A mast cell degranulation screening assay for the identification of novel mast cell activating agents†
Herman F.
Staats
*abc,
Shaun M.
Kirwan
a,
Hae Woong
Choi
a,
Christopher P.
Shelburne
a,
Soman N.
Abraham
abde,
Gulice Y. C.
Leung
f and
David Y.-K.
Chen
*g
aDepartment of Pathology, Duke University Medical Center, Box 3712, Durham, NC 27710, USA. E-mail: herman.staats@duke.edu
bDepartment of Immunology, Duke University Medical Center, Durham, NC 27710, USA
cDepartment of Medicine, Duke University Medical Center, Durham, NC 27710, USA
dDepartment of Molecular Genetics & Microbiology, Duke University Medical Center, Durham, NC 27710, USA
eProgram in Emerging Infectious Diseases, Duke-National University of Singapore Graduate Medical School, Singapore 169547
fOrganic Chemistry @ Helios, Institute of Chemical and Engineering Sciences (ICES), Agency for Science, Technology and Research (A*STAR), 11 Biopolis Way, The Helios Block, #03-08, Singapore 138667
gDepartment of Chemistry, Seoul National University, Gwanak-1 Gwanak-ro, Gwanak-gu, Seoul 151-742, South Korea. E-mail: davidchen@snu.ac.kr
First published on 21st June 2012
Abstract
The development and use of vaccines and their ability to prevent infection/disease is a shining example of the benefit of biomedical research. Modern vaccines often utilize subunit immunogens that exhibit minimal immunogenicity and require the use of adjuvants to maximize the induction of protective immune responses. We recently described a novel class of vaccine adjuvants, mast cell (MC) activators, that exhibit safe and effective vaccine adjuvant activity when administered by intranasal or intradermal routes. A compound library containing 580 functionalized benzopyrans, a structural motif found in a diverse array of natural and designed bioactive compounds, was screened using a MC degranulation assay to identify novel MC activating compounds for future evaluation as novel vaccine adjuvants. This approach identified 12 novel MC degranulating compounds. Therefore, MC degranulation can be used to reliably detect novel compounds for evaluation as adjuvants for use in mucosal vaccine strategies.
Introduction
The emergence of new infectious agents, as well as current biological threats, has created a great need for the rapid development of safe and efficacious vaccines. However, current strategies to create such vaccines are unable to meet the need for broad prophylactic immune protection, especially along mucosal portals such as the lung or gastrointestinal tract used by pathogens to gain access to deeper tissues. A defect in these strategies is a lack of effective adjuvant compounds, the immuno-stimulatory component of vaccines that are able to evoke protective immune responses along mucosal sites. For instance, alum, one of the few licensed adjuvants for human use, is only effective in the induction of systemic immune responses.1–7 Therefore, there is a need to identify new mucosal adjuvant compounds that are effective, safe and economical for future vaccine design.Recently, several studies have elucidated a role for mast cells (MCs), which are granulated tissue resident leukocytes in the development of adaptive immunity. MC release of pro-inflammatory mediators such as TNF or IL-1 was found to affect the development of both humoral and cellular components of immunity in mice after challenge with diverse stimuli including bacteria, the TLR7 ligand imiquimod or contact sensitizers such as FITC.8–10 Moreover, targeted activation of MCs with the small molecule MC activator compound 48/80 (c48/80) was found to induce safe and robust systemic and mucosal immune responses against various protein antigens when co-administered with vaccine antigens at mucosal surfaces.11,12 In all cases, MCs were suggested to affect adaptive immunity by enhancing the recruitment of dendritic cells (DCs) and lymphocytes into local lymphoid tissues, a prerequisite for optimal immune responses. These data suggest that MCs may be a common denominator in the modulation of immunity to many diverse agents and molecules capable of activating MCs are potentially highly effective adjuvants. Therefore, MCs might be used as sensors to screen chemical libraries to detect novel compounds with mucosal adjuvant activity.
In this study, we describe the development of a novel screen using the MC degranulation response as a sensitive monitor to detect potential mucosal adjuvant compounds. We report that this methodology successfully detected 12 novel MC degranulating compounds from a library containing 580 functionalized benzopyrans, a privileged structural motif found in a diverse array of natural and designed bioactive compounds.13–18 Therefore, MC degranulation can be used to reliably detect novel compounds which may greatly expand our current lexicon of potential adjuvant compounds for use in mucosal vaccine strategies.
Results
Development of an in vitro mucosal adjuvant screen based on MC activation
A scheme for the use of readily available MC lines to screen for novel mucosal adjuvant compounds is outlined in Fig. 1. For this screening assay, we selected the mouse mast cell line MC/9.19–24 The MC/9 mouse mast cell line expresses IgE Fc receptors and degranulates and produces cytokines in response to IgE–antigen stimulation.25 The MC/9 mouse mast cell line has been described as a cell line representative of a “mucosal mast cell”.22,25 The MC/9 cell line was specifically selected for our screening assay because the cells are not readily activated by agonists and therefore would minimize false-positive reactions. Indeed, others have reported that stimulation of MC/9 cells for 15 minutes with 0.5 μg ml−1 of compound 48/80 induced minimal degranulation of MC/9 cells based on β-hexosaminidase release (1.5% of total) and tryptase (1.2% of total) while primary peritoneal mast cells released 16% of total β-hexosaminidase and 3.5% of total tryptase when stimulated with 0.5 μg ml−1 compound 48/80.26,27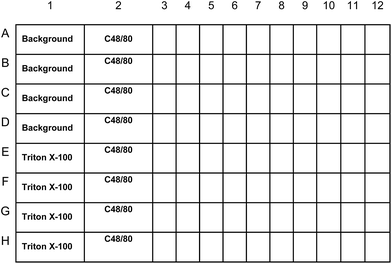 | ||
| Fig. 1 MC screening assay plate layout. Internal plate controls will include MC/9 cells treated with Tyrode's buffer alone (background), a maximum value control treated with the detergent Triton-X 100 (Triton-X 100), and MC/9 cells treated with the known MC activator c48/80. Each compound was evaluated in duplicate. Percent (%) degranulation will be calculated using the following equation: percent (%) degranulation = (experimental β-hex release − vehicle control β-hex release)/(Triton-X-100 β-hex release − vehicle control β-hex release) × 100. Compounds that elicit 5% degranulation of MC/9 cells will be selected for further evaluation. | ||
In this scheme, chemical compounds are mixed with the murine MC/9 MC line,19–24 and evaluated for their ability to induce the release of MC granule contents, a process called degranulation. Degranulation, a reliable marker of MC activation, is assayed by examining cell culture supernatants for the presence of β-hexosaminidase (β-hex), an enzyme located in MC granules. Release of this enzyme implies that MCs have released their granule components, whereas its absence suggests that the chemical compounds do not have the ability to activate MCs. To perform this assay, we modified a procedure described by others.26,28 MC/9 cells were aliquoted in sterile Tyrode's buffer and added to each well of a sterile V-bottom plate at 5 × 105 cells per well in a 100 μl volume. For each 96-well plate, internal plate controls included MCs treated with the positive control degranulating agent compound 48/80 (c48/80; doses of 49.6, 198.3 and 793.5 μM corresponding to 31.25, 125 and 500 μg ml−1). C48/80 is a cationic mast cell stimulating chemical compound first reported for its ability to induce histamine secretion.29 Compound 48/80 appears to activate mast cells by a receptor-independent mechanism that activates G-proteins required for mast cell exocytosis.30–35 Other studies, however, suggest that Mas-related gene (Mrg) receptors on mast cells are involved in the mast cell activating activity of compound 48/80.36 Additional assay controls included a vehicle control (DMSO) to evaluate background β-hex release and a maximum β-hex release control, which is cells treated with the detergent Triton X-100. Potential adjuvant compounds are added to each well in 100 μl volume at a final concentration of 100 μM.
Quality control results for the MC activating screen
An important component of this assay is the development of quality control criteria ensuring the reproducibility of the assay between multiple plates. Such reproducibility is required to establish a rank summary of the chemical compounds and their ability to induce β-hex release from MC/9 cells. These quality control criteria were established through iterative testing and determined to be: (1) baseline β-hex activity in vehicle control wells (spontaneous release measurements) should be consistent and below 0.2; (2) maximum β-hex activity in Triton X-100 wells (or maximum release measurements) should be consistent and above 1.0; and (3) the calculated β-hex release from c48/80 treated samples should be consistent between assays with results varying less than 3 standard deviations from the mean. Results from plates that do not meet these quality control criteria are discarded and repeated. Using these quality control criteria, we evaluated MC activating capabilities of 580 functionalized benzopyrans which were prepared in accordance to the protocols developed by Nicolaou and co-workers.13–15 More specifically, this “benzopyran library” shared a common 2,2-dimethylbenzopyran core structure, which was synthesized through a Claisen rearrangement of hydroxy benzaldehyde derived propargyl ethers followed by further functionalization. In this manner, a diverse array of haloaryls, ethers, esters, amides, sulfonates and heterocycles could be readily accessed with high chemical yield and purity (Fig. 2). | ||
| Fig. 2 General synthetic scheme for the construction of the benzopyran library. | ||
The compounds were examined for their ability to induce β-hex release from MC/9 cells in a total of 17 plates. As shown in Fig. 3, our quality control results from these plates generated consistent minimal background β-hex release (average mean = 0.096 ± 0.02) (Fig. 3A) and maximal β-hex release measurements (average mean = 2.26 ± 0.50) (Fig. 3A) which are well within the quality control limits. Moreover, we found that treatment of MC/9 cells with the positive control mast cell degranulator c48/80 generated consistent results within three standard deviations from the mean of all assays (Fig. 3B). Collectively, these data demonstrate the reproducibility of the assay and that a result from any given plate should be comparable with results on a separate plate.
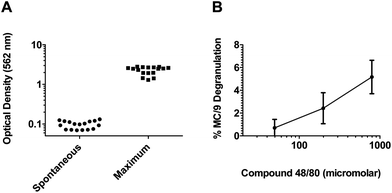 | ||
| Fig. 3 The MC activation screen for novel adjuvants is a consistent high throughput assay. (A) Background β-hex release values for all 17 plates evaluated in this assay. Note that all values fall below the quality control criteria of 0.2. Maximal β-hex release values from MC/9 cells treated with the detergent Triton-X 100 from all 17 plates evaluated in the assay. Note that all values are greater than the quality control criteria of 1.0. (B) Average percent β-hex release from MC/9 cells treated with the MC activator c48/80 (doses of 49.6, 198.3 and 793.5 μM corresponding to 31.25, 125 and 500 μg ml−1) from all 17 plates evaluated in the assay. The individual values represent the calculated % degranulation from each assay plate and demonstrate that the assay performed reproducibly since the calculated % degranulation for each plate was within 3 standard deviations of the mean for each concentration of c48/80 tested. | ||
Detection of novel adjuvant compounds using the MC activating screen
Using these quality control criteria, we found that our screen was able to detect 12 compounds (2%) of the total chemical library that were able to induce at least 5% release of β-hex from MC/9 cells. The results from the top 50 compounds are listed in Table 1. Of these, only compound number 10 induced greater than 10% β-hex release from MC/9 cells, while the rest induced β-hex release from MC/9 cells in a range between 5.5 and 8.4%. Intriguingly, structure–activity relationship analysis of these compounds revealed three notable chemical classes that induce MC degranulating capability (Fig. 4).| Rank | Compound number | % Degranulation | Rank | Compound number | % Degranulation |
|---|---|---|---|---|---|
| 1 | 10 | 10.1 | 26 | 117 | 3.7 |
| 2 | 293 | 8.4 | 27 | 471 | 3.6 |
| 3 | 360 | 7.8 | 28 | 428 | 3.3 |
| 4 | 272 | 7.4 | 29 | 327 | 3.2 |
| 5 | 43 | 7.4 | 30 | 511 | 3.2 |
| 6 | 90 | 7.3 | 31 | 260 | 3.1 |
| 7 | 254 | 6.7 | 32 | 177 | 3.1 |
| 8 | 309 | 6.5 | 33 | 280 | 3.1 |
| 9 | 479 | 6.4 | 34 | 348 | 3.0 |
| 10 | 290 | 5.7 | 35 | 289 | 3.0 |
| 11 | 379 | 5.6 | 36 | 16 | 3.0 |
| 12 | 37 | 5.5 | 37 | 302 | 2.9 |
| 13 | 133 | 4.9 | 38 | 242 | 2.9 |
| 14 | 433 | 4.9 | 39 | 459 | 2.9 |
| 15 | 278 | 4.7 | 40 | 319 | 2.8 |
| 16 | 376 | 4.6 | 41 | 358 | 2.8 |
| 17 | 328 | 4.3 | 42 | 369 | 2.8 |
| 18 | 340 | 4.2 | 43 | 95 | 2.7 |
| 19 | 38 | 4.2 | 44 | 324 | 2.7 |
| 20 | 427 | 4.0 | 45 | 337 | 2.7 |
| 21 | 113 | 3.9 | 46 | 291 | 2.7 |
| 22 | 380 | 3.8 | 47 | 274 | 2.6 |
| 23 | 356 | 3.8 | 48 | 89 | 2.6 |
| 24 | 11 | 3.8 | 49 | 152 | 2.6 |
| 25 | 326 | 3.7 | 50 | 245 | 2.6 |
 | ||
| Fig. 4 Notable structural classes of the benzopyran library found to elicit degranulation of MC/9 cells. | ||
Structural analysis of the benzopyran library in correlation to MC activation
Recognizing that the bezopyrans are privileged structures reported to elicit a wide spectrum of biological responses, the structural diversity represented by the benzopyran library described herein was a crucial selection criterion at the outset of this study to maximize the likelihood of identifying a MC activator. Indeed, careful assessment of the MC activation screen revealed benzylamine, cyanostilbene and chalcone as the three structural classes of interest and which warrants further discussion and future investigation (Fig. 4). Furthermore, a preliminary structure–activity relationship could be established within each structural class (Fig. 5–7), which proved to be path-pointing for further chemical modifications of lead candidates to improve their MC activation capabilities.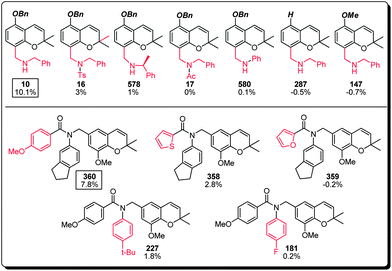 | ||
| Fig. 5 Structure–activity relationship of the benzylamine subset of the benzopyran library (compound identification number over % MC/9 degranulation). | ||
(a) The benzylamines represent the major sub-group within the top 50 MC activating compounds (34%, 17 compounds) with compound 10 and compound 360 the most active members of this family. Further structural analysis of compound 10 suggested the necessity of its benzyl ether and secondary benzylamine bearing an unsubstituted methylene linker, since compounds 287, 147, and 16, 17, 578 and 580 all showed significant loss of MC activation capability. For lead compound 360, both the para-methoxy benzamide (cf. compounds 358 and 359) and the indanyl substitutions (cf. compounds 181 and 227) appear to be critical, where the carefully balanced steric environment is likely to be responsible for the observed MC activation capability (Fig. 5).
(b) The cyanostilbenes represent 14% (7 compounds) of the top 50 MC activating compounds with compound 272 being the most active member of this family (Fig. 6). Structure–activity analysis indicated that the regiochemistry of the cyanostilbene substitution was crucial for the observed activity (cf. compound 349), together with marginal tolerability in the substitution of the two aromatic systems (cf. compounds 7, 32, 36 and 37).
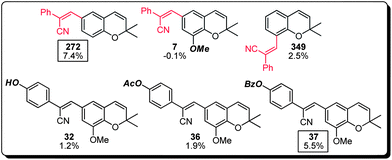 | ||
| Fig. 6 Structure–activity relationship of the cyanostilbene subset of the benzopyran library (compound identification number over % MC/9 degranulation). | ||
(c) Lastly, the chalcone family of compounds within the benzopyran library also displayed a MC activation property that warrants additional attention (Fig. 7). This class appeared to demonstrate a wider structure–activity window, in which compounds 293, 309, 479 and 290 all exhibited comparable potencies. Substitution at the non-benzopyran bearing aromatic ring appears to be an essential element for the observed activity (cf. compound 2), and a variety of substituents including but not limited to halogen, hydroxyl, methoxy and methylmercaptan groups (cf. compounds 293, 309, 290 and 289) were well tolerated. Interestingly, the chalcone olefin could be replaced with an analogous conformation restricting pyrazole functionality with good retention of MC activation capability (cf. compound 479), suggesting that suitable functional handles at this location could be used to further improve the potency and pharmacological properties of this family of compounds.
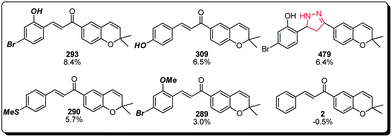 | ||
| Fig. 7 Structure–activity relationship of the chalcone subset of the benzopyran library (compound identification number over % MC/9 degranulation). | ||
Finally, the major MC activating compounds shortlisted and discussed herein showed little cross-structural resemblance. Although we have demonstrated the utility of the benzopyran library in the bioassay-guided identification of bioactives, it was not possible to formulate a unified structure–activity relationship proposal. Nevertheless, the structural diversity exhibited by benzylamine, cyanostilbene and chalcone presents new avenues for further structural modifications and biological investigations.
Discussion
The development of new vaccines has long centered on the development of vaccine antigens that would appropriately target the pathogen of interest. However, with the development of vaccine regimens that use peptides or purified proteins as antigens has come the realization that new adjuvant compounds, the immuno-stimulatory components of vaccines, must also be developed to fully realize the potency of different vaccines. Currently, many potential vaccines are limited by the dearth of potent adjuvants, both in terms of effectiveness and in the induction of protective antibody responses along mucosal surfaces. For instance, vaccine regimens against the hepatitis B virus do not confer effective immuno-protection in all adults. This vaccine and many others use alum, one of only two approved adjuvants for use in the United States (the other is ASO4, a combination of the adjuvants alum and monophosphoryl lipid A (MPL), a detoxified derivative of lipopolysaccharide from Gram negative organisms). Therefore, the inability of these adjuvants to confer widespread protection, especially along mucosal surfaces, suggests that the adjuvant repertoire must be expanded.In this study we sought to develop a novel strategy to facilitate the detection of potential mucosal adjuvant compounds from a privileged-structure inspired chemical library based on their ability to activate MCs. MCs have recently been described to play a role in the development of adaptive immunity in several pro-inflammatory scenarios. Importantly, targeted activation of MCs using the small molecule MC activator c48/80 was found to induce robust systemic and mucosal immune responses in mice without any harmful side effects. Therefore, we reasoned that MCs might be used to identify novel agents that might be effective adjuvant compounds in vivo. The MC based screen is a high throughput strategy that has the capacity to screen thousands of chemical compounds from virtually any source. The high reproducibility of the assay based on internal plate controls that evaluate total β-hex, baseline β-hex release and a positive control using the MC activator compound 48/80 ensures that results from plate to plate can be compared and ranked based on MC degranulating activity as we described herein.
Others have used a semi-high-throughput screening assay37 to identify possible adjuvant compounds based on induction of interleukin 8 (IL-8) by bovine epithelial cells. Screening a natural compound library containing 280 compounds revealed 3 compounds that consistently induced IL-8 production by bovine intestinal epithelial cells. Our mast cell degranulation screening assay also utilizes live cells but has an important difference when compared to the epithelial cell screening assay used by others.37 The epithelial screening assay depends upon the incubation of the cells with the test compound for 24 hours followed by testing of the culture supernatant for IL-8 produced in response to the test compound. Our mast cell degranulation is a rapid assay that tests the ability of the test compound to induce mast cell degranulation and release of granule contents into the tissue culture after a short incubation time of 30 minutes.
Evaluating novel compounds for cytotoxicity in cell-based screens is a common step in screening assays. However, we intentionally did not monitor cell viability due to the recent observations that alum, a US FDA approved vaccine adjuvant, involves cell death in its mechanism of adjuvant activity.38 Alum has been suggested to be an inflammasome activator. The inflammasome is an intracellular multiprotein complex that is activated by a variety of microbial pathogens or alum that may result in “pyroptotic cell death”.39 Cell death via the pyroptotic pathway results in the release of the cytosolic contents as well as interleukin 1β (IL-1β) secretion and both events are expected to be inflammatory.39 While cytotoxicity is typically viewed as an adverse event and an outcome that should be avoided, the recent observation that alum provides effective and safe vaccine adjuvant activity via a mechanism that requires cell death38 suggests that controlled cell death may be an effective strategy to mimic infection that ultimately provides safe and effective adjuvant activity. For this reason, cell viability was not measured since cell death may be an indicator of inflammasome activation and also an indicator of effective adjuvant activity. Mast cells have recently been reported to produce IL-1β via an inflammasome-dependent mechanism.40 Therefore, additional studies beyond the scope of this screening assay will be required to determine if the mast cell activating compounds identified in this screen: (1) provide effective adjuvant activity in vivo and (2) activate the inflammasome to provide IL-1β production and/or pyroptotic cell death.
Cumulatively, in this screen, we evaluated 580 compounds from a benzopyran-based chemical library and successfully identified 12 compounds that could induce at least 5% MC/9 degranulation. Based on their structures and functionalities, these compounds fall into three distinct categories namely, the benzylamine derivatives, the cyanostilbenes and the chalcones. Although comparative analysis mindful of c48/80 (Fig. 8) revealed several essential structural features within each of the categories, a definitive structure–activity relationship conclusion cannot be reached at this juncture. This finding may suggest that these compounds mediate MC activation by engaging different receptors along the relevant biochemical pathway, and the structural diversity presented by the three classes of compounds opens up new avenues for further molecular interrogations in the search for small molecule(s) with mucosal adjuvant activity.
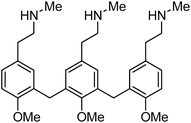 | ||
| Fig. 8 Chemical structure of compound 48/80. | ||
Materials and methods
Reagents
All chemicals were purchased from Sigma-Aldrich (St. Louis, MO). All cell culture reagents were purchased from Invitrogen unless otherwise noted. The chemical library of 650 compounds was prepared with an adaptation of reported literature procedures.16–18 All compounds used in biological testing were greater than 95% purity by 1H NMR analysis.Cell culture
The MC/9 mast cell line was purchased from ATCC (http://www.atcc.org/) and cultured according to their guidelines. All cells were cultured at 37 °C in a humidified water-jacketed incubator under 5% CO2/95% air atmosphere.β-Hexosaminidase assays
MC/9 cells were used in the MC degranulation as described by others,26,28 except that the assay was performed in 96-well V bottom plates at a density of 5 × 105 cells per well in 100 μl sterile Tyrode's buffer. C48/80 (positive control) or library compounds were added to each well in a 100 μl volume to the indicated final concentrations. Each sample was run in duplicate. Controls included MC/9 cells mixed with 0.2% Triton-X 100 to measure total β-hex release, and samples mixed with Tyrode's buffer (vehicle control) as a measure of background β-hex release. After 30 minutes of incubation with controls or test samples, each well is assayed for the presence of β-hex by combining 30 μl of supernatant from each well with 10 μl of substrate solution, p-nitrophenyl-N-acetyl-β-D-glucosaminide (NAG; SIGMA cat. #N9376). Following a 30 minute incubation of 37 °C, a color change was induced by the addition of 100 μl carbonate buffer per well followed by reading the absorbance at 405 nm. Percent (%) degranulation was calculated using the following equation: (experimental β-hex release − vehicle control β-hex release)/(Triton-X-100 β-hex release − vehicle control β-hex release) × 100. Quality control criteria outlined in the text were used to grade the performance of each plate. Plates that did not meet these criteria were discarded and the assay was repeated.Acknowledgements
This work was supported by awards from NIH U01-AI082107; R21-DA029731; and R01 DK077159. The authors wish to thank A*STAR (Agency for Science, Technology and Research), Singapore, for financial support.References
- R. K. Gupta and G. R. Siber, Vaccine, 1995, 13, 1263–1276 CrossRef CAS.
- J. C. Cox and A. R. Coulter, Vaccine, 1997, 15, 248–256 CrossRef CAS.
- M. Singh and D. O'Hagan, Nat. Biotechnol., 1999, 17, 1075–1081 CrossRef CAS.
- C. S. Schmidt, W. J. Morrow and N. A. Sheikh, Expert Rev. Vaccines, 2007, 6, 391–400 CrossRef CAS.
- N. Petrovsky and J. C. Aguilar, Immunol. Cell Biol., 2004, 82, 488–496 CrossRef CAS.
- A. Pashine, N. M. Valiante and J. B. Ulmer, Nat. Med., 2005, 11, S63–S68 CrossRef CAS.
- B. Pulendran and R. Ahmed, Cell, 2006, 124, 849–863 CrossRef CAS.
- P. J. Bryce, M. L. Miller, I. Miyajima, M. Tsai, S. J. Galli and H. C. Oettgen, Immunity, 2004, 20, 381–392 CrossRef CAS.
- V. Heib, M. Becker, T. Warger, G. Rechtsteiner, C. Tertilt, M. Klein, T. Bopp, C. Taube, H. Schild, E. Schmitt and M. Stassen, Blood, 2007, 110, 946–953 CrossRef CAS.
- C. P. Shelburne, H. Nakano, A. L. St John, C. Chan, J. B. McLachlan, M. D. Gunn, H. F. Staats and S. N. Abraham, Cell Host Microbe, 2009, 6, 331–342 CAS.
- A. L. McGowen, L. P. Hale, C. P. Shelburne, S. N. Abraham and H. F. Staats, Vaccine, 2009, 27, 3544–3552 CrossRef CAS.
- J. B. McLachlan, C. P. Shelburne, J. P. Hart, S. V. Pizzo, R. Goyal, R. Brooking-Dixon, H. F. Staats and S. N. Abraham, Nat. Med., 2008, 14, 536–541 CrossRef CAS.
- K. C. Nicolaou, J. A. Pfefferkorn, S. Barluenga, H. J. Mitchell, A. J. Roecker and G. Q. Cao, J. Am. Chem. Soc., 2000, 122, 9968–9976 CrossRef CAS.
- K. C. Nicolaou, J. A. Pfefferkorn, H. J. Mitchell, A. J. Roecker, S. Barluenga, G. Q. Cao, R. L. Affleck and J. E. Lillig, J. Am. Chem. Soc., 2000, 122, 9954–9967 CrossRef CAS.
- K. C. Nicolaou, J. A. Pfefferkorn, A. J. Roecker, G. Q. Cao, S. Barluenga and H. J. Mitchell, J. Am. Chem. Soc., 2000, 122, 9939–9953 CrossRef CAS.
- K. Nicolaou, J. Pfefferkorn, F. Schuler, A. Roecker, G. Cao and J. Casida, Chem. Biol., 2000, 7, 979–992 CrossRef CAS.
- K. C. Nicolaou, R. M. Evans, A. J. Roecker, R. Hughes, M. Downes and J. A. Pfefferkorn, Org. Biomol. Chem., 2003, 1, 908–920 CAS.
- K. C. Nicolaou, A. J. Roecker, S. Barluenga, J. A. Pfefferkorn and G. Q. Cao, ChemBioChem, 2001, 2, 460–465 CrossRef CAS.
- N. W. Lukacs, C. M. Hogaboam, S. L. Kunkel, S. W. Chensue, M. D. Burdick, H. L. Evanoff and R. M. Strieter, J. Leukocyte Biol., 1998, 63, 746–751 CAS.
- M. W. Musch and M. I. Siegel, Biochem. Biophys. Res. Commun., 1985, 126, 517–525 CrossRef CAS.
- M. W. Musch, R. W. Bryant, C. Coscolluela, R. F. Myers and M. I. Siegel, Prostaglandins Other Lipid Mediators, 1985, 29, 405–430 CrossRef CAS.
- P. R. Burd, H. W. Rogers, J. R. Gordon, C. A. Martin, S. Jayaraman, S. D. Wilson, A. M. Dvorak, S. J. Galli and M. E. Dorf, J. Exp. Med., 1989, 170, 245–257 CrossRef CAS.
- S. J. Galli, A. M. Dvorak, J. A. Marcum, G. Nabel, J. M. Goldin, R. D. Rosenberg, H. Cantor and H. F. Dvorak, Monogr. Allergy, 1983, 18, 166–170 CAS.
- G. Nabel, S. J. Galli, A. M. Dvorak, H. F. Dvorak and H. Cantor, Nature, 1981, 291, 332–334 CrossRef CAS.
- A. M. Dvorak and S. J. Galli, Am. J. Pathol., 1987, 126, 535–545 CAS.
- I. Bot, S. C. de Jager, A. Zernecke, K. A. Lindstedt, T. J. van Berkel, C. Weber and E. A. Biessen, Circulation, 2007, 115, 2516–2525 CrossRef CAS.
- I. Bot, S. C. de Jager, M. Bot, S. H. van Heiningen, P. de Groot, R. W. Veldhuizen, T. J. van Berkel, J. H. von der Thusen and E. A. Biessen, Circ. Res., 2010, 106, 89–92 CrossRef CAS.
- N. Sudo, K. Tanaka, Y. Koga, Y. Okumura, C. Kubo and K. Nomoto, J. Immunol., 1996, 156, 3970–3979 CAS.
- W. D. Paton, Br. J. Pharmacol., 1951, 6, 499–508 CrossRef CAS.
- M. Mousli, C. Bronner, J. Bockaert, B. Rouot and Y. Landry, Immunol. Lett., 1990, 25, 355–357 CrossRef CAS.
- M. Mousli, J. L. Bueb, C. Bronner, B. Rouot and Y. Landry, Trends Pharmacol. Sci., 1990, 11, 358–362 CrossRef CAS.
- M. Aridor, G. Rajmilevich, M. A. Beaven and R. Sagi-Eisenberg, Science, 1993, 262, 1569–1572 CAS.
- X. Ferry, S. Brehin, R. Kamel and Y. Landry, Peptides, 2002, 23, 1507 CrossRef CAS.
- I. Shefler and R. Sagi-Eisenberg, J. Immunol., 2001, 167, 475–481 CAS.
- M. Kulka, C. H. Sheen, B. P. Tancowny, L. C. Grammer and R. P. Schleimer, Immunology, 2008, 123, 398–410 CrossRef CAS.
- K. Tatemoto, Y. Nozaki, R. Tsuda, S. Konno, K. Tomura, M. Furuno, H. Ogasawara, K. Edamura, H. Takagi, H. Iwamura, M. Noguchi and T. Naito, Biochem. Biophys. Res. Commun., 2006, 349, 1322–1328 CrossRef CAS.
- D. Buckner, S. Wilson, S. Kurk, M. Hardy, N. Miessner and M. A. Jutila, J. Biomol. Screening, 2006, 11, 664–671 CrossRef CAS.
- T. Marichal, K. Ohata, D. Bedoret, C. Mesnil, C. Sabatel, K. Kobiyama, P. Lekeux, C. Coban, S. Akira, K. J. Ishii, F. Bureau and C. J. Desmet, Nat. Med., 2011, 17, 996–1002 CrossRef CAS.
- E. A. Miao, J. V. Rajan and A. Aderem, Immunol. Rev., 2011, 243, 206–214 CrossRef CAS.
- Y. Nakamura, N. Kambe, M. Saito, R. Nishikomori, Y. G. Kim, M. Murakami, G. Nunez and H. Matsue, J. Exp. Med., 2009, 206, 1037–1046 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c2md20073b |
| This journal is © The Royal Society of Chemistry 2013 |
