Dendrimer-entrapped gold nanoparticles modified with folic acid for targeted gene delivery applications†
Tongyu Xiaoa, Wenxiu Houa, Xueyan Caoa, Shihui Wena, Mingwu Shena and Xiangyang Shi*ab
aCollege of Chemistry, Chemical Engineering and Biotechnology, Donghua University, Shanghai 201620, People's Republic of China. E-mail: xshi@dhu.edu.cn
bCQM-Centro de Química da Madeira, Universidade da Madeira, Campus da Penteada, 9000-390 Funchal, Portugal
First published on 8th August 2013
Abstract
We report a new use of dendrimer-entrapped gold nanoparticles (Au DENPs) modified with folic acid (FA) as a non-viral vector for targeted gene delivery applications. In this study, amine-terminated generation 5 poly(amidoamine) dendrimers modified with FA via covalent conjugation were used as templates to synthesize gold nanoparticles with an Au salt/dendrimer molar ratio of 25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. The synthesized FA-modified Au DENPs (Au DENPs-FA) were used as a non-viral vector for the delivery of plasmid DNA (pDNA) into a model cancer cell line (HeLa cells) overexpressing high-affinity FA receptors (FAR). The DNA compaction ability of the formed Au DENPs-FA was systematically characterized using a gel retardation assay, zeta potential, and dynamic light scattering. We show that similar to the Au DENPs vector without FA, the Au DENPs-FA vector was able to compact the pDNA encoding enhanced green fluorescent protein (EGFP) at an N/P ratio of 0.5. Transfection results show that the Au DENPs-FA vector enables much higher luciferase and EGFP gene expression in HeLa cells overexpressing FAR than the Au DENPs without FA, demonstrating the role played by FA-mediated targeting for enhanced gene transfection in target cells. With a lower cytotoxicity than that of the Au DENPs without FA proven by a cell viability assay, the developed FA-modified Au DENPs may be used as a promising non-viral vector for safe and targeted gene therapy applications.
1. The synthesized FA-modified Au DENPs (Au DENPs-FA) were used as a non-viral vector for the delivery of plasmid DNA (pDNA) into a model cancer cell line (HeLa cells) overexpressing high-affinity FA receptors (FAR). The DNA compaction ability of the formed Au DENPs-FA was systematically characterized using a gel retardation assay, zeta potential, and dynamic light scattering. We show that similar to the Au DENPs vector without FA, the Au DENPs-FA vector was able to compact the pDNA encoding enhanced green fluorescent protein (EGFP) at an N/P ratio of 0.5. Transfection results show that the Au DENPs-FA vector enables much higher luciferase and EGFP gene expression in HeLa cells overexpressing FAR than the Au DENPs without FA, demonstrating the role played by FA-mediated targeting for enhanced gene transfection in target cells. With a lower cytotoxicity than that of the Au DENPs without FA proven by a cell viability assay, the developed FA-modified Au DENPs may be used as a promising non-viral vector for safe and targeted gene therapy applications.
Introduction
Gene therapy has been used to introduce a piece of genetic material to target cells for curing a large number of inherited and acquired diseases.1–3 The prerequisite for successful clinical applications in gene therapy is the presence of safe and effective gene delivery carriers.4 An ideal gene delivery system can transfer exogenous genetic materials (DNA or RNA) into target cells selectively and efficiently without causing any side-effects.5 Despite the advantages of efficient transfection, high-level and permanent gene expression,6 the high toxicity, oncogenicity, and immunogenicity of viral vectors somewhat limit their practical applications.7,8 Therefore, much effort has been devoted to the development of various non-viral vectors for gene delivery applications.Among the many commonly used cationic polymers such as polyethyleneimine (PEI), polylysine, poly(2-(dimethylamino)ethyl methacrylate), dendrimers, poly(ether ester amide)s, and chitosan9–13 that are able to condense plasmid DNA (pDNA) effectively for intracellular gene transfection, poly(amidoamine) (PAMAM) dendrimers14 have been used as one of the most promising vectors for gene delivery applications due to their well-defined molecular architecture, monodispersity, and defined surface functionalities.15 However, the existence of abundant terminal amine groups gives PAMAM dendrimers high cytotoxicity. And in some cases, the gene delivery efficiency of PAMAM dendrimers is still low. Therefore, it is essential to modify the dendrimer surface functionalities or architecture to overcome these issues.
It has been shown that by appropriately modifying the dendrimer surface amines such as by PEGylation,16–18 acetylation,19 alkylation,20,21 and peptide or amino acid conjugation,22–24 the cytotoxicity of PAMAM dendrimers can be decreased, while the gene delivery efficiency can be improved or at least not compromised. On the other hand, by appropriately modifying the dendrimer structure, the gene delivery efficiency of dendrimer vectors can also be improved with decreased cytotoxicity. For instance, Kim and his coworkers demonstrated the use of a triblock copolymer PAMAM–PEG–PAMAM as an efficient gene delivery carrier with improved biocompatibility.25 In another study, Lee et al. modified the interior tertiary amine groups of PAMAM-OH dendrimers by methylation to make the dendrimers more cationic, enabling efficient DNA condensation.26 The hydroxyl-terminal groups of the dendrimers rather afforded decreased cytotoxicity of the vectors when compared with PAMAM dendrimers with amine termini. In our previous work,27 we have shown that dendrimer-entrapped gold nanoparticles (Au DENPs) prepared using generation 5 (G5) PAMAM dendrimers as templates can be used as highly efficient nonviral gene delivery vectors with decreased cytotoxicity. The enhanced gene delivery efficiency is speculated to be due to the fact that the entrapment of the rigid Au core NPs within the interior of dendrimers helps reserve the 3-dimensional spherical shape of G5 dendrimers, which is important to render the dendrimers with enhanced DNA compaction ability.28 With the adaptable surface functionalization features of PAMAM dendrimers that can be linked with various cancer targeting ligands29–34 such as folic acid (FA), RGD peptide, lactobionic acid etc., it is expected that highly efficient Au DENP-based nonviral gene delivery vectors may be developed for targeted gene delivery applications, especially for targeted gene therapy of cancer.
In this present study, we report the use of folic acid (FA)-modified Au DENPs (Au DENPs-FA) for targeted gene delivery applications with high transfection efficiency. First, amine-terminated G5 dendrimers (G5.NH2) were covalently modified with FA. The formed FA-modified G5 dendrimers (G5.NH2-FA) were then used as templates to synthesize Au DENPs-FA (Scheme 1a). For comparison Au DENPs without FA were also synthesized (Scheme 1b). The formed Au DENPs-FA vector was characterized via different techniques. An agarose gel retardation assay was used to examine the binding ability of the vectors to plasmid DNA. The Au DENPs-FA/pDNA polyplexes were also analyzed by dynamic light scattering (DLS) and zeta potential measurements. A thiazoyl blue tetrazolium bromide (MTT) assay was used to evaluate the cytotoxicity of the vectors. Finally, the gene transfection efficiency and specificity using Au DENPs-FA as a vector were evaluated by transfecting both the luciferase (Luc) reporter gene and the enhanced green fluorescent protein (EGFP) gene to HeLa cells. To our knowledge, this is the first report related to the use of FA-targeted Au DENPs as a nonviral vector for targeted gene delivery applications.
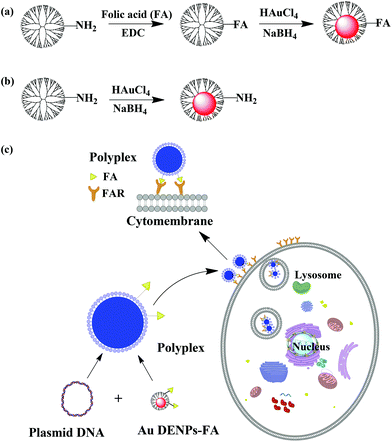 | ||
| Scheme 1 Schematic illustration of the synthesis of Au DENPs-FA (a), Au DENPs without FA (b), and the targeted gene delivery process (c). | ||
Experimental section
Materials
Ethylenediamine cored amine-terminated G5.NH2 dendrimer was purchased from Dendritech (Midland, MI). FA, 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide hydrochloride (EDC), MTT, Hoechst 33342, and ethidium bromide were purchased from Aldrich (St. Louis, MO). Agarose was from Shanghai Sangon Biological Engineering Technology & Services Co., Ltd (Shanghai, China). The Luc assay kit and pGL4 Luciferase reporter vector (pGL4.12 luc2CP/SV40) were purchased from Promega (Fitchburg, WI). The bicinchoninic acid (BCA) protein quantitation kit was purchased from Biomiga (Sandiago, CA). The Label IT® Tracker™ Nucleic Acid Labeling kit containing Cy3™ reagent (MIR 7020) was from Mirus Bio LLC (Madison, WI). Lipofectamine 2000 was obtained from Invitrogen (Carlsbad, CA). HeLa cells were purchased from Institute of Biochemistry and Cell Biology (the Chinese Academy of Sciences, Shanghai, China). Dulbecco's modified Eagle's medium (DMEM), fetal bovine serum (FBS), penicillin, and streptomycin were purchased from Hangzhou Jinuo Biomedical Technology (Hangzhou, China). All other chemicals are of reagent grade. All materials were used as received. Water used in all experiments was purified using a Thermo Scientific Barnstead Easypure II RF water purification system (Thermo Scientific, USA) with a resistivity higher than 18.2 MΩ cm. Regenerated cellulose dialysis membranes (MWCO = 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000) were acquired from Fisher (Pittsburgh, PA).
000) were acquired from Fisher (Pittsburgh, PA).Synthesis and characterization of FA-modified Au DENPs
The method of modification of G5.NH2 with FA to form G5.NH2-FA was adapted from our previous report.35 Briefly, FA (2.02 mg) dissolved in 6.0 mL DMSO was activated by EDC (8.80 mg) pre-dissolved in DMSO (1.5 mL) under vigorous magnetic stirring for 3 h. After that, activated FA was added dropwise into the G5.NH2 solution (23.84 mg, in 6 mL DMSO) under vigorous magnetic stirring. The reaction was continued for 3 d. Then, the reaction mixture was extensively dialyzed against PBS buffer (3 times, 4 L) and water (3 times, 4 L) for 2 d to remove the DMSO and excess of reactants and byproducts using cellulose dialysis membranes (MWCO = 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000), followed by lyophilization to obtain the G5.NH2-FA conjugate.
000), followed by lyophilization to obtain the G5.NH2-FA conjugate.For the synthesis of Au DENPs-FA, the synthesized G5.NH2-FA conjugates were used as templates to entrap Au NPs with the dendrimer/gold salt molar ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 25.33,36,37 Briefly, the G5.NH2-FA conjugate (20.0 mg) was dissolved in 6 mL water. An HAuCl4 solution (10 mg mL−1, 0.633 mL in water) was then added to the dendrimer aqueous solution under vigorous stirring. After 30 min, an ice cold NaBH4 solution (3.45 mg mL−1, 1 mL in water–methanol, v/v = 2
25.33,36,37 Briefly, the G5.NH2-FA conjugate (20.0 mg) was dissolved in 6 mL water. An HAuCl4 solution (10 mg mL−1, 0.633 mL in water) was then added to the dendrimer aqueous solution under vigorous stirring. After 30 min, an ice cold NaBH4 solution (3.45 mg mL−1, 1 mL in water–methanol, v/v = 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) with 5 molar equivalents of the gold salt was added to the gold salt–dendrimer mixture solution under vigorous stirring. The reaction mixture turned deep-red within a few seconds after the addition and the stirring process was continued for 1 h to complete the reaction. Finally, the reaction mixture was purified and lyophilized according to the procedure used for the purification of G5.NH2-FA to obtain the {(Au0)25-G5.NH2-FA} product (Au DENPs-FA). The control {(Au0)25-G5.NH2} DENPs without FA conjugation were also synthesized using G5.NH2 dendrimers as templates under similar experimental conditions. Similar to our previous study,27 the Au DENPs were characterized to have a Au core size of 1.9 nm via transmission electron microscopy (TEM).
1) with 5 molar equivalents of the gold salt was added to the gold salt–dendrimer mixture solution under vigorous stirring. The reaction mixture turned deep-red within a few seconds after the addition and the stirring process was continued for 1 h to complete the reaction. Finally, the reaction mixture was purified and lyophilized according to the procedure used for the purification of G5.NH2-FA to obtain the {(Au0)25-G5.NH2-FA} product (Au DENPs-FA). The control {(Au0)25-G5.NH2} DENPs without FA conjugation were also synthesized using G5.NH2 dendrimers as templates under similar experimental conditions. Similar to our previous study,27 the Au DENPs were characterized to have a Au core size of 1.9 nm via transmission electron microscopy (TEM).
Characterization techniques
1H NMR spectra were recorded using a Bruker DRX 400 nuclear magnetic resonance spectrometer. Samples were dissolved in D2O before measurements. UV-Vis spectra were collected using a Lambda 25 UV-Vis spectrometer (Perkin Elmer, USA). Samples were dissolved in water before the experiments. The size and morphology of the Au DENPs-FA were characterized using a JEOL 2010F analytical electron microscope (JEOL, Japan) operating at 200 kV. TEM samples were prepared by depositing a diluted particle suspension (1 mg mL−1, 5 μL) onto a carbon-coated copper grid and air-dried before measurements.Determination of the number of primary amines of Au DENPs and Au DENPs-FA
Au DENPs and Au DENPs-FA were separately dissolved in water at a concentration of 1 mg mL−1. The mean number of their surface amines was quantified using a Megazyme Primary Amino Nitrogen (K-PANOPA) Assay Kit. The assay was carried out according to the manufacturer's instructions.Polyplexes preparation
The polyplexes of Au DENPs-FA/pDNA and Au DENPs/pDNA were prepared at different N/P ratios (the molar ratio of primary amines of the vectors to phosphates at the pDNA's backbone). Phosphate buffered saline (PBS) was utilized to dilute both vectors and pDNA to a final volume of 50 μL, while the concentration of the vectors was varied due to the varying N/P ratios. Then the vector and pDNA solutions were mixed together, gently vortexed for seconds, and incubated at room temperature for at least 20 min to obtain the polyplex with a final volume of 100 μL.Agarose gel retardation assay
An agarose gel retardation assay was used to evaluate the vectors’ binding ability to pDNA at various N/P ratios. Agarose gel (1.0% w/v) was prepared in Tris-borate-EDTA buffer. Polyplex solutions were prepared using 1 μg pDNA (pEGFP) per well with different N/P ratios. Naked pDNA was used as a control. Gel electrophoresis was typically carried out for 30 min at 120 V. The band locations of pDNA were analyzed using a gel image analysis system (Shanghai FURI Science & Technology, Shanghai, China).Hydrodynamic size and zeta potential measurements
The hydrodynamic size and zeta potential of the polyplexes formed at various N/P ratios were measured using a Zetasizer Nano ZS system (Malvern, UK) equipped with a standard 633 nm laser. For each sample, 5 μg pDNA (pEGP) was used to prepare the polyplex solutions at the N/P ratios of 1, 2.5, and 5, respectively. The polyplex was then diluted to a final volume of 1 mL by adding 0.9 mL PBS buffer before measurements.Cytotoxicity assay
The cytotoxicity of the vectors was investigated by an MTT cell viability assay. HeLa cells were seeded onto a 96-well plate at a density of 5 × 103 cells per well and cultivated in 200 μL DMEM containing 10% FBS, 100 U mL−1 penicillin, and 100 U mL−1 streptomycin under 37 °C and 5% CO2 for 24 h. After that, the medium was replaced with fresh DMEM containing vectors with different concentrations ranging from 50 nM to 3000 nM and the cells were incubated under 37 °C and 5% CO2 for 24 h. Then, MTT (5.0 mg mL−1, 20 μL) was added into each sample well and the cells were incubated at 37 °C and 5% CO2 for 4 h. Thereafter, the medium was removed and 200 μL dimethylsulfoxide (DMSO) was added to dissolve the formazan crystals. The assays were performed according to the manufacturer's instructions using a Thermo Scientific Multiskan MK3 ELISA reader (Thermo Scientific, USA) at 570 nm. The mean and standard deviation for five parallel wells were reported for each sample.Gene transfection
To further prove the targeted gene delivery efficiency, HeLa cells with low FA receptor (FAR) expression were also transfected with the Luc gene under similar conditions for comparison. The HeLa cells grown in FA-containing medium (5 μM) expressed low-level FAR.
Cellular uptake of Au DENPs-FA/pDNA polyplexes
The intracellular uptake of Au DENPs-FA/pDNA polyplexes was evaluated by flow cytometry. Cy3-labeled pEGFP DNA (2 μg per well) was complexed with Au DENPs-FA at an N/P ratio of 1, 2.5, and 5, respectively. The Cy3-labeled pEGFP DNA was prepared according to the protocol of a Label IT® Tracker™ intracellular nucleic acid localization kit (Mirus Bio LLC, USA). The carrier–cargo polyplexes were prepared by mixing vectors with Cy3-labeled pEGFP DNA and incubated at room temperature for 30 min. HeLa cells were seeded at a density of 1 × 105 cells per well in 12-well plates and cultivated at 37 °C and 5% CO2 overnight. Prior to transfection, the medium was exchanged with fresh basic medium without FBS. Then the polyplex solutions were added and the cells were transfected for 4 h. After that, the medium was removed and the cells were washed with PBS for 3 times. Cells were trypsinated, centrifuged, washed twice with PBS buffer, and resuspended into 1 mL PBS before flow cytometry analysis. The cells were gated (G1) on a forward scatter/side scatter (FSC-H/SSC-H) dot plot and the fluorescence of each cell sample was measured in the FL2-H channel. For each sample, 1 × 104 cells were counted and the experiment was repeated 3 times.Statistical analysis
One-way ANOVA statistical analysis was performed to evaluate the experimental data. The p value of 0.05 was selected as the significance level, and the data were indicated with (*) for p < 0.05, (**) for p < 0.01, and (***) for p < 0.001, respectively.Results and discussion
Synthesis and characterization of FA-modified Au DENPs
Our previous work has shown that Au DENPs are promising non-viral vectors for highly efficient gene delivery applications.27 In this work, we attempted to prepare FA-modified Au DENPs for targeted gene delivery to cancer cells overexpressing FAR. This is very important for future targeted gene therapy applications, because FAR has been known to be expressed in several human carcinomas including breast, ovary, endometrium, kidney, lung, head and neck, brain, and myeloid cancers.38–40The conjugation of FA moieties onto G5 dendrimers was confirmed via1H NMR spectroscopy (Fig. S1, ESI†). In the 1H NMR spectrum, the chemical shift between 2.25 and 3.34 ppm is assigned to the –CH2– proton peaks of G5 PAMAM dendrimers, while the characteristic proton peaks at 6.6, 7.5, 8.5 and 1.9 ppm are assigned to the attached FA moiety. According to 1H NMR integration, the number of FA moieties attached to each dendrimer molecule was estimated to be 5.0.
The synthesized FA-modified G5 dendrimers (G5.NH2-FA) were used as templates to entrap Au NPs. Since our previous work has shown that Au DENPs formed with a G5 dendrimer/Au salt molar ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 25 give rise to the highest gene delivery efficiency, in this study we selected the same dendrimer/Au salt molar ratio to prepare FA-modified Au DENPs. The final formed Au DENPs-FA vector was characterized by UV-Vis spectrometry (Fig. S2, ESI†). The surface plasmon resonance (SPR) band at around 512 nm indicates the formation of Au NPs. Compared with the Au DENPs without FA, the Au DENPs-FA vector shows the apparent absorption peak at 280 nm, which is the typical absorption peak of FA. This clearly suggests the successful conjugation of FA moieties. The size and morphology of Au DENPs-FA were characterized with TEM (Fig. 1). It is clear that all Au DENPs-FA have a close-to-spherical shape, a small diameter (2.0 nm), and a uniform size distribution (Fig. 1). This indicates that the FA modified dendrimer is effective to restrict the growth of Au NPs within the dendrimer interior, similar to the one without FA modification.
25 give rise to the highest gene delivery efficiency, in this study we selected the same dendrimer/Au salt molar ratio to prepare FA-modified Au DENPs. The final formed Au DENPs-FA vector was characterized by UV-Vis spectrometry (Fig. S2, ESI†). The surface plasmon resonance (SPR) band at around 512 nm indicates the formation of Au NPs. Compared with the Au DENPs without FA, the Au DENPs-FA vector shows the apparent absorption peak at 280 nm, which is the typical absorption peak of FA. This clearly suggests the successful conjugation of FA moieties. The size and morphology of Au DENPs-FA were characterized with TEM (Fig. 1). It is clear that all Au DENPs-FA have a close-to-spherical shape, a small diameter (2.0 nm), and a uniform size distribution (Fig. 1). This indicates that the FA modified dendrimer is effective to restrict the growth of Au NPs within the dendrimer interior, similar to the one without FA modification.
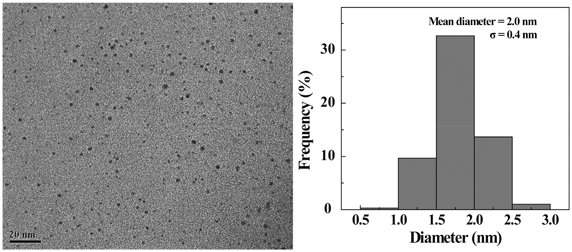 | ||
| Fig. 1 TEM image and size distribution histogram of the Au DENPs-FA. | ||
The mean number of primary amines of Au DENPs and Au DENPs-FA vectors was determined by the PANOPA assay. Table 1 lists the parameters of the two vectors. The results show that both vectors possess 67 primary amines per dendrimer on their surface, indicating that the modification of 5 FA moieties per dendrimer does not appreciably change the density of dendrimer surface amines.
| Samples | Au DENPs | Au DENPs-FA |
|---|---|---|
| Calculated Mw | 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 900 900 | 33![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 107 107 |
| Mean number of primary amines per dendrimer | 67 | 67 |
Agarose gel retardation assay
An agarose gel retardation assay was used to confirm the ability of the vectors to compact pDNA. In general, negatively charged pDNA migrates during the electrophoresis process. The formation of polyplexes between cationic vectors and pDNA via electrostatic interaction is able to neutralize the negatively charged pDNA, consequently resulting in a retardation of the polyplexes in agarose gel. As shown in Fig. 2, with an increase of the N/P ratio between vectors and pDNA, the pDNA band is less brilliant and then disappears. Both vectors regardless of the FA modification are able to compact the pDNA at an N/P ratio of 0.5 or above, suggesting that the FA modification onto the dendrimer surface does not compromise the vector's compaction ability. The strong compaction ability of Au DENPs-FA ensured effective gene delivery applications.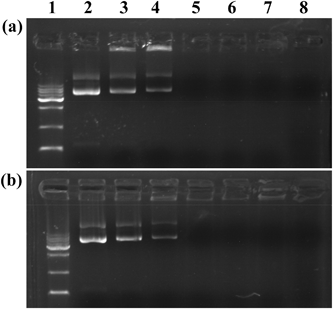 | ||
Fig. 2 Gel retardation assay of pDNA complexed with Au DENPs (a) and Au DENPs-FA (b) at different N/P ratios. Lane 1: DNA marker; lane 2: pDNA alone; lane 3: N/P = 0.125![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1; lane 4: N/P = 0.25 1; lane 4: N/P = 0.25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1; lane 5: N/P = 0.5 1; lane 5: N/P = 0.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1; lane 6: N/P = 1 1; lane 6: N/P = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1; lane 7: N/P = 2 1; lane 7: N/P = 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1; lane 8: N/P = 5 1; lane 8: N/P = 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. 1. | ||
Hydrodynamic size and zeta potential measurements
Hydrodynamic size and zeta potential of the formed polyplexes are known to be important factors affecting the gene delivery process. DLS and zeta potential measurements were used to determine the polyplexes’ hydrodynamic size and surface potential, respectively (Fig. 3). It is clear that for all of the N/P ratios, the sizes of both polyplexes are much smaller than those of the vector alone (Fig. 3a). For both vectors, at the N/P ratio of 2.5, the polyplexes have the smallest size. It seems that the FA-modified Au DENPs are able to better compact the pDNA than the Au DENPs without FA at similar N/P ratios. The formed Au DENPs-FA/pDNA polyplexes have a size ranging from 200 to 390 nm at the N/P ratios of 1, 2.5, and 5.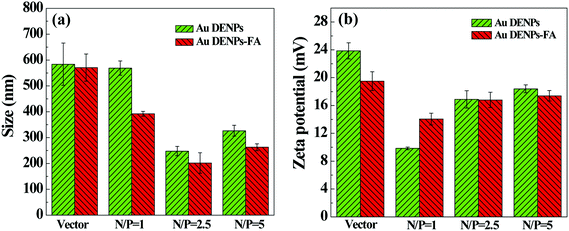 | ||
| Fig. 3 Mean particle size (a) and surface potential (b) of the polyplexes at three different N/P ratios. | ||
For effective intracellular uptake of the vector/DNA polyplexes via electrostatic interaction, it is important to render the formed polyplexes with positive surface potentials. Zeta potential measurements show that the surface potentials of both vectors decrease after the formation of polyplexes, and the surface potentials of polyplexes increase with the N/P ratio. With the small hydrodynamic sizes and quite positive surface potential, the formed Au DENPs-FA/pDNA polyplex should be able to be used for gene delivery applications.
Cytotoxicity assay
Before applying the vectors for gene delivery applications, the cytotoxicity of the vectors was tested by the MTT viability assay of HeLa cells (Fig. 4). It can be clearly seen that the cell viability gradually decreases with an increase of vector concentration for both vectors. Interestingly, Au DENPs-FA seems to be less toxic than Au DENPs, especially at high concentrations. This is presumably due to the fact that the surface modification of FA moieties onto the dendrimer surface may be able to alleviate the strong electrostatic interaction between the particles and the cells, although both vectors were measured to have a similar number of primary amines on the particle surfaces. At the tested highest concentration of 3000 nM, there are still more than 50% cells alive, suggesting that Au DENPs-FA is a safer gene transfection vector than the Au DENPs without FA.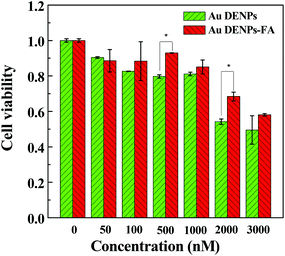 | ||
| Fig. 4 Cell viability of HeLa cells treated with Au DENPs and Au DENPs-FA at different concentrations (mean ± SD, n = 5). | ||
Gene transfection
We next used both firefly Luc and EGFP gene expression assays to investigate the targeted gene transfection efficiency of Au DENPs-FA vector in FAR-overexpressing HeLa cells. The Luc gene transfection efficiency values were reported as relative Luc units per mg protein (RLU mg−1). Fig. 5a shows the gene transfection efficiency of both Au DENPs and Au DENPs-FA vectors as a function of the N/P ratio. It can be seen that both vectors have much higher transfection efficiency than that of the control cells without transfection treatment and cells transfected with naked pDNA; however, the transfection efficiency of both vectors is lower than that of the Lipofectamine 2000 vector. We also show that the modification of FA onto Au DENPs affords Au DENPs-FA with much higher gene delivery efficiency compared to the Au DENPs vector without FA at the N/P ratios of 1, 2.5, and 5 (p < 0.001). The gene transfection efficiency of both vectors is largely dependent on the N/P ratio of the polyplexes. At the low N/P ratio of 1, both Au DENPs-FA and Au DENPs vectors had a similar Luc expression efficiency, which is probably due to the incomplete packaging of pDNA. At the N/P ratios of 2.5 and 5, the Luc expression value for the Au DENPs-FA vector reached 2.19 × 106 and 1.99 × 106, respectively, significantly higher than that of Au DENPs without FA and the N/P ratio of 2.5 gave rise to the highest Luc transfection efficiency for the FA-targeted Au DENPs. These results suggest that the FA-modification onto the Au DENPs is able to render the vector with a very high gene transfection efficiency to FAR-overexpressing HeLa cells, although both Au DENPs with or without FA show no appreciable difference in the binding affinity to pDNA. It should be noted that entrapment of Au core NPs within dendrimers is essential to have an improved gene delivery efficiency. Using G5.NH2-FA dendrimers without the core Au NPs as a vector, the gene delivery efficiency is much less than that using the Au DENPs-FA vector. This further suggests that the entrapment of Au core NPs is able to render the 3-dimensional spherical shape of dendrimers, significantly improving the gene delivery efficiency, in agreement with our previous results.27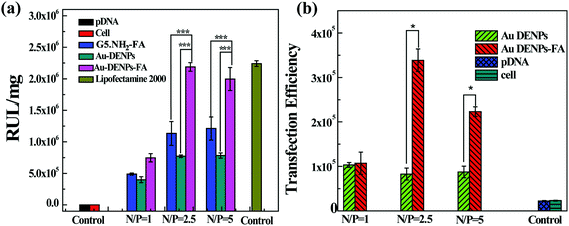 | ||
Fig. 5 (a) Luciferase gene transfection efficiency of Au DENPs/pDNA and Au DENPs-FA/pDNA polyplexes determined in HeLa cells in the N/P ratios of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 2.5 1, 2.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, and 5 1, and 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, respectively. (b) EGFP gene transfection efficiency of Au DENPs/pDNA and Au DENPs-FA/pDNA polyplexes determined in HeLa cells in the N/P ratios of 1 1, respectively. (b) EGFP gene transfection efficiency of Au DENPs/pDNA and Au DENPs-FA/pDNA polyplexes determined in HeLa cells in the N/P ratios of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 2.5 1, 2.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, and 5 1, and 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, respectively. 1, respectively. | ||
The targeted gene transfection efficiency of the Au DENPs-FA vector was also confirmed by the pEGFP expression assay. In this case, the gene delivery efficiency was determined by flow cytometry and is reported as mean fluorescence intensity, which represents the amount of EGFP expression. Cells without treatment and cells transfected by naked pDNA alone were used as controls. As shown in Fig. 5b, at the N/P ratios of 2.5 and 5, the Au DENPs-FA vector has a significantly higher gene delivery efficiency than the Au DENPs vector without FA modification, corroborating the Luc transfection assay data. The pEGFP expression was also visually confirmed by confocal microscopic observation of the HeLa cells (Fig. 6). In contrast to the control cells that did not show any green fluorescence signals, both Au DENP vectors enabled the generation of green fluorescence signals within the cells. However, Au DENPs-FA vector-transfected HeLa cells show more prominent green fluorescence features than those transfected using the Au DENPs vector without FA at the same N/P ratios, indicating the targeting role played by the modified FA ligands onto the Au DENP surfaces.
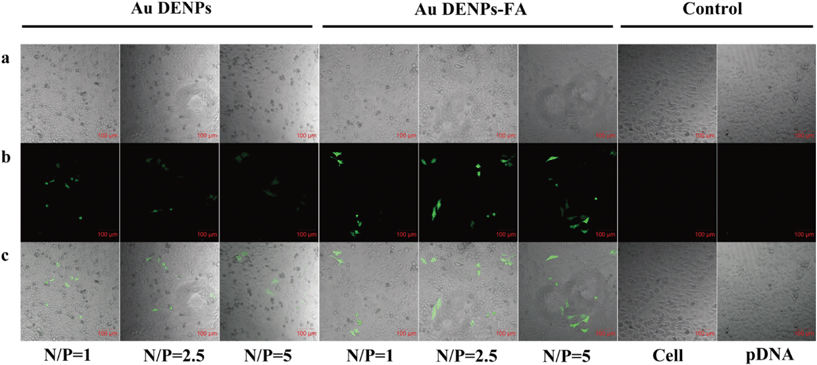 | ||
| Fig. 6 Confocal microscopic images (100×) of EGFP gene expression in HeLa cells. Images were taken 24 h after transfection under similar instrumental conditions. (a) Bright field image; (b) green fluorescence image; and (c) merged bright field image and green fluorescence image. | ||
To further prove the FAR-mediated targeted gene delivery efficacy, HeLa cells were pretreated with free FA to block the surface FAR overexpression, and then transfected with the Luc gene using both Au DENPs-FA and Au DENPs vectors. Fig. 7 shows the Luc gene delivery efficiency in the FAR-blocked HeLa cells using both vectors at different N/P ratios. It can be seen that the RLU mg−1 values of Au DENPs-FA/pDNA were almost similar to those of Au DENPs/pDNA at the same N/P ratios, indicating that both vectors with or without FA do not show a significant difference in the Luc gene transfection efficiency. Taken together with the Luc and EGFP gene delivery results, our results clearly suggest that the developed Au DENPs-FA vector is able to specifically deliver genes to FAR-overexpressing cancer cells via a receptor-mediated targeting pathway.
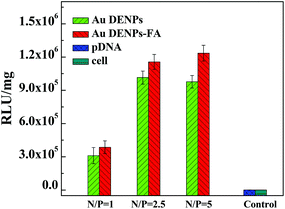 | ||
| Fig. 7 Luciferase gene transfection efficiency in FAR-blocked HeLa cells at different N/P ratios. | ||
Cellular uptake of vector cargo polyplexes
To further understand the gene delivery process, Au DENPs-FA and Au DENPs vectors were complexed with Cy3-labeled pEGFP at different N/P ratios and the transfection was performed for 4 h according to the same protocols used to transfect both Luc and EGFP genes. Then, flow cytometry was used to investigate the intracellular uptake of the polyplexes (Fig. 8). It can be seen that the HeLa cells transfected using the Au DNEPs-FA vector have significantly higher Cy3-derived red fluorescence intensity than those transfected using the Au DENPs vector without FA at the same N/P ratios (p < 0.01). The much higher red fluorescence intensity of cells transfected with the Au DENPs-FA vector should be due to the FAR-mediated enhanced cellular uptake of the polyplexes. Therefore, the targeted gene delivery using the Au DENPs-FA vector may be closely associated with the enhanced cellular uptake of the polyplexes via the FAR-mediated active targeting pathway.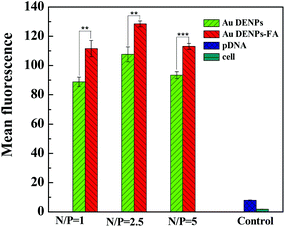 | ||
| Fig. 8 Flow cytometry analysis of the intracellular uptake of vector/Cy3-labeled pDNA polyplexes at three different N/P ratios in HeLa cells. Cells without treatment and cells treated with free Cy3-labeled pDNA were used as controls. | ||
Conclusion
In summary, we developed the use of FA-modified Au DENPs as a nonviral vector for targeted gene delivery to cancer cells overexpressing FAR. The gel retardation assay, DLS, and zeta-potential experiments reveal that the formed Au DENPs-FA vector is able to effectively compact pDNA and the formed polyplex has a small size and positive potential, which is suitable for gene delivery applications. The transfection assays using two different pDNAs encoding Luc and EGFP demonstrate that the modification of FA moieties onto Au DENPs enables the vector to deliver genes to cancer cells overexpressing FAR with high specificity. With a lower cytotoxicity than that of the Au DENPs without FA modification, the formed Au DNEPs-FA may be used as a new non-viral vector for targeted gene therapy of different types of FAR-overexpressing cancer cells.Acknowledgements
Wenxiu Hou and Tongyu Xiao equally contributed to this work. This research was financially supported by the National Natural Science Foundation of China (81101150 and 21273032), the Program for New Century Excellent Talents in University, State Education Ministry, the Fund of the Science and Technology Commission of Shanghai Municipality (11nm0506400 for X. S. and 12520705500 for M. S.), and the Fundamental Research Funds for the Central Universities (for M. S. and X. S.). X. S. gratefully acknowledges the Fundação para a Ciência e a Tecnologia (FCT) and Santander Bank for the Invited Chair in Nanotechnology, and FCT through the project PTDC/CTM-NAN/1748/2012 and through the Strategic Plan PEst-OE/QUI/UI0674/2011.Notes and references
- I. M. Verma and M. D. Weitzman, Annu. Rev. Biochem., 2005, 74, 711–738 CrossRef CAS.
- P. R. Lowenstein, Curr. Opin. Mol. Ther., 2008, 10, 428 Search PubMed.
- A. Fischer and M. Cavazzana-Calvo, Lancet, 2011, 371, 2044–2047 CrossRef.
- F. Theodore, The development of human gene therapy, CSHL Press, Cold Spring Harbor, NY, 1999 Search PubMed.
- S. Li and L. Huang, Gene Ther., 2000, 7, 31–34 CrossRef CAS.
- P. Mancheño-Corvo and P. Martin-Duque, Clin. Transl. Oncol., 2006, 8, 858–867 CrossRef.
- P. Smaglik, Nature, 1999, 402, 707–707 CrossRef.
- C. E. Thomas, A. Ehrhardt and M. A. Kay, Nat. Rev. Genet., 2003, 4, 346–358 CrossRef CAS.
- I. M. Verma and N. Somia, Nature, 1997, 389, 239–242 CrossRef CAS.
- G. Navarro and I. C. Tros, Nanomedicine, 2009, 5, 287 CrossRef CAS.
- S. C. De Smedt, J. Demeester and W. E. Hennink, Pharm. Res., 2000, 17, 113–126 CrossRef CAS.
- M. D. Brown, A. G. Schätzlein and I. F. Uchegbu, Int. J. Pharm., 2001, 229, 1–21 CrossRef CAS.
- J. Wu, D. Yamanouchi, B. Liu and C.-C. Chu, J. Mater. Chem., 2012, 22, 18983–18991 RSC.
- D. A. Tomalia, H. Baker, J. Dewald, M. Hall, G. Kallos, S. Martin, J. Roeck, J. Ryder and P. Smith, Polym. J., 1985, 17, 117–132 CrossRef CAS.
- A. U. Bielinska, C. Chen, J. Johnson and J. R. Baker Jr., Bioconjugate Chem., 1999, 10, 843–850 CrossRef CAS.
- K. Fant, E. K. Esbjörner, A. Jenkins, M. C. Grossel, P. Lincoln and B. Nordén, Mol. Pharmaceutics, 2010, 7, 1734–1746 CrossRef CAS.
- H. He, Y. Li, X. R. Jia, J. Du, X. Ying, W. L. Lu, J. N. Lou and Y. Wei, Biomaterials, 2011, 32, 478–487 CrossRef CAS.
- R. Qi, Y. Gao, Y. Tang, R. R. He, T. L. Liu, Y. He, S. Sun, B. Y. Li, Y. B. Li and G. Liu, AAPS J., 2009, 11, 395–405 CrossRef CAS.
- R. B. Kolhatkar, K. M. Kitchens, P. W. Swaan and H. Ghandehari, Bioconjugate Chem., 2007, 18, 2054–2060 CrossRef CAS.
- J. L. Santos, H. Oliveira, D. Pandita, J. Rodrigues, A. P. Pêgo, P. L. Granja and H. Tomás, J. Controlled Release, 2010, 144, 55–64 CrossRef CAS.
- A. Shakhbazau, I. Isayenka, N. Kartel, N. Goncharova, I. Seviaryn, S. Kosmacheva, M. Potapnev, D. Shcharbin and M. Bryszewska, Int. J. Pharm., 2010, 383, 228–235 CrossRef CAS.
- J. L. Santos, D. Pandita, J. Rodrigues, A. P. Pêgo, P. L. Granja, G. Balian and H. Tomás, Mol. Pharmaceutics, 2010, 7, 763–774 CrossRef CAS.
- L. Han, A. Zhang, H. Wang, P. Pu, X. Jiang, C. Kang and J. Chang, Hum. Gene Ther., 2010, 21, 417–426 CrossRef CAS.
- J. S. Choi, K. Nam, J. Park, J. B. Kim and J. K. Lee, J. Controlled Release, 2004, 99, 445–456 CrossRef CAS.
- T. Kim, H. J. Seo, J. S. Choi, H. S. Jang, J. Baek, K. Kim and J. S. Park, Biomacromolecules, 2004, 5, 2487–2492 CrossRef CAS.
- J. H. Lee, Y. Lim, J. S. Choi, Y. Lee, T. Kim, H. J. Kim, J. K. Yoon, K. Kim and J. Park, Bioconjugate Chem., 2003, 14, 1214–1221 CrossRef CAS.
- Y. Shan, T. Luo, C. Peng, R. Sheng, A. Cao, X. Cao, M. Shen, R. Guo, H. Tomás and X. Shi, Biomaterials, 2012, 33, 3025–3035 CrossRef CAS.
- J. Chen, X. Cao, R. Guo, M. Shen, C. Peng, T. Xiao and X. Shi, Analyst, 2012, 137, 223–228 RSC.
- Q. Chen, K. Li, S. Wen, H. Liu, C. Peng, H. Cai, M. Shen, G. Zhang and X. Shi, Biomaterials, 2013, 34, 5200–5209 CrossRef CAS.
- R. Guo and X. Shi, Curr. Drug Metab., 2012, 13, 1097–1109 CrossRef CAS.
- R. Guo, Y. Yao, G. Cheng, S. H. Wang, Y. Li, M. Shen, Y. Zhang, J. R. Baker Jr., J. Wang and X. Shi, RSC Adv., 2012, 2, 99–102 RSC.
- J. F. Kukowska-Latallo, K. A. Candido, Z. Cao, S. S. Nigavekar, I. J. Majoros, T. P. Thomas, L. P. Balogh, M. K. Khan and J. R. Baker Jr., Cancer Res., 2005, 65, 5317–5324 CrossRef CAS.
- X. Shi, S. Wang, S. Meshinchi, M. E. Van Antwerp, X. Bi, I. Lee and J. R. Baker Jr., Small, 2007, 3, 1245–1252 CrossRef CAS.
- R. Shukla, E. Hill, X. Shi, J. Kim, M. C. Muniz, K. Sun and J. R. Baker Jr., Soft Matter, 2008, 4, 2160–2163 RSC.
- Y. Wang, R. Guo, X. Y. Cao, M. W. Shen and X. Y. Shi, Biomaterials, 2011, 32, 3322–3329 CrossRef CAS.
- X. Shi, S. Wang, H. Sun and J. R. Baker Jr., Soft Matter, 2007, 3, 71–74 RSC.
- R. Guo, H. Wang, C. Peng, M. Shen, M. Pan, X. Cao, G. Zhang and X. Shi, J. Phys. Chem. C, 2009, 114, 50–56 Search PubMed.
- I. G. Campbell, T. A. Jones, W. D. Foulkes and J. Trowsdale, Cancer Res., 1991, 51, 5329–5338 CAS.
- J. F. Ross, P. K. Chaudhuri and M. Ratnam, Cancer, 1994, 73, 2432–2443 CrossRef CAS.
- D. Weitman, R. H. Lark, L. R. Coney, D. W. Fort, V. Frasca, V. R. Surawski and B. A. Kamen, Cancer Res., 1992, 52, 3396–3401 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c3bm60138b |
| This journal is © The Royal Society of Chemistry 2013 |
