Research highlights
Šeila
Selimović
ab,
Gulden
Camci-Unal
ab,
Mehmet R.
Dokmeci
ab and
Ali
Khademhosseini
*abcd
aCenter for Biomedical Engineering, Department of Medicine, Brigham and Women's Hospital, Harvard Medical School, Cambridge, Massachusetts 02139, USA. E-mail: alik@rics.bwh.harvard.edu
bHarvard-MIT Division of Health Sciences and Technology, Massachusetts Institute of Technology, Cambridge, Massachusetts 02139, USA
cWyss Institute for Biologically Inspired Engineering, Harvard University, Boston, Massachusetts 02115, USA
dWorld Premier International – Advanced Institute for Materials Research (WPI- AIMR), Tohoku University, Sendai 980-8577, Japan
First published on 19th November 2012
Microtagging using QR codes
Some medical products, such as drugs, and certain restricted chemicals can be readily modified and counterfeited. Codes imprinted on the packaging can be easily removed or altered. Thus, assigning labels (e.g. expiration date) onto the packaging in the form of a unique code is insufficient to ensure reliable tracking and identification of each drug batch.1 Instead, each drug or chemical batch may be microtagged by including microscale Quick Response (QR) codes.QR codes are two-dimensional matrix patterns that encode information. They are superior to barcodes due to the large amount of programmed information (more than 7000 alphanumeric characters), and the inclusion of error correction patterns. In addition, location markers facilitate scanning and automatic analysis of QR codes. To render QR tagging compatible with small quantities of drugs contained inside tablets or capsules, or small quantities of chemicals, and to generate miniaturized QR patterns, Han et al.2 have recently implemented flow lithography, an optofluidic technique.
In this work, the group encoded information (name, source, expiration date) about a mock drug into QR patterns with capacities of up to 174 characters. They projected the patterns onto a UV-sensitive prepolymer solution flowing inside a poly(dimethylsiloxane) channel, using a maskless photolithography setup (Fig. 1A–C). The solution contained a diacrylated poly(ethylene glycol) (PEG) and a fluorescent, rhodamine based acrylic monomer to make the microtag visible against the drug (Fig. 1D). The choice of polymer and fluorescent tag was dictated by non-toxicity and biocompatibility requirements.
 | ||
| Fig. 1 A PEG-based microtag (scale bar: 200 μm) is patterned with a QR code (A), whose structure is detailed in (B), using flow lithography (C). The microtag and its fluorescence line profile are shown in (D). Figure reprinted with permission from Han et al.2 | ||
To generate the patterns, the researchers exploited the diffraction behavior of light inside the microfluidic channel. Namely, two light spots projected onto the flowing prepolymer are resolved and appear as two distinct spots if the Rayleigh criterion is met; otherwise they appear as a single feature. This means that any bright patterns (e.g. the bright frames within the position detection patterns, Fig. 1B) appear distinct from the surrounding pattern, if the Rayleigh criterion holds. However, complete resolution means that these frames detach from the QR code, rendering it useless. In this case it is advantageous to adjust the setup dimensions, such that the polymer separating the bright frame from the surrounding pattern is weakly crosslinked – then the criterion holds and the frame remains attached to the microtag.
The generated QR microtags were damaged on purpose to test the error correction mechanism implicit in the code. In most cases, the information encoded into damaged patterns could be retrieved using a common smartphone QR reader, unless the extent of the damage was large (>30%) or the position detection patterns were missing. Similarly, QR microtags embedded inside drug capsules could be recaptured, washed to remove the drug powder, scanned with a cell-phone camera and analyzed on the spot.
The proposed method for fabricating QR microtags is simple, affordable and amenable to the large scale production of drugs and chemicals. As such, it could be implemented with any (solid) material that needs to be traced, or whose quality needs to be maintained (e.g. precious stones). As long as the polymer and the fluorescent labels are non-toxic and chemically compatible with the cargo, any potential risks associated with this approach are tied to the QR-reading software, which could be tampered with to include malicious code.
Microfluidic isoelectric focusing
Gel electrophoresis – the movement of charged particles through a crosslinked gel matrix in an electric field – is a widely used technique for the separation of biological molecules (DNA, RNA, proteins).3 However, characterization of dynamic events, such as the migration of bands in protein separations, is difficult using traditional gel electrophoresis. In addition, capillary electrophoresis does not allow the monitoring of dynamic processes in real-time due to its single point detection capabilities.4 In this context, microfluidic separations are used as alternatives to analyze dynamic events in proteins. Advantages of the microfluidic approach include high performance, improved readouts, enhanced assay speed and reduced sample size. For example, microfluidic isoelectric focusing (IEF)-based separations could be powerful tools for studying kinetic, dynamic, and photophysical behavior of fluorescent proteins. Particularly, the dynamics of proton transfer processes in photoswitching fluorescent proteins has become important for applications ranging from membrane transport to high resolution imaging.Hughes et al. have recently addressed some limitations of traditional electrophoresis.5 The research group studied the photoswitching behavior of a wild-type bioluminescent jellyfish, Aequorea victoria, avGFP and the E222G mutant, Aequorea coerulescens acGFP, utilizing a microfluidic IEF device (Fig. 2). IEF is used to separate proteins based on their isoelectric points (pI) upon exposure to an electric field along a pH gradient on the gel. This technique yields highly resolved protein bands and can detect isoform variations as small as a single charge. In this work, glass microfluidic chips were used, with straight channels etched into them and acrylate-functionalized before introducing the gel solution for electrophoresis. The GFP isoform dynamics were analyzed during the reversible photoswitching process in real-time, utilizing the microfluidic IEF assay in conjunction with a fluorescence microscope.
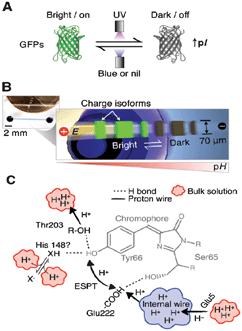 | ||
| Fig. 2 Reversible photoswitching and dynamic analysis of fluorescent proteins. (A) UV exposure induces formation of dark GFP isoforms and pI increase. (B) Microfluidic device for real-time monitoring of dynamic photoswitching in protein isoforms. (C) Sketch of avGFP chromophore and proton transfer. Figure reprinted with permission from Hughes et al.5 Copyright 2012 American Chemical Society. | ||
In addition to IEF, in situ immunoblotting was integrated into the experimental set-up, enabling the analysis of both fluorescent (bright) and non-fluorescent (dark) GFPs. Both types of GFP demonstrated different charges and pIs. Exposure to blue or UV light resulted in reversible photobleaching by interconversion of electrostatic charges. In addition, dark isoform populations with enhanced pI values could be detected upon exposing the focused protein to UV light, due to reversible photobleaching. When subsequently exposed to blue light, however, bright isoform populations were induced by triggering the dynamic photoswitching mechanism.
This study presents an elegant approach to dynamically characterize proton transfers and photophysics in GFPs from purified recombinant Aequorea sp., and the reported high-resolution microfluidic technique can be considered a significant improvement compared to conventional electrophoresis in determining the kinetics of charge transfer processes. The proposed set-up can also be used for high-throughput screening of photoswitchable proteins, reflecting their potential to interface with light-sensitive constructs. Such constructs could be used as substrate coatings to influence hydrophobicity or adhesion behavior of the resulting smart biomaterials, or they could be applied in tissue engineering to monitor cellular fate.
Balancing energy and entropy using micro- and nanofluidics
The recent developments in nanofabrication can be coupled with microfluidics to create devices with enhanced control. Compared to traditional microfluidics, the inclusion of nanoscale structures may enable additional modifications to the flow profiles, and the effects of various forces on liquids can be exploited to achieve even more control over the liquid sample.6A recent example of combining micro- with nanoscale fluidic channels has been offered by Lam et al.,7 who utilized fluidic elements on both scales to unfold bacterial artificial chromosomes (BAC) and analyze the genome sequences. This approach was predicated on the energy and entropy differences between coiled and stretched DNA states, such that the DNA could be coaxed to unfold and assume this energetically less favorable state.
The approach developed by the authors involved patterning thousands of nm sized (45 nm deep, 120 nm wide), long, parallel channels on a silicon wafer using interference lithography, and molding a fluidic device from this master. The device also contained a series of cylindrical microscale barriers that gradually reduced in size and were placed in front of the nanochannels. Without the barriers, the DNA strands would clog the entrance to the nanochannels and could not enter these small pathways in their preferred coiled state. The DNA was fluorescently labeled, so that a strongly fluorescent band could be observed in front of the nanochannels. With the barriers, however, the packing of the DNA was not dense enough to cause clogging, yet the molecules were sufficiently confined that they had to unfold and flow into the nanochannels under the influence of an externally applied electrical pulse. In this case, individual, almost fully stretched DNA strands were observed as blue lines inside the nanofluidic region.
Specific sites on the DNA, so-called ‘nick sites’, were labeled with green fluorescent dye. These sites were always placed at nucleotide 3′ which was a set distance away from the main sequence motif. An automated imaging protocol was applied to record and analyze the length of the captured DNA strands and nick site positions. Then, the data was grouped based on similarities in patterns of nick site locations. Since the orientation of a trapped, linearized DNA molecule could not be known, two sets of peaks (with opposite orientations) were used to represent a single molecule and generate a histogram depicting the locations of the nick sites and, further, maps of sequence motifs.
The novelty in the described study of genome mapping lies in utilizing the properties of micro- and nanofluidic device structures to linearize and confine DNA molecules for an extended period of time. This approach allows a high level of uniformity in the uncoiled molecules, which contributes to accurate and precise measurements of the nick site locations. Aside from genome sequencing, the proposed microfluidic structure could also be applied to study the folding and unfolding mechanisms in DNA, RNA, and proteins. Along these lines, utilizing this type of micro- and nanofluidic device could ultimately lead to better understanding of the formation and function of biological structures.
References
- L. Cozzella, C. Simonetti and G. S. Spagnolo, Opt. Lasers Eng., 2012, 50, 1359–1371 Search PubMed.
- S. Han, H. J. Bae, J. Kim, S. Shin, S.-E. Choi, S. H. Lee, S. Kwon and W. Park, Adv. Mater., 2012 DOI:10.1002/adma.201201486.
- N. C. Stellwagen, Electrophoresis, 2009, 30, S188–S195.
- A. Petrov, V. Okhonin, M. Berezovski and S. N. Krylov, J. Am. Chem. Soc., 2005, 127, 17104–17110 CrossRef CAS.
- A. J. Hughes, A. M. Tentori and A. E. Herr, J. Am. Chem. Soc., 2012, 134(42), 17582–17591 Search PubMed.
- (a) J. Tegenfeldt, C. Prinz, H. Cao, R. Huang, R. Austin, S. Chou, E. Cox and J. Sturm, Anal. Bioanal. Chem., 2004, 378, 1678–1692 CrossRef CAS; (b) G. Hu and D. Li, Chem. Eng. Sci., 2007, 62, 3443–3454 Search PubMed.
- E. T. Lam, A. Hastie, C. Lin, D. Ehrlich, S. K. Das, M. D. Austin, P. Deshpande, H. Cao, N. Nagarajan, M. Xiao and P.-Y. Kwok, Nat. Biotechnol., 2012, 30, 771–777 Search PubMed.
| This journal is © The Royal Society of Chemistry 2013 |
