Amphiphilic random copolymer vesicle induces differentiation of mouse C2C12 myoblasts†
Sumit K. Dey‡a, Krishna Dan‡b, Mahua R. Dasa, Shekhar Sahaa, Provas Dasa, Suhrit Ghosh*b and Siddhartha S. Jana*a
aDepartment of Biological Chemistry, Indian Association for the Cultivation of Science, 2A & 2B Raja S. C. Mullick Road, Kolkata, 700032, India. E-mail: bcssj@iacs.res.in; Fax: +91-33-2483-6561; Tel: +91-33-2473-4971 (Ext 1519)
bPolymer Science Unit, Indian Association for the Cultivation of Science, 2A & 2B Raja S. C. Mullick Road, Kolkata, 700032, India. E-mail: psusg2@ iacs.res.in; Fax: +91-33-2483-6561; Tel: +91-33-2473-4971 (Ext 1563)
First published on 1st October 2013
Abstract
Herein we report that vesicular assembly from a simple non-ionic amphiphilic random copolymer initiates extremely efficient myotube formation from C2C12 myoblast cells in standard growth media lacking horse serum. Control experiments with structurally related polymers and a small molecule suggest the absolute necessity of the vesicular assembly as well as the particular hydrophilic functionality in mediating such high yielding muscle cell generation. The LDH assay indicates that the membrane integrity is retained during cell–cell fusion. Expression of various myogenic factors such as MyoD, myogenin and P-21 has been examined in the presence of the polymersome and control molecules to rationalize this serendipitous discovery.
Various nano-structured assemblies from amphiphilic copolymers1 have been investigated with great interest due to their relevance in biological applications2 such as delivery vehicles for therapeutics and uses in tissue engineering as well as their important interactions with biomacromolecules.3 Several issues such as the nature of the aggregates, specifically their stability, biocompatibility, and degradability and the interaction of the macromolecular assemblies with biomacromolecules are of paramount importance in this interdisciplinary area of research where chemistry meets biology. In the last three decades several block copolymer scaffolds have been explored both in terms of understanding the effect of their structure on their self-assembly as well as their biological applications.2,4 Although nano-structured assemblies from block copolymers are fascinating beyond any doubt, issues have been raised related to difficulties in their synthesis and the ability to control the functional groups that are displayed on their surfaces, which are extremely important points in terms of achieving specific targeting in biological applications. We envisaged investigating the assemblies of some classical polymers, such as random copolymers,5 in light of developments in the field of self-assembly, to provide important relevant information. In the recent past, we have shown the vesicular assembly and thermo responsive morphological transition of a remarkably simple amphiphilic random copolymer P1 (Scheme 1).6 To explore its use in biological applications, the cell viability of CHO, Neuro-2a and C2C12 myoblast cells was checked in the presence of P1 (0–500 μM) by MTT assay. In all cases, more than 90% of the cells were alive in the presence of 100 μM polymer (Fig. S1†). Even at 500 μM of P1, almost 50% of the cells were found to be alive after 24 h. The morphology of the cells (Fig. S2†) such as their attachment to the surface and cell size remained unchanged in the presence of P1 for 24 h. When the cells were cultured for an additional 72 h, we observed that P1 showed an astonishingly good ability to mediate the formation of multinucleated cells (Fig. 1) from C2C12 cells, which might have enormous importance7 to tissue engineering in terms of muscle cell regeneration.
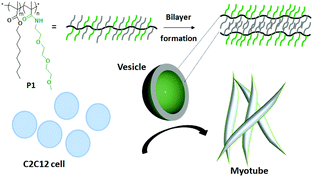 | ||
Scheme 1 Schematic representation of cell fusion mediated by a P1 (Mn = 51![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 g mol−1; PDI = 1.25) vesicle. 000 g mol−1; PDI = 1.25) vesicle. | ||
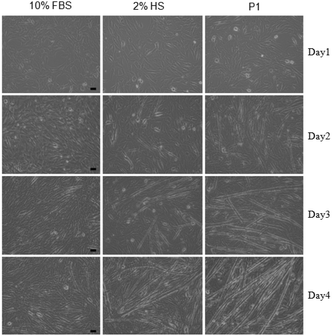 | ||
| Fig. 1 Differentiation of C2C12-myoblast to myotube; brightfield images of cells treated with 10% FBS (left), 2% HS (middle) and P1 (100 μM) (right) for 4 days. Scale bar = 50 μm. | ||
To estimate the efficacy of the process qualitatively, the myotube formation was compared after day 4 with a 2% HS (horse serum) treated sample, which is commonly used as a positive control for such studies.8 We found (Fig. 1) that multinucleated cell formation was more effective in the presence of P1 than in the presence of even 2% HS. Before we examined this intriguing observation in further detail, cell viability was checked over an extended time, ∼80% of the cells were found to be alive after 4 days in the presence of 100 μM P1 (Fig. 2a).
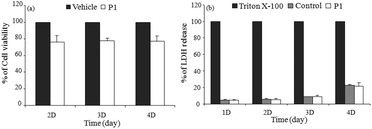 | ||
| Fig. 2 (a) C2C12 cell viability results of P1 (100 μM) by MTT assay at different time intervals. Controls (vehicle) are 100% cell viable; (b) amount of LDH released in culture supernatant from P1 or untreated (control) C2C12 cells estimated using an enzyme assay as described in materials and methods.† % of LDH release was calculated considering 0.1% triton X-100 treated cells as 100%. The experiment was repeated three times. Data are presented as mean ± SD. | ||
Further, we checked the cellular plasma membrane integrity by using a permeability assay based on the release of LDH (lactate dehydrogenase)9 into the cultured medium in the presence of 100 μM of P1 over 4 days. As a control, cells were also treated with 0.1% triton X-100, which gave a maximum release of LDH due to lysis. The percentage of released LDH from the polymer treated cells was calculated considering the triton X-100 treated LDH-release as 100%. It can be seen in Fig. 2b that even after 4 days, the release of LDH was negligible in the presence of P1 suggesting that the membrane integrity was not lost during the cell–cell fusion process.
Now we examined the phenomenon in further detail in terms of quantification of multinucleated cells in the presence of structurally related polymeric and small molecule substrates, which are shown in Scheme 2. P2 contains the same hydrophilic functional group as P1 but no hydrophobic chain so it remains as a unimer. M1 replicates only the repeat unit of P2 while PEG (polyethylene glycol) is a commonly available water-soluble polymer. P3 is an amphiphilic diblock copolymer, which forms micellar particles in aqueous solution.10 We used 2% HS and 10% fetal bovine serum (FBS) as the positive and negative control, respectively, for multinucleated cell formation. We quantified the length, width and fusion index of the multinucleated cells using Image J analysis.11 To visualize the myotubes (Fig. 3), we stained the antibody against myosin heavy chains. The fusion index was estimated using Image J analysis.11 It was found (Fig. 4) that multinucleated cells, generated by polymersome P1, exhibited an average length (∼734 μm), width (∼32 μm) and fusion index (∼42%) that were even greater than the positive control 2% HS (length, width and fusion index were estimated to be ∼641 μm, ∼24 μm and ∼28%, respectively) suggesting a very high efficacy for P1. On the contrary, PEG, M1, P2 and P3 (Scheme 2) were unable to induce the formation of multinucleated cells. The fusion index, length and width of the multinucleated cells in these cases were found (Fig. 4) to be similar to those of the negative control (10% FBS).
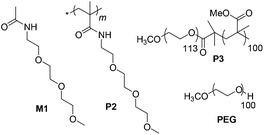 | ||
| Scheme 2 Structure of control molecules. | ||
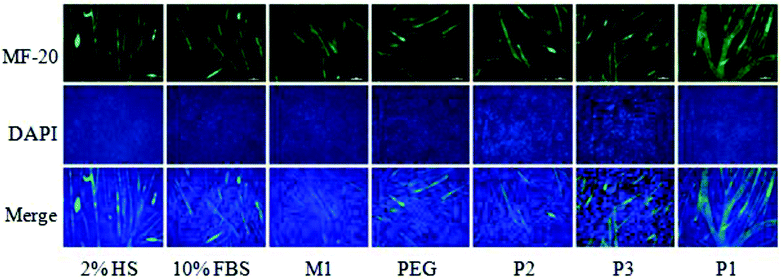 | ||
| Fig. 3 Differentiation of C2C12-myoblast to myotube. Fluorescence images of cells treated with 2% HS, 10% FBS, M1, PEG, P2, P3 and P1 (100 μM) for 4 days. The myotubes and nuclei were stained with MF20 (green) and DAPI (blue), respectively. Both green and blue images were merged to quantify length, width and fusion index. Scale bar = 50 μm. | ||
![Fusion index (top) [which was calculated using the formula: (number of nuclei in multinucleated cells/number of total nuclei present in a given field) × 100], length (middle) and width (bottom) of multinucleated cells. *, p < 0.05, P1vs.P2, P3, M1, PEG or 10% FBS. Data presented here are mean ± SEM from three independent experiments.](/image/article/2013/BM/c3bm60180c/c3bm60180c-f4.gif) | ||
| Fig. 4 Fusion index (top) [which was calculated using the formula: (number of nuclei in multinucleated cells/number of total nuclei present in a given field) × 100], length (middle) and width (bottom) of multinucleated cells. *, p < 0.05, P1vs.P2, P3, M1, PEG or 10% FBS. Data presented here are mean ± SEM from three independent experiments. | ||
Several studies have been carried out to establish12 the role of membrane fluidity, surface charge and the presence of cholesterol in cell–cell fusion. Herein, we used different compounds with similar hydrophilic groups and also an amphiphilic block copolymer known to form micelles.
We noticed enhanced cell–cell fusion only in the case of P1. So it is apparent that the vesicular organization of the polymer, rather than just its chemical composition, plays a key role in this process.
Myoblast fusion is known to be the combination of multiple sequential biochemical processes: (i) cell–cell recognition,13 (ii) change of membrane dynamics14 and (iii) intracellular signaling proteins,15 which control the fusion process.
Perturbation of any of these processes interferes with the cell–cell fusion. Although C2C12 cells possess a high cell–cell recognition ability due to their unique enrichment of phosphatidyl serine (PS) and phosphatidyl ethanol amine (PE) on their surface,13b 10% FBS alone could not induce the fusion (Fig. 1). On the other hand, the addition of P1 in normal growth media, containing 10% FBS, induced cell–cell fusion more rapidly than even 2% HS, which is known to increase intracellular signaling proteins in C2C12 cells, suggesting that P1 was most likely to influence step (ii) and (iii) rather than the cell–cell recognition step.
A number of signaling pathway components and co-transcriptional regulators are involved in myoblast differentiation, in many cases through up-regulation of myogenic factors16 such as MyoD, myogenin, and P-21. We thus checked the expression profiles of these proteins by using immunoblots in multinucleated cells generated in the presence of P1 and compared them with control molecules. Quantification of immunoblots revealed (Fig. 5) that P1 could induce 5.0 fold more expression of P-21, 1.5 fold of MyoD and 2.0 fold of myogenin compared with 2% HS suggesting that the polymersome can be used to induce more efficiently the formation of multinucleated cells than the commonly used 2% HS in an in vitro system. It is important to note that P2, PEG or M1 could also induce expression of myogenic factors but to a lesser extent compared to P1. This may indicate that the fusion process starts only above a certain threshold limit for the expression of the myogenic factors, which was only attained when using P1 but not the other control molecules.
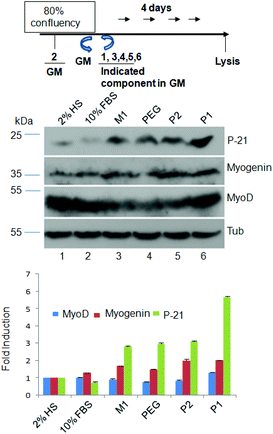 | ||
| Fig. 5 Immunoblots of myogenic factors. Top – schematic diagram of the experiments. Middle – cell lysates of 2% HS, 10% FBS, M1, PEG, P2, and P1 (100 μM) treated C2C12 cells fractionated on SDS-PAGE, and probed with an antibody against MyoD and myogenin and P-21 as indicated. Tubulin was used as a loading control. Bottom – quantification of band intensity by Image J analysis. Fold induction in each case was calculated considering the value for 2% HS as “1”. Data presented here are mean ± SD from three independent experiments. | ||
Conclusions
In summary, we have shown that a simple amphiphilic random copolymer vesicle could induce the formation of myotubes from C2C12 myoblast cells. Although P1 could induce myoblast fusion in C2C12 cells, it failed to induce other cells like CHO and Neuro-2a cells. Several reports17 exist on tuning myotube formation by substrate modification in standard differentiation media containing 2% HS. Contrary to this, herein we have reported a simple polymeric scaffold that forms vesicles in growth media and consequently initiates extremely efficient myotube formation, which is unprecedented to the best of our knowledge. Removal of growth serum is known to be responsible for cell cycle arrest and initiation of differentiation. But in the present case, P1 could induce the differentiation even without the removal of the growth serum. Thus, this structurally simple, nontoxic amphiphilic random copolymer may be useful for muscle cell regeneration in diseases7 such as centronuclear myopathy or myotonic dystrophy, which arise due to a defect in myoblast fusion. Currently, we are exploring other structurally related polymers to gain more insight into this unprecedented polymersome induced cell–cell fusion as well as in vivo applications.Notes and references
- (a) I. W. Hamley, Block Copolymers in Solution: Fundamentals and Applications, John Wiley & Sons, Ltd, 2005 Search PubMed; (b) M. Moffitt, K. Khougaz and A. Eisenberg, Acc. Chem. Res., 1996, 29, 95 CrossRef CAS; (c) D. J. Pochan, Z. Chen, H. Cui, K. Hales, K. Qi and K. L. Wooley, Science, 2004, 306, 94 CrossRef CAS PubMed; (d) C. L. McCormick and A. B. Lowe, Acc. Chem. Res., 2004, 37, 312 CrossRef CAS PubMed; (e) T. S. Kale, A. Klaikherd, B. Popere and S. Thayumanavan, Langmuir, 2009, 25, 9660 CrossRef CAS PubMed; (f) J. Dua and R. K. O'Reilly, Soft Matter, 2009, 5, 3544 RSC; (g) R. P. Brinkhuis, F. P. J. T. Rutjes and J. C. M. van Hest, Polym. Chem., 2011, 2, 1449 RSC.
- (a) A. P. Esser-Kahn, S. A. Odom, N. R. Sottos, S. R. White and J. S. Moore, Macromolecules, 2011, 44, 5539 CrossRef CAS; (b) H. Cabral and K. Kataoka, Sci. Technol. Adv. Mater., 2010, 11, 1 CrossRef; (c) D. Roy, J. N. Cambre and B. S. Sumerlin, Prog. Polym. Sci., 2010, 35, 278 CrossRef CAS; (d) S. R. S. Ting, G. Chen and M. H. Stenzel, Polym. Chem., 2010, 1, 1392 RSC and references therein; (e) C. R. Becer, M. I. Gibson, J. Geng, R. Ilyas, R. Wallis, D. A. Mitchell and D. M. Haddleton, J. Am. Chem. Soc., 2010, 132, 15130 CrossRef CAS PubMed.
- L. S. Nair and C. T. Laurencin, Adv. Biochem. Eng./Biotechnol., 2006, 102, 47 CrossRef CAS PubMed.
- (a) F. S. Bates, Science, 1991, 251, 898 CrossRef CAS PubMed; (b) D. Y. Ryu, K. Shin, E. Drockenmuller, C. J. Hawker and T. P. Russell, Science, 2005, 308, 236 CrossRef CAS PubMed; (c) L. Zhang and A. Eisenberg, J. Am. Chem. Soc., 1996, 118, 3168 CrossRef CAS; (d) S. Motala-Timol, D. Jhurry, J. W. Zhou, A. Bhaw-Luximon, G. Mohun and H. Ritter, Macromolecules, 2008, 41, 5571 CrossRef CAS; (e) O. Colombani, M. Ruppel, F. Schubert, H. Zettl, D. V. Pergushov and A. H. E. Muller, Macromolecules, 2007, 40, 4338 CrossRef CAS; (f) P. Bhargava, Y. F. Tu, J. X. Zheng, H. M. Xiong, R. P. Quirk and S. Z. D. Cheng, J. Am. Chem. Soc., 2007, 129, 1113 CrossRef CAS PubMed; (g) J. F. Gohy, S. Creutz, M. Garcia, B. Mahltig, M. Stamm and R. Jerome, Macromolecules, 2000, 33, 6378 CrossRef CAS.
- (a) F. Ilhan, T. H. Galow, M. Gray, G. Clavier and V. M. Rotello, J. Am. Chem. Soc., 2000, 122, 5895 CrossRef CAS; (b) J. M. Pollino, L. P. Stubbs and M. Weck, J. Am. Chem. Soc., 2003, 126, 563 CrossRef PubMed; (c) J. F. Lutz, S. Pfeifer, M. Chanana, A. F. Thünemann and F. Bienert, Langmuir, 2006, 22, 7411 CrossRef CAS PubMed; (d) K. Feng, N. Xie, B. Chen, L. P. Zhang, C. H. Tung and L. Z. Wu, Macromolecules, 2012, 45, 5596 CrossRef CAS; (e) K. Kaushlendra and S. K. Asha, Langmuir, 2012, 28, 12731 CrossRef CAS PubMed.
- K. Dan, N. Bose and S. Ghosh, Chem. Commun., 2011, 47, 12491 RSC.
- (a) E. H. Chen and E. N. Olson, Science, 2005, 308, 369 CrossRef CAS PubMed; (b) E. Farkas-Bargeton, J. P. Barbet, S. Dancea, R. Wehrle, A. Checouri and O. Dulac, J. Neurol. Sci., 1988, 83, 145 CrossRef CAS PubMed; (c) L. Wockel, U. P. Ketelsen, M. Stotter, S. Laule, R. Meyermann and A. Bornemann, Acta Neuropathol., 1998, 95, 547 CrossRef CAS PubMed.
- K.-J. Kim, O.-H. Lee and B.-Y. Lee, Br. J. Nutr., 2011, 106, 1836 CrossRef CAS PubMed.
- A. Feldman-Salit, S. Hering, H. Messiha, N. Veith, V. Cojocaru, A. Sieg, H. Westerhoff, B. Kreikemeyer, R. C. Wade and T. Fiedler, J. Biol. Chem., 2013, 288, 21295 CrossRef CAS PubMed.
- P3 (Mn = 15
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 g mol−1; PDI = 1.2) shows micellar aggregation with Critical Micellar Concentration (CMC) = 0.01 mM and average hydrodynamic radius = 142 nm. For more detail see: D. Basak and S. Ghosh, ACS Macro Lett., 2013, 2, 799 CrossRef CAS.
000 g mol−1; PDI = 1.2) shows micellar aggregation with Critical Micellar Concentration (CMC) = 0.01 mM and average hydrodynamic radius = 142 nm. For more detail see: D. Basak and S. Ghosh, ACS Macro Lett., 2013, 2, 799 CrossRef CAS. - (a) N. Sangaj, P. Kyriakakis, D. Yang, C.-W. Chang, G. Arya and S. Varghese, Biomacromolecules, 2010, 11, 3294 CrossRef CAS PubMed; (b) T. Samson, C. Will, A. Knoblauch, L. Sharek, K. von der Mark, K. Burridge and V. Wixler, J. Biol. Chem., 2007, 282, 15730 CrossRef CAS PubMed.
- (a) J. Prives and M. Shinitzky, Nature, 1977, 268, 761 CrossRef CAS PubMed; (b) D. Papahadjopoulos, G. Poste and B. E. Schaeffer, Biochim. Biophys. Acta, Biomembr., 1973, 323, 23 CrossRef CAS; (c) F. Martin and R. MacDonald, Nature, 1974, 252, 161 CrossRef CAS PubMed.
- (a) G.-U. Bae, U. Gaio, Y.-J. Yang, H.-J. Lee, J.-S. Kang and R. S. Krauss, J. Biol. Chem., 2008, 283, 8301 CrossRef CAS PubMed; (b) A. Sessions and A. F. Horwitz, FEBS Lett., 1981, 134, 75 CrossRef CAS PubMed.
- N. Kalderon and N. B. Gilula, J. Cell Biol., 1979, 81, 411 CrossRef CAS PubMed.
- L. A. Sabourin and M. A. Rudnicki, Clin. Genet., 2000, 57, 16 CrossRef CAS PubMed.
- (a) S. J. Odelberg, A. Kollhoff and M. T. Keating, Cell, 2000, 103, 1099 CrossRef CAS PubMed; (b) S. Dedieu, G. Mazeres, P. Cottin and J.-J. Brustis, Int. J. Dev. Biol., 2002, 46, 235 CAS.
- (a) I. E. Palama, S. D'Amone, A. M. L. Coluccia and G. Gigli, Biotechnol. Bioeng., 2013, 110, 586 CrossRef CAS PubMed; (b) J. M. Dugan, R. F. Collins, J. E. Gough and S. J. Eichhorn, Acta Biomater., 2013, 9, 4707 CrossRef CAS PubMed; (c) Y. Chandorkar, G. Madras and B. Basu, J. Mater. Chem. B, 2013, 1, 865 RSC; (d) S. Romanazzo, G. Forte, M. Ebara, K. Uto, S. Pagliari, T. Aoyagi, E. Traversa and A. Taniguchi, Sci. Technol. Adv. Mater., 2012, 13, 064211 CrossRef; (e) I. Jun, S. Jeong and H. Shin, Biomaterials, 2009, 30, 2038 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Synthesis of polymers, materials, methods and protocol details for all biological studies. See DOI: 10.1039/c3bm60180c |
| ‡ Both authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2013 |
