporter gene from the coaT operator–promoter when examined in E. coli (Fig. 10).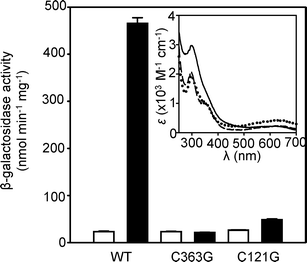 | ||
| Fig. 10 Expression from the coaT promoter in E. coli cells containing either pET3acoa or mutated variants (CoaR C363G and C121G), cultured in the absence (open bars) or presence (closed bars) of 100 μM CoCl2. Data are means of triplicate assays with standard deviation. Inset shows apo-subtracted difference spectra following addition of Co(II) (two molar equivalents) to C363G CoaR (8.6 μM) (dotted line), C121G CoaR (9.5 μM) (dashed line) and wild-type CoaR (9.5 μM) (solid line). | ||
Discussion
In this research we observe binding of the cobalt responsive transcriptional activator CoaR to Co(II) and to DNA (Fig. 1), but unexpectedly estimate its KCo(II) to be the weakest among a sub-set of metal-sensors from Synechocystis PCC 6803: a sub set which includes one sensor from each family encoded by this genome (Fig. 2). This relatively weak KCo(II) is supported by analyses of metal-dependent quenching of intrinsic CoaR fluorescence in the presence of zinc-sensing Zn1Zur (estimated to have the tightest KCo(II) of the set of sensors) (Fig. 3). Quenching of CoaR fluorescence upon addition of one molar equivalent of Co(II) is lost in the presence of equimolar Zn1Zur, consistent with Co(II) binding to the tighter sites of the Zn(II)-sensor, but quenching is regained if the sensory sites of Zn1Zur are first saturated with a molar equivalent of Zn(II) (Fig. 3). These data indicate that KZn(II) Zn1Zur is tighter than KZn(II) CoaR, consistent with the former but not the latter detecting Zn(II) in vivo, and in contrast with the inverse correlation between KCo(II) and Co(II)-sensing. The results for Co(II) also contrast with the order of affinity for KNi(II) of the same set of proteins in which the cytosolic Ni(II)-sensor, InrS, has the tightest KNi(II).17 Ni(II) and potentially Zn(II), are thus expected to partition to the sensor of tightest affinity, triggering expression of efflux, preventing the buffered cytosolic concentration of Ni(II) or Zn(II) from rising to a sufficiently high level to bind to other sensors of weaker KNi(II) and KZn(II). In contrast, the specific detection of Co(II) by the weakest Co(II)-binding sensor CoaR is enigmatic.The possibility that the KCo(II) of CoaR may be enhanced by conformational changes in the CoaR–DNA adduct was explored, since this adduct is the active species. Co(II)–CoaR is inferred to induce transcription from the coa operator–promoter by under winding DNA in a mechanism analogous to Hg(II)–MerR.12,13,27 In the presence of excess coa operator–promoter at a concentration at least an order of magnitude greater than KDNA CoaR (Fig. 1A), CoaR failed to compete with Fura-2 for Co(II) (Fig. 4). Both ZiaR and Zn1Zur do compete with Fura-2 for Co(II) (Fig. 2 and 4). The possibility that selectivity may not relate to Co(II) binding but may instead be due to Co(II) failing to trigger the allosteric mechanisms of ZiaR and Zur was investigated next. However, Co(II) does drive DNA-dissociation of ZiaR and encourages DNA-association by Zn1Zur (Fig. 5).
CoaR, ZiaR and Zur show perfect discrimination between Co(II) and Zn(II) after prolonged exposure (48 h) of cells to maximum non-inhibitory metal concentrations but crucially this selectivity is not detected for ZiaR after short term (1 h) exposure (Fig. 7). While the latter observation is consistent with in vitro data for ZiaR (Fig. 5), the former is not. The relative abundance of CoaR and ZiaR in the cell has not been determined but the mode of action of CoaR implies that mass action could not provide a bias in favour of CoaR detecting cobalt. The precedent is that metal-bound forms of MerR-like activators have slightly weaker KDNA than apo-forms and hence a larger number of CoaR molecules would further dis-favour activation of the coaT operator–promoter in response to Co(II).27 Failure to respond to cobalt at 48 h is inferred to reflect an in vivo regime under which CoaR somehow gains access to surplus Co(II) in preference to ZiaR. This kinetic contribution must be sufficient to overcome a dis-favourable thermodynamic gradient of at least two orders of magnitude (Fig. 2F).
A requirement for 0.1% w/v n-dodecyl β-D-maltoside to purify CoaR suggests association with membranes via its precorrin isomerise-like domain. Enzymes of vitamin B12 biosynthesis are known to be membrane associated, and one of the homologues of precorrin isomerase in Synechocystis PCC 6803 (Slr1467) has been shown by proteomics to be membrane associated.28–30 Slr1467 shares sequence similarity (45%) with the precorrin isomerase-like domain of CoaR and the two proteins have analogous predicted hydrophobic carboxyl-terminal regions (Fig. S1, ESI†). Negative charge associated with the inner leaflet of plasma-membranes would be attractive to cations such as Co(II), raising the possibility that Co(II) and CoaR encounter at the membrane whereas Co(II) and ZiaR or Zur encounter in the cytosol. Ignoring any contribution from thylakoid membranes, if the radius of the cell is ∼ three orders of magnitude greater than that of CoaR (Fig. S9, ESI†), a three dimensional search between Co(II) and ZiaR or Zur would take approximately two orders of magnitude longer than a two dimensional search for CoaR (associated with one of the multiple chromosome copies) at the membrane (Fig. S9, ESI†).
The pathway for vitamin B12 biosynthesis is also known to be membrane associated in other bacteria with intermediates channelled between enzymes.28–31 The precorrin isomerase-like domain of CoaR might be expected to associate with this machinery, in common with the precorrin isomerase enzyme that acts in the pathway. Previously, bacterial two-hybrid interactions were not detected between the cobalt-exporter CoaT and the cobalt-chelatase for B12 biosynthesis CbiX,32 but interaction with the cobalt importer HupE33 or with CoaR is formally possible, and might enable Co(II) to be channelled to CoaR.
Mutation of a candidate tetrapyrrole binding site in CoaR suggests that B12, one of its precursors or a related tetrapyrrole, is required for Co(II)-dependent transcriptional activation (Fig. 8). Notably, this requirement was observed in E. coli which does not possess a complete B12 biosynthetic pathway and imports B12. The siroheme ferrochelatase (CysG) of E. coli can however introduce Co(II) into tetrapyrroles,34 but CoaR remains responsive to Co(II) in ΔcysG E. coli (Fig. S10, ESI†), implying that CoaR does not detect a Co(II)-containing product of this enzyme. A hyper-responsive phenotype was previously observed in Synechocystis PCC 6803 ΔcbiE and it was inferred that this mutant is missing a tetrapyrrole that represses CoaR.12 However, cobalt is inserted into the corrin ring sooner than previously anticipated in this organism via the co-called anaerobic pathway,35 and an explanation consistent with current data (Fig. 8), is that there is hyper-accumulation of a Co(II)-containing activator of CoaR in ΔcbiE.12,36 In this alternative scenario the tetrapyrrole may either be the sole activator, potentially channelled to CoaR via the B12 assembly pathway (albeit it is unclear how a Co(II)-cofactor is generated in E. coli), or a co-activator with additional channelling of cobalt to a site involving Cys121 associated with the MerR-like domain, and carboxyl-terminal Cys363 (Fig. 10). Potentially, KCo(II) may be enhanced on binding tetrapyrroles.
In conclusion, these studies highlight the merit in considering selectivity in metal sensing as an integrated system: a combined function of a cells set of sensors (a metallomic approach), rather than solely a property of individual sensors acting in isolation. For Ni(II),17 and provisionally Zn(II), specificity matches the thermodynamic gradient among the set of sensors. However for Co(II) this is not the case. This uncovers a requirement for additional studies to explain the mechanism of action of CoaR and its interplay with vitamin B12 biosynthesis.
Methods
General reagents, bacterial cultures and cloning procedures
All reagents and chemicals were sourced from standard suppliers. E. coli strains DH5α, JM101 and BL21(DE3) were grown in standard Luria Bertini (LB) medium. Constructs (in pET29a) for overexpression of wild-type CoaR, ZiaR, Zur and InrS were produced previously.17,21,23 ZiaR and CoaR variants were generated by QuikChange mutagenesis (Stratagene) with pET29a overexpression plasmids as template. Primers for each are shown in Table S1 (ESI†). A coaT reporter–promoter construct was generated by XhoI/BsaI release of a coaR–promoter–lacZ fragment from plasmid pLACCOA (produced previously)12 followed by ligation to BsaI and SalI sites to form pET3acoa (the derivative of pET3a also contained cbiX controlled by an IPTG-responsive promoter which was not activated in these studies). Mutated variants of coaR were produced by QuikChange mutagenesis (primers in Table S1, ESI†). Due to the large size of pET3acoa, nicks were first sealed using T4 DNA ligase (2 μl 10× T4 DNA ligase buffer (Promega), 1 μl T4 DNA ligase (Promega), 15 μl H2O, 2 μl DNA, 15 °C for 4 h) followed by enzyme inactivation at 65 °C for 20 min. All gene variants were confirmed by sequencing.Protein expression and purification
Expression and purification of ZiaR, Zn1Zur, CoaR InrS have been described previously.17,21,23 Mutated variants of ZiaR and Δα3N ZiaR (C71S/C73S), Δα5 ZiaR (H116R), and Δα3NΔα5 ZiaR variants, were overexpressed in E. coli BL21(DE3) and purified using the same procedures as for wild-type ZiaR,23 with the exception that HiTrap heparin affinity chromatography (GE Healthcare) replaced Ni(II)-affinity chromatography for Δα3NΔα5 ZiaR (which binds weakly to Ni(II)-affinity matrices). C121G and C363G CoaR variants were expressed and purified as described previously for wild-type.17 S167A and Y287F CoaR variants were expressed as described previously,17 but purified by refolding protein recovered from inclusion bodies, re-suspended in 4 ml of 500 mM NaCl, 10 mM Hepes (pH 7.8), 5 mM DTT, 8 M urea, 1% w/v SDS. Post incubation (37 °C, 1 h), supernatant (13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 400 × g, 10 min), was diluted 100-fold into 500 mM NaCl, 5 mM DTT, 10 mM Hepes (pH 7.8), 0.5% w/v n-dodecyl β-D-maltoside, further incubated (1 h) and supernatant (13
400 × g, 10 min), was diluted 100-fold into 500 mM NaCl, 5 mM DTT, 10 mM Hepes (pH 7.8), 0.5% w/v n-dodecyl β-D-maltoside, further incubated (1 h) and supernatant (13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 400 × g, 10 min) diluted 50-fold into 100 mM NaCl, 1 mM DTT, 10 mM Hepes (pH 7.8), 100 mM imidazole, 1 mM EDTA, 0.05% w/v n-dodecyl β-D-maltoside. This solution was applied to a 1 ml HiTrap heparin affinity column (GE Healthcare) pre-equilibrated in the same buffer and subsequently eluted in buffer containing 1M NaCl and fractions analysed by SDS-PAGE.
400 × g, 10 min) diluted 50-fold into 100 mM NaCl, 1 mM DTT, 10 mM Hepes (pH 7.8), 100 mM imidazole, 1 mM EDTA, 0.05% w/v n-dodecyl β-D-maltoside. This solution was applied to a 1 ml HiTrap heparin affinity column (GE Healthcare) pre-equilibrated in the same buffer and subsequently eluted in buffer containing 1M NaCl and fractions analysed by SDS-PAGE.
ZiaR, Zur, CoaR and InrS (plus variants of ZiaR and CoaR) were quantified as described previously,17 with the exception of Y287F CoaR which has an altered theoretical extinction coefficient (ε = 25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 940 M−1 cm−1). Proteins were confirmed to be >95% apo by ICP-MS analysis. Anaerobic stocks of protein were confirmed to be >95% reduced by reaction with 5,5′-dithiobis-(2-nitrobenzoic acid) (DTNB) as described previously.17,21,23 CoaR showed inconsistent thiol-reactivity although DNA-binding and metal-binding remained consistent.
940 M−1 cm−1). Proteins were confirmed to be >95% apo by ICP-MS analysis. Anaerobic stocks of protein were confirmed to be >95% reduced by reaction with 5,5′-dithiobis-(2-nitrobenzoic acid) (DTNB) as described previously.17,21,23 CoaR showed inconsistent thiol-reactivity although DNA-binding and metal-binding remained consistent.
UV-visible absorption spectroscopy
Proteins were diluted into chelex-treated, N2-purged buffer (containing 400 mM KCl, 100 mM NaCl, 10 mM Hepes (pH 7.8) and, in the case of CoaR, 0.1% w/v n-dodecyl β-D-maltoside) in an anaerobic chamber. UV-visible spectra were collected under anaerobic conditions, using sealed, gas tight quartz cuvettes, using Cary 4E UV-visible (Varian, UK) or Perkin Elmer λ35 spectrophotometers. Concentrations of ZnCl2 and CoCl2 stocks used in titrations were verified by ICP-MS.Fluorescence anisotropy
Fluorescent znu-O/P, nrsD-O/P and zia-O/P probes have been described previously.17,21,23 Oligonucleotides (primers 25 and 26, Table S1, ESI†) containing a region of the coa operator–promoter including the 13-6-13 CoaR binding site were annealed as described previously to produce 36 bp hexachlorofluorescein-labelled coa-O/P.12,37 Wild-type and mutant forms of apo-CoaR protein were titrated against 10 nM coa-O/P in buffer containing 150 mM NaCl, 5 mM DTT, 1 mM EDTA, 200 mM imidazole, 10 mM Hepes (pH 7.8). For anaerobic dissociation experiments with ZiaR and InrS, protein–DNA complexes were pre-formed by adding protein to a final concentration of 1 μM in a sealed quartz cuvette containing 10 nM DNA in chelex-treated, N2-purged buffer containing either 120 mM KCl, 30 mM NaCl, 10 mM Hepes (pH 7.8) (for ZiaR) or 240 mM KCl, 60 mM NaCl, 10 mM Hepes (pH 7.8) (for InrS). Protein–DNA complexes were titrated with Co(II) under anaerobic conditions. A solution of Zn1Zur (100 nM) and znu-O/P (10 nM) was titrated with Co(II) aerobically in chelex-treated buffer containing 120 mM KCl, 30 mM NaCl, 1 mM TCEP 10 mM Hepes (pH 7.8). Data were collected either using an 8100 Fluorometer (SLM-Aminco, Urbana, USA) or a modified Cary Eclipse Fluorescence Spectrophotometer (Agilent Technologies) as previously.17,37Protein–Fura-2 Co(II) competition
Experiments were performed under anaerobic conditions in chelex-treated, N2-purged buffer containing 400 mM KCl, 100 mM NaCl and 10 mM Hepes (pH 7.8) (with 0.1% w/v n-dodecyl β-D-maltoside in experiments with CoaR). Fura-2 was quantified by measurement of absorption intensity at 363 nm (ε = 28![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 M−1 cm−1). In competition experiments Fura-2 was incubated with Co(II) for 10 min before addition of protein. The fluorescence emission at 510 nm (λex = 360 nm) was monitored to equilibrium, determining [Co(II)–Fura-2] in conjunction with Co(II)-Fura-2 standards. KCo(II) was calculated as described in Table S2 (ESI†). For ZiaR, Zn1Zur and InrS the parameters required to determine KCo(II) are shown in Tables S2–S4 (ESI†). To analyse the effect of DNA binding on the KCo(II) of CoaR, a mixture CoaR (1 μM), Fura-2 (1 μM) and coa-O/P DNA (1.2 μM) (prepared using primer 26 and an unlabelled version of primer 25, Table S1, ESI†) was titrated with Co(II) under anaerobic conditions and the equilibrium fluorescence intensities (λex = 360 nm, λem = 500 nm) recorded. Comparative titrations were performed for ZiaR and Zn1Zur (1.5 μM protein, 1.5 μM Fura-2) (no DNA was present). The affinity of Fura-2 under the buffer conditions used in these analyses (400 mM KCl, 100 mM NaCl, 10 mM Hepes pH 7.8) was determined directly by competition with the nitrilotriacetic acid (NTA) which binds Co(II) with KCo(II) = 3.59 × 10−9 M at pH 7.8 (Fig. S4, ESI†).
000 M−1 cm−1). In competition experiments Fura-2 was incubated with Co(II) for 10 min before addition of protein. The fluorescence emission at 510 nm (λex = 360 nm) was monitored to equilibrium, determining [Co(II)–Fura-2] in conjunction with Co(II)-Fura-2 standards. KCo(II) was calculated as described in Table S2 (ESI†). For ZiaR, Zn1Zur and InrS the parameters required to determine KCo(II) are shown in Tables S2–S4 (ESI†). To analyse the effect of DNA binding on the KCo(II) of CoaR, a mixture CoaR (1 μM), Fura-2 (1 μM) and coa-O/P DNA (1.2 μM) (prepared using primer 26 and an unlabelled version of primer 25, Table S1, ESI†) was titrated with Co(II) under anaerobic conditions and the equilibrium fluorescence intensities (λex = 360 nm, λem = 500 nm) recorded. Comparative titrations were performed for ZiaR and Zn1Zur (1.5 μM protein, 1.5 μM Fura-2) (no DNA was present). The affinity of Fura-2 under the buffer conditions used in these analyses (400 mM KCl, 100 mM NaCl, 10 mM Hepes pH 7.8) was determined directly by competition with the nitrilotriacetic acid (NTA) which binds Co(II) with KCo(II) = 3.59 × 10−9 M at pH 7.8 (Fig. S4, ESI†).
CoaR–Zur Co(II) competition
Recombinant, wild-type CoaR and Zn1Zur were each diluted separately to 5 μM in chelex-treated, N2-purged buffer containing 400 mM KCl, 100 mM NaCl, 10 mM Hepes (pH 7.8) and 0.1% w/v n-dodecyl β-D-maltoside in an anaerobic chamber. Fluorescence emission spectra (λex = 295 nm for all experiments) were collected in sealed quartz cuvettes for apo-proteins and following addition of Co(II). CoaR and Zn1Zur were competed for Co(II) by diluting both to 5 μM in a quartz cuvette. Fluorescence emission spectra were collected for apo-proteins, one molar equivalent of Co(II) was then added in an anaerobic chamber and the emission spectrum re-recorded. In an analogous experiment 0.9 molar equivalents Zn(II) was added followed by 1.1 molar equivalents of Co(II).RNA-isolation and reverse transcriptase PCR
Cells were inoculated to an OD595 nm of 0.1 and cultured under standard conditions with maximum non-inhibitory concentrations of ZnSO4 (16 μM) or CoCl2, (2 μM) (concentrations determined in growth experiments to give <10% growth inhibition), for either 1 h or 48 h. Nucleic acid was isolated from logarithmically growing Synechocystis PCC 6803 using an established method.38 Nucleic acids were treated with DNase I (Sigma) and 1 μg used to produce cDNA using an Im-PromII reverse transcription kit with random hexameric primers (Promega). Reverse transcriptase was omitted from negative control reactions, which in all cases confirmed the absence of contaminating DNA. PCR primers for ziaA, rps1, znuA (described previously)21,23 and coaT (primers 23 and 24, Table S1, ESI†) were used to amplify ∼300 bp of each gene. Cycling conditions included denaturation (95 °C for 2 min) then 25–30 cycles of 95 °C 1 min, 60 °C 1 min, 72 °C 20 s, followed by a final extension step at 72 °C for 5 min. Products were analysed on 1% w/v agarose gels.Structural modelling
A dimeric model of the precorrin isomerase-like domain of CoaR bound to hydrogenobyrinic acid (HBA) was produced by threading residues 162–358 onto the co-crystal structure from P. denitrificans (PDB code: 1I1H) using SwissModel (http://swissmodel.expasy.org/). The resulting monomer was converted to a dimer and aligned with CobH in WinCoot108,39 and the dimer analysed in PyMol and Swiss PDB Viewer.β-Galactosidase assays
JM101 cells containing pET3acoa (and variants), grown at 37 °C overnight were diluted 100-fold into fresh LB medium and cultured in the absence or presence of 100 μM CoCl2 (maximum non-inhibitory concentrations used previously)12 at 37 °C until an OD595 nm of 0.2–0.3 was reached. Cells were assayed in triplicate for β-galactosidase activity as described previously.40Acknowledgements
This work was supported by Biotechnology and Biological Research Council Grants BB/H006052/2 and BB/E001688/1. We acknowledge related (data not included) and helpful collaborations with Natasha Cant, Simon Moore and Martin Warren.References
- K. J. Waldron and N. J. Robinson, How do bacterial cells ensure that metalloproteins get the correct metal?, Nat. Rev. Microbiol., 2009, 7, 25–35 CrossRef CAS.
- D. P. Giedroc and A. I. Arunkumar, Metal sensor proteins: nature's metalloregulated allosteric switches, Dalton Trans., 2007, 3107–3120 RSC.
- J. W. Lee and J. D. Helmann, Functional specialization within the Fur family of metalloregulators, Biometals, 2007, 20, 485–499 CrossRef CAS.
- M. P. Schmitt, E. M. Twiddy and R. K. Holmes, Purification and characterization of the diphtheria toxin repressor, Proc. Natl. Acad. Sci. U. S. A., 1992, 89, 7576–7580 CrossRef CAS.
- J. S. Iwig and P. T. Chivers, Coordinating intracellular nickel-metal-site structure-function relationships and the NikR and RcnR repressors, Nat. Prod. Rep., 2010, 27, 658–667 RSC.
- N. L. Brown, J. V. Stoyanov, S. P. Kidd and J. L. Hobman, The MerR family of transcriptional regulators, FEMS Microbiol. Rev., 2003, 27, 145–163 CrossRef CAS.
- D. Osman and J. S. Cavet, Bacterial metal-sensing proteins exemplified by ArsR-SmtB family repressors, Nat. Prod. Rep., 2010, 27, 668–680 RSC.
- T. Liu, A. Ramesh, Z. Ma, S. K. Ward, L. Zhang, G. N. George, A. M. Talaat, J. C. Sacchettini and D. P. Giedroc, CsoR is a novel Mycobacterium tuberculosis copper-sensing transcriptional regulator, Nat. Chem. Biol., 2007, 3, 60–68 CrossRef CAS.
- D. Strausak and M. Solioz, CopY is a copper-inducible repressor of the Enterococcus hirae copper ATPases, J. Biol. Chem., 1997, 272, 8932–8936 CrossRef CAS.
- K. J. Waldron, J. C. Rutherford, D. Ford and N. J. Robinson, Metalloproteins and metal sensing, Nature, 2009, 460, 823–830 CrossRef CAS.
- S. Tottey, D. R. Harvie and N. J. Robinson, Understanding how cells allocate metals using metal sensors and metallochaperones, Acc. Chem. Res., 2005, 38, 775–783 CrossRef CAS.
- J. C. Rutherford, J. S. Cavet and N. J. Robinson, Cobalt-dependent transcriptional switching by a dual-effector MerR-like protein regulates a cobalt-exporting variant CPx-type ATPase, J. Biol. Chem., 1999, 274, 25827–25832 CrossRef CAS.
- A. Z. Ansari, M. L. Chael and T. V. O'Halloran, Allosteric underwinding of DNA is a critical step in positive control of transcription by Hg–MerR, Nature, 1992, 355, 87–89 CrossRef CAS.
- J. Wu and B. P. Rosen, Metalloregulated expression of the ars operon, J. Biol. Chem., 1993, 268, 52–58 CAS.
- C. Thelwell, N. J. Robinson and J. S. Cavet, An SmtB-like repressor from Synechocystis PCC 6803 regulates a zinc exporter, Proc. Natl. Acad. Sci. U. S. A., 1998, 95, 10728–10733 CrossRef CAS.
- J. S. Iwig, J. L. Rowe and P. T. Chivers, Nickel homeostasis in Escherichia coli – the rcnR–rcnA efflux pathway and its linkage to NikR function, Mol. Microbiol., 2006, 62, 252–262 CrossRef CAS.
- A. W. Foster, C. J. Patterson, R. Pernil, C. R. Hess and N. J. Robinson, Cytosolic Ni(II) sensor in cyanobacterium: nickel detection follows nickel affinity across four families of metal sensors, J. Biol. Chem., 2012, 287, 12142–12151 CrossRef CAS.
- S. I. Patzer and K. Hantke, The ZnuABC high-affinity zinc uptake system and its regulator Zur in Escherichia coli, Mol. Microbiol., 1998, 28, 1199–1210 CrossRef CAS.
- H. B. Pakrasi, T. Ogawa and M. Bhattacharyya-Pakrasi, in Regulatory Aspects of Photosynthesis, ed. E. M Aro and B. A. Anderson, Kluer Academic, Dordrecht, 2001, pp. 253–264 Search PubMed.
- D. C. Brown and K. D. Collins, Dihydroorotase from Escherichia coli. Substitution of Co(II) for the active site Zn(II), J. Biol. Chem., 1991, 266, 1597–1604 CAS.
- S. Tottey, C. J. Patterson, L. Banci, I. Bertini, I. C. Felli, A. Pavelkova, S. J. Dainty, R. Pernil, K. J. Waldron, A. W. Foster and N. J. Robinson, Cyanobacterial metallochaperone inhibits deleterious side reactions of copper, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 95–100 CrossRef CAS.
- J. S. Iwig, S. Leitch, R. W. Herbst, M. J. Maroney and P. T. Chivers, Ni(II) and Co(II) sensing by Escherichia coli RcnR, J. Am. Chem. Soc., 2008, 130, 7592–7606 CrossRef.
- S. J. Dainty, C. J. Patterson, K. J. Waldron and N. J. Robinson, Interaction between cyanobacterial copper chaperone Atx1 and zinc homeostasis, JBIC, J. Biol. Inorg. Chem., 2010, 15, 77–85 CrossRef CAS.
- S. J. Moore and M. J. Warren, The anaerobic biosynthesis of vitamin B12, Biochem. Soc. Trans., 2012, 40, 581–586 CrossRef CAS.
- L. W. Shipman, D. Li, C. A. Roessner, A. I. Scott and J. C. Sacchettini, Crystal structure of precorrin-8× methyl mutase, Structure, 2001, 9, 587–596 CrossRef CAS.
- A. Changela, K. Chen, Y. Xue, J. Holschen, C. E. Outten, T. V. O'Halloran and A. Mondragón, Molecular basis of metal-ion selectivity and zeptomolar sensitivity by CueR, Science, 301, 1383–1387 CrossRef CAS.
- J. Parkhill, A. Z. Ansari, J. G. Wright, N. L. Brown and T. V. O'Halloran, Construction and characterization of a mercury-independent MerR activator (MerRAC): transcriptional activation in the absence of Hg(II) is accompanied by DNA distortion, EMBO J., 1993, 12, 413–421 CAS.
- C. L. Zayas, K. Claas and J. C. Escalante-Semerena, The CbiB protein of Salmonella enterica is an integral membrane protein involved in the last step of the de novo corrin ring biosynthetic pathway, J. Bacteriol., 2007, 189, 7697–7708 CrossRef CAS.
- L. A. Maggio-Hall, K. R. Claas and J. C. Escalante-Semerena, The last step in coenzyme B12 synthesis is localized to the cell membrane in bacteria and archaea, Microbiology, 2004, 150, 1385–1395 CrossRef CAS.
- A. Mata-Cabana, F. J. Florencio and M. Lindahl, Membrane proteins from the cyanobacterium Synechocystis sp. PCC 6803 interacting with thioredoxin, Proteomics, 2007, 7, 3953–3963 CrossRef CAS.
- E. Deery, S. Schroeder, A. D. Lawrence, S. L. Taylor, A. Seyedarabi, J. Waterman, K. S. Wilson, D. Brown, M. A. Geeves, M. J. Howard, R. W. Pickersgill and M. J. Warren, An enzyme-trap approach allows isolation of intermediates in cobalamin biosynthesis, Nat. Chem. Biol., 2012, 8, 933–940 CAS.
- H. K. Leech, E. Raux, K. J. McLean, A. W. Munro, N. J. Robinson, G. P. Borrelly, M. Malten, D. Jahn, S. E. Rigby, P. Heathcote and M. J. Warren, Characterisation of the cobaltochelatase CbiXL: evidence for a 4Fe–4S center housed within an MXCXXC motif, J. Biol. Chem., 2003, 278, 41900–41907 CrossRef CAS.
- D. Hoffmann, K. Gutekunst, M. Klissenbauer, R. Schulz-Friedrich and J. Appel, Mutagenesis of hydrogenase accessory genes of Synechocystis sp. PCC 6803, additional homologues of hypA and hypB are not active in hydrogenase maturation, FEBS J., 2006, 273, 4516–4527 CrossRef CAS.
- J. B. Spencer, N. J. Stolwich, C. A. Roessner and I. Scott, The Escherichia coli cysG gene encodes the multifunctional protein, siroheme synthase, FEBS J., 1993, 335, 57–60 CrossRef CAS.
- E. Raux, A. Lanois, M. J. Warren, A. Rambach and C. Thermes, Cobalamin (vitamin B12) biosynthesis: identification and characterization of a Bacillus megaterium cobI operon, Biochem. J., 1998, 335, 159–166 CAS.
- S. J. Moore and M. J. Warren, The anaerobic biosynthesis of vitamin B12, Biochem. Soc. Trans., 2012, 40, 581–586 CrossRef CAS.
- D. R. Harvie, C. Andreini, G. Cavallaro, W. Meng, B. A. Connolly, K. Yoshida, Y. Fujita, C. R. Harwood, D. S. Radford, S. Tottey, J. S. Cavet and N. J. Robinson, Predicting metals sensed by ArsR-SmtB repressors: allosteric interference by a non-effector metal, Mol. Microbiol., 2006, 59, 1341–1356 CrossRef CAS.
- J. W. Huckle, A. P. Morby, J. S. Turner and N. J. Robinson, Isolation of a prokaryotic metallothionein locus and analysis of transcriptional control by trace metal ions, Mol. Microbiol., 1993, 7, 177–187 CrossRef CAS.
- P. Emsley, B. Lohkamp, W. G. Scott and K. Cowtan, Features and development of Coot, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2010, 66, 486–501 CrossRef CAS.
- A. P. Morby, J. S. Turner, J. W. Huckle and N. J. Robinson, SmtB is a metal-dependent repressor of the cyanobacterial metallothionein gene smtA: identification of a Zn inhibited DNA-protein complex, Nucleic Acids Res., 1993, 21, 921–925 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c3mt20241k |
| This journal is © The Royal Society of Chemistry 2013 |
