The design and synthesis of 5- and 6-isoxazolylbenzimidazoles as selective inhibitors of the BET bromodomains†‡
Duncan
Hay
a,
Oleg
Fedorov
a,
Panagis
Filippakopoulos
a,
Sarah
Martin
a,
Martin
Philpott
a,
Sarah
Picaud
a,
David S.
Hewings
b,
Sagar
Uttakar
a,
Tom D.
Heightman§
a,
Stuart J.
Conway
b,
Stefan
Knapp
a and
Paul E.
Brennan
*a
aStructural Genomics Consortium, Nuffield Department of Clinical Medicine, University of Oxford, Old Road Campus Research Building, Roosevelt Drive, Oxford OX3 7DQ, UK. E-mail: paul.brennan@sgc.ox.ac.uk
bDepartment of Chemistry, Chemistry Research Laboratory, University of Oxford, Mansfield Road, Oxford, OX1 3TA, UK
First published on 13th August 2012
Abstract
Simple 1-substituted 5- and 6-isoxazolyl-benzimidazoles have been shown to be potent inhibitors of the BET bromodomains with selectivity over the related bromodomain of CBP. The reported inhibitors were prepared from simple starting materials in two steps followed by separation of the regioisomers or regioselectively in three steps.
Bromodomains are discrete protein domains that selectively recognize acetyl lysine in proteins.1 There are 61 bromodomains in proteins that have a variety of functions including histone acetyl transferases such as CBP (cyclic AMP response element-binding protein, binding protein), methyl transferases, transcriptional regulators such as BRD4 (bromodomain-containing protein 4) and chromatin remodelling complexes.2 The BET family of bromodomain containing proteins is comprised of BRDT, BRD2, BRD3 and BRD4 each of which has two bromodomains that bind to acetylated histone tails.3 Recently BET inhibitors have been shown to have potential for use in inflammatory disease, atherosclerosis, NUT midline carcinoma, acute leukaemia and lymphoma.4–9
Triazoloazepines such as (+)-JQ1 1, iBET762 (structure not shown) and isoxazoles such as compound 2 have been identified as potent BET inhibitors (Fig. 1).4,9,10 Since then, a number of other templates incorporating the privileged isoxazole moiety such as in compounds 3 and 4 have been identified by researchers in the EpiNova group at GlaxoSmithKline.6,11,12 As most known BET inhibitors are complex stereogenic molecules it would be advantageous to find simple, rapidly accessible inhibitors that would be selective for the BET family as exemplified by BRD4 over proteins containing similar bromodomains, such as CBP.
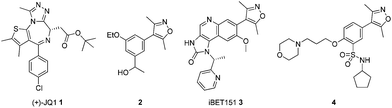 | ||
| Fig. 1 BET bromodomain inhibitors. | ||
It was thought that fusing a 5-membered ring to the 4-aryl-3,5-dimethylisoxazole moiety of compound 2 (ref. 10) would give access to previously unexploited substitution patterns in known isoxazole-containing bromodomain inhibitors. Simple 5,6-bicyclic bromides 5a–c were transformed into isoxazoles by either direct arylation of 3,5-dimethylisoxazole or Suzuki reaction of the isoxazolylboronic acid to give compounds 6–8 (Scheme 1).10,13 When tested in an AlphaScreen® assay using isolated bromodomains, compounds 6 and 7 were modest inhibitors of the first bromodomain of BRD4 (BRD4(1)) with no affinity for the CBP bromodomain whereas compound 8 had comparable affinity for both bromodomains (Table 1).14
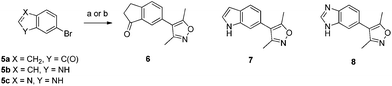 | ||
| Scheme 1 Synthesis of indanone, indole and benzimidazole containing inhibitors. Reagents and conditions: (a) 5a, 3,5-dimethylisoxazole, PdCl2, KOAc, N,N-dimethylacetamide, 130 °C, 50%; (b) 3,5-dimethylisoxazolylboronic acid, Pd(PPh3)4, Na2CO3, DMF, H2O, 140 °C μwave, 5b (12% yield) or 5c (13% yield). | ||
The indanone 6 presented an attractive intermediate for further derivitization (Scheme 2). Reduction to the racemic indanol followed by alkylation with benzyl bromide or 2-bromomethyl quinolone gave compounds 9 and 10. The amines 10–14 were prepared by SN1 alkylation of the indanol with 3-bromo-n-propanol followed by bromide substitution. The basic centres of varying pKa in compounds 10–14 were designed with the potential to interact with an acidic residue on the edge of the BRD4(1) binding pocket, D145.
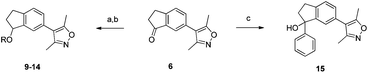 | ||
| Scheme 2 Derivitization of indanone 6. Reagents and conditions: (a) NaBH4, EtOH, 90%; (b) ArCH2Br, NaH, 76–81% or (i) Br(CH2)3OH, pTsOH, CH2Cl2, reflux, 45%; (ii) 2° amine, TEA, THF, reflux, 22–100%; (c) Mg, PhBr, Et2O, 60%. | ||
Addition of an O-benzyl group in compound 9 did not increase the affinity for either BRD4(1) or CBP bromodomains compared to the indanone 6 (Table 2). In this series only the piperazine derived compound 14 with the piperazine group was more potent than compound 9. A large increase in BRD4(1) affinity, without a corresponding increase in CBP affinity, was obtained when the indanone 6 was reacted with phenylmagnesium bromide to give the racemic tertiary indanol 15 (Scheme 2).
The increase in affinity of compound 15 for BRD4(1) was explained when it was co-crystallized with this bromodomain (Fig. 2). As expected from previous X-ray structures of BRD4(1)-bound isoxazole ligands,6,10 the isoxazole oxygen atom formed one hydrogen bond to the conserved asparagine (N140) and the nitrogen atom formed a water-mediated hydrogen bond to Y97. The aryl ring occupied the hydrophobic groove of the WPF shelf, defined by a W81, M149 and I146. Interaction with these three hydrophobic residues is important for the affinity of all known BET inhibitors (Fig. 1).
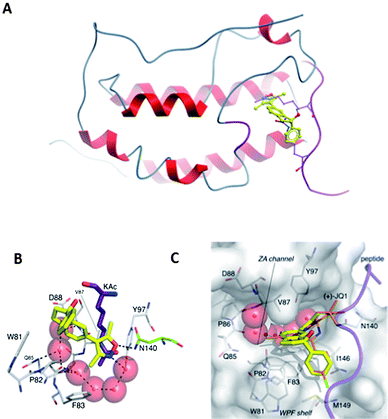 | ||
| Fig. 2 Compound 15 bound to BRD4(1). (A) Overlay of compound 15 (yellow, PDB ID: 4GPJ) with H4K5AcK8Ac (purple, PDB ID: 3UVW);1 (B) residues and conserved waters in BRD4(1) binding to compound 15; (C) surface view of BRD4(1) in the protein–ligand complex overlaid with (+)-JQ1 (orange, PDB ID: 3MXF). | ||
Encouraged by the excellent potency, selectivity and simple synthesis of indanol 15, further effort was made to optimize the compound. A range of substituted aryl Grignard reagents were reacted with the ketone, but yields were generally low and the products appeared to decompose during or following isolation. Re-examination of stock solutions of the first indanol 15 by LCMS showed decomposition of the parent compound and a mass loss of 18 amu from the molecular ion peak. It is likely that the doubly benzylic tertiary alcohols dehydrate to give the indenes 16 (Scheme 3).15
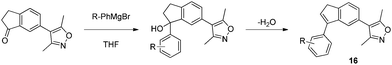 | ||
| Scheme 3 Decomposition of aryl indanols. | ||
In an effort to prepare compounds that were as potent as 15 but without the chemical instability problem, attention returned to the benzimidazole 8. To mimic the potency enhancement of 15 compared to 6, an aryl group was added to N1 of the benzimidazole (Scheme 4). Ullman coupling with phenyl iodide gave a mixture of arylated benzimidazoles 17a and 17b that could be separated although the identity of the regioisomers could not be confirmed. The benzyl substituted compounds 18 were prepared in a similar manner by alkylation of N1 with benzyl bromide but could not be separated. Isoxazole coupling furnished the first substituted benzimidazoles 19a,b and 20. The mixture of regioisomers 20 was tested against both BRD4(1) and CBP and was shown to be at least three-fold more potent than the either of the directly arylated compounds 19a or 19b.
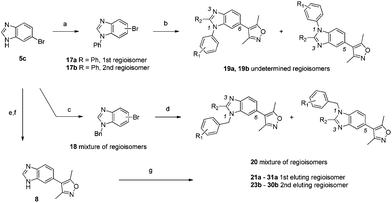 | ||
| Scheme 4 Synthesis of substituted benzimidazoles. Reagents and conditions: (a) PhI, KOH, TBAB, CuI, 110 °C 44% 17a and 22% 17b; (b) 3,5-dimethylisoxazolylboronic acid, Pd(dppf)Cl2, NaHCO3, DME, 120 °C, 50% (19a) or 26% (19b); (c) BnBr, K2CO3, MeCN, reflux, 52%; (d) 3,5-dimethylisoxazole, PdCl2, KOAc, N,N-dimethyl-acetamide, 120 °C, 52% (20); (e) Boc2O, TEA, CH2Cl2, 100%; (f) 3,5-dimethylisoxazolylboronic acid, Pd(PPh3)4, Na2CO3, 1,4-dioxane, reflux, 48%; (g) RBr, K2CO3, MeCN, reflux, 18–74%. | ||
Analogues of 20 were prepared using the non-selective alkylation chemistry (21a–31a, 23b–30b). By coupling the isoxazole group first, diversification was possible in the last step. The low yield of compound 8 in Scheme 1 was improved by Boc-protection of the benzimidazole followed by Suzuki reaction and in situ deprotection. In the case of the substituted benzyl compounds separation was achieved by silica gel chromatography. Comparison of the 1H-NMRs of the first and second eluting regioisomers showed consistent chemical shift changes between the two isomers. This observation confirmed that if the absolute identity of a single regioisomeric pair could be determined, the structures of the others could be correlated by elution order from silica gel.16
A regioselective route to both N-benzylbenzimidazoles was developed in order to determine the identity of the regioisomers and prepare additional analogues.17 Substitution of 2,4- and 2,5-dibromonitrobenzene 36a and 36b with benzyl amines was followed by reduction of the nitro group and cyclization to the benzimidazoles 37a and 37b with formic acid. Coupling of the 3,5-dimethylisoxazole group completed the synthesis. Compounds 23a, and 23b were synthesized by both non-selective and regioselective routes in order to correlate the elution order with the identity of the regioisomers.18
Table 3 summarizes the affinity data for the target compounds against BRD4(1) and CBP. In general all compounds were more potent against BRD4(1) than CBP. The 6-isoxazolyl-benzimidazoles were more potent against BRD4(1) than the 5-substituted (see compounds 23a,b, 24a,b, 27a,b and 30a,b). This observation is consistent with an expected binding mode where the benzyl group fills the same pocket as the phenyl group of compound 15. The opposite is true of CBP potency, where the 5-substitued regioisomers were more potent. The nature of the benzyl substituent did not have a dramatic effect on the potency with both polar compounds (25a, 27a, 28a, 31a–33a) and non-polar compounds (21a–24a, 26a, 30a) showing similar potency. A methyl group was incorporated at the 2-position of the benzimidazole but it had little effect on either BRD4(1) or CBP potency as shown by comparing compounds 25a and 35a.
| Cpd | R1 | pIC50a | |||
|---|---|---|---|---|---|
| R2 | BRD4(1) | CBP | Routec | ||
| a Mean pIC50 ± standard error of the mean (number of determinations). b The regioisomeric identity of compounds 19a and 19b could not be confirmed. c Synthetic route sep: separation of regioisomers as per Scheme 4; reg: regioselective synthesis as per Scheme 5. | |||||
| 19a | H | H | 5.0 ± 0.3 (3) | 5.4 ± 0.1 (2) | NA |
| 19b | H | H | 5.5 ± 0.2 (2) | <4.6 (2) | NA |
| 20 | H | H | 6.0 ± 0.1 (2) | ND | NA |
| 21a | 3-F | H | 6.2 ± 0.1 (2) | 4.9 ± 0.3 (2) | sep |
| 22a | 3-Cl | H | 5.9 ± 0.1 (2) | 4.7 ± 0.4 (2) | sep |
| 23a | 4-Cl | H | 6.1 ± 0.1 (9) | <4.7 (2) | sep/reg |
| 23b | 4-Cl | H | 5.6 ± 0.1 (2) | 5.7 ± 0.2 (2) | sep/reg |
| 24a | 3-Br | H | 6.0 ± 0.1 (3) | <5.0 (2) | sep |
| 24b | 3-Br | H | 5.8 ± 0.4 (2) | 5.6 ± 0.3 (3) | sep |
| 25a | 4-MeO | H | 6.1 ± 0.3 (2) | <5.0 (2) | sep |
| 26a | 2-Cl | H | 6.1 ± 0.5 (2) | <5.0 (2) | sep |
| 27a | 2-NO2 | H | 6.2 ± 0.4 (2) | <4.0 (2) | sep |
| 27b | 2-NO2 | H | 4.9 ± 1.0 (2) | 4.8 ± 0.9 (3) | sep |
| 28a | 4-CN | H | 6.7 ± 0.3 (4) | <4.0 (2) | sep |
| 29a | 2-CN | H | 6.0 ± 0.6 (2) | 5.9 ± 0.5 (2) | sep |
| 30a | 3,4-Cl2 | H | 5.4 ± 0.2 (2) | <4.0 (2) | reg |
| 30b | 3,4-Cl2 | H | 5.0 ± 0.2 (2) | 5.9 ± 0.0 (2) | reg |
| 31a | 4-CO2Me | H | 5.9 ± 0.0 (2) | <4.0 (2) | sep |
| 32a | 3-CON(Me)2 | H | 6.2 ± 0.3 (3) | 5.2 ± 0.2 (2) | reg |
| 33a | 4-CON(Me)2 | H | 5.7 ± 0.1 (2) | 5.2 ± 0.2 (2) | reg |
| 34b | 4-OH | Me | 5.7 ± 0.1 (2) | 5.1 ± 0.7 (2) | reg |
| 35a | 4-MeO | Me | 5.8 ± 0.2 (2) | 4.6 ± 0.3 (2) | reg |
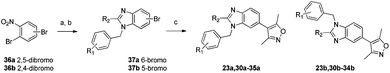 | ||
| Scheme 5 Regioselective synthesis of benzimidazoles. Reagents and conditions: (a) R1PhCH2NH2, Na2CO3, EtOH, reflux 18–75%; (b) Fe, HCO2H or (MeO)3CMe, 33–76%; (c) 3,5-dimethylisoxazolylboronic acid, Pd(PPh3)4, Na2CO3, 1,4-dioxane, reflux, 22–43%. | ||
The most potent inhibitor 28a was further screened against a panel of diverse bromodomains using a differential scanning fluorimetry (DSF) assay (Table 4).14 Compound 28a showed no stabilization of seven other bromodomains and only minimal stabilization of CBP, confirming its selectivity for BRD4(1).
Conclusions
Simple 3,5-dimethylisoxazole substituted benzimidazoles are potent and selective inhibitors of the first bromodomain of BRD4 over the bromodomain of CREB-binding protein. The most potent compound, 28a, has a BRD4(1) pIC50 of 6.7 (IC50 = 180 nM) and is at least 100-fold selective over CBP. The addition of the bicyclic benzimidazole ring system to the previously reported phenylisoxazole (compound 2) has improved both potency for BRD4(1) and selectivity over CBP without loss of selectivity over other bromodomains. With a simple two-step synthesis and regioisomer separation, or three-step regioselective synthesis and multiple positions for modification, this is an attractive template for bromodomain lead discovery projects.Acknowledgements
The SGC is a registered charity (number 1097737) that receives funds from the Canadian Institutes for Health Research, Genome Canada, GlaxoSmithKline, Lilly Canada, the Novartis Research Foundation, Pfizer, Takeda, AbbVie, the Canada Foundation for Innovation, the Ontario Ministry of Economic Development and Innovation, and the Wellcome Trust [092809/Z/10/Z]. The authors would like to thank Yue Zhu at Changchun Discovery Sciences, Ltd.Notes and references
- P. Filippakopoulos, S. Picaud, M. Mangos, T. Keates, J.-P. Lambert, D. Barsyte-Lovejoy, I. Felletar, R. Volkmer, S. Müller, T. Pawson, A.-C. Gingras, C. H. Arrowsmith and S. Knapp, Cell, 2012, 149, 214–231 CrossRef CAS.
- S. Muller, P. Filippakopoulos and S. Knapp, Expert Rev. Mol. Med., 2011, 13, e29 CrossRef.
- P. Filippakopoulos and S. Knapp, FEBS Lett., 2012, 586, 2692–2704 CrossRef CAS.
- E. Nicodeme, K. L. Jeffrey, U. Schaefer, S. Beinke, S. Dewell, C.-W. Chung, R. Chandwani, I. Marazzi, P. Wilson, H. Coste, J. White, J. Kirilovsky, C. M. Rice, J. M. Lora, R. K. Prinjha, K. Lee and A. Tarakhovsky, Nature, 2010, 468, 1119–1123 CrossRef CAS.
- J. Zuber, J. Shi, E. Wang, A. R. Rappaport, H. Herrmann, E. A. Sison, D. Magoon, J. Qi, K. Blatt, M. Wunderlich, M. J. Taylor, C. Johns, A. Chicas, J. C. Mulloy, S. C. Kogan, P. Brown, P. Valent, J. E. Bradner, S. W. Lowe and C. R. Vakoc, Nature, 2011, 478, 524–528 CrossRef CAS.
- M. A. Dawson, R. K. Prinjha, A. Dittmann, G. Giotopoulos, M. Bantscheff, W.-I. Chan, S. C. Robson, C.-W. Chung, C. Hopf, M. M. Savitski, C. Huthmacher, E. Gudgin, D. Lugo, S. Beinke, T. D. Chapman, E. J. Roberts, P. E. Soden, K. R. Auger, O. Mirguet, K. Doehner, R. Delwel, A. K. Burnett, P. Jeffrey, G. Drewes, K. Lee, B. J. P. Huntly and T. Kouzarides, Nature, 2011, 478, 529–533 CrossRef CAS.
- O. Mirguet, Y. Lamotte, F. Donche, J. Toum, F. Gellibert, A. Bouillot, R. Gosmini, V.-L. Nguyen, D. Delannée, J. Seal, F. Blandel, A.-B. Boullay, E. Boursier, S. Martin, J.-M. Brusq, G. Krysa, A. Riou, R. Tellier, A. Costaz, P. Huet, Y. Dudit, L. Trottet, J. Kirilovsky and E. Nicodeme, Bioorg. Med. Chem. Lett., 2012, 22, 2963–2967 CrossRef CAS.
- J. E. Delmore, G. C. Issa, M. E. Lemieux, P. B. Rahl, J. Shi, H. M. Jacobs, E. Kastritis, T. Gilpatrick, R. M. Paranal, J. Qi, M. Chesi, A. C. Schinzel, M. R. McKeown, T. P. Heffernan, C. R. Vakoc, P. L. Bergsagel, I. M. Ghobrial, P. G. Richardson, R. A. Young, W. C. Hahn, K. C. Anderson, A. L. Kung, J. E. Bradner and C. S. Mitsiades, Cell, 2011, 146, 904–917 CrossRef CAS.
- P. Filippakopoulos, J. Qi, S. Picaud, Y. Shen, W. B. Smith, O. Fedorov, E. M. Morse, T. Keates, T. T. Hickman, I. Felletar, M. Philpott, S. Munro, M. R. McKeown, Y. Wang, A. L. Christie, N. West, M. J. Cameron, B. Schwartz, T. D. Heightman, N. La Thangue, C. A. French, O. Wiest, A. L. Kung, S. Knapp and J. E. Bradner, Nature, 2010, 468, 1067–1073 CrossRef CAS.
- D. S. Hewings, M. Wang, M. Philpott, O. Fedorov, S. Uttarkar, P. Filippakopoulos, S. Picaud, C. Vuppusetty, B. Marsden, S. Knapp, S. J. Conway and T. D. Heightman, J. Med. Chem., 2011, 54, 6761–6770 CrossRef CAS.
- C.-W. Chung, A. W. Dean, J. M. Woolven and P. Bamborough, J. Med. Chem., 2011, 55, 576–586 CrossRef.
- P. Bamborough, H. Diallo, J. D. Goodacre, L. Gordon, A. Lewis, J. T. Seal, D. M. Wilson, M. D. Woodrow and C.-W. Chung, J. Med. Chem., 2011, 55, 587–596 CrossRef.
- Y. Fall, C. Reynaud, H. Doucet and M. Santelli, Eur. J. Org. Chem., 2009, 4041–4050 CrossRef CAS.
- M. Philpott, J. Yang, T. Tumber, O. Fedorov, S. Uttarkar, P. Filippakopoulos, S. Picaud, T. Keates, I. Felletar, A. Ciulli, S. Knapp and T. D. Heightman, Mol. BioSyst., 2011, 7, 2899–2908 RSC.
- Although the identity of compound 15 as the active compound in the AlphaScreen® assay conditions could not be unequivically confirmed, the presence of the compound in the co-crystal structure with BRD4(1) is evidence for its role as the pharmacologically active component.
- 1H-NMR resonance differences of the two isoxazole methyl groups and the benzylic methylene were consistently shifted in all pairs of regioisomers and were used to make compound assignments.
- E. H. Sessions, M. Smolinski, B. Wang, B. Frackowiak, S. Chowdhury, Y. Yin, Y. T. Chen, C. Ruiz, L. Lin, J. Pocas, T. Schröter, M. D. Cameron, P. LoGrasso, Y. Feng and T. D. Bannister, Bioorg. Med. Chem. Lett., 2010, 20, 1939–1943 CrossRef CAS.
- Final compounds were purified by flash chromatography (25
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 CH2Cl2/EtOAc). Correlating the identity of compounds 23a, 23b, 30a and 30b made via the separation and regioselective routes showed that the 6-isoxazolyl-substituted compounds 23a and 30a eluted before 5-isoxazolyl-substituted compounds 23b and 30b.
1 CH2Cl2/EtOAc). Correlating the identity of compounds 23a, 23b, 30a and 30b made via the separation and regioselective routes showed that the 6-isoxazolyl-substituted compounds 23a and 30a eluted before 5-isoxazolyl-substituted compounds 23b and 30b.
Footnotes |
| † This article is part of a MedChemComm ‘New Talents’ issue highlighting the work of outstanding rising scientists in medicinal chemistry research. |
| ‡ Electronic supplementary information (ESI) available: Synthetic details for selected compounds and crystallographic data for compound 15. See DOI: 10.1039/c2md20189e |
| § Current address: Astex Pharmaceuticals, 436 Cambridge Science Park, Cambridge CB4 0QA, UK, E-mail: tom.heightman@astx.com. |
| This journal is © The Royal Society of Chemistry 2013 |