Enhanced intracellular drug delivery of pH-sensitive doxorubicin/poly(ethylene glycol)-block-poly(4-vinylbenzylphosphonate) nanoparticles in multi-drug resistant human epidermoid KB carcinoma cells†
Masao
Kamimura‡
a,
Tatsuhiko
Furukawa
b,
Shin-ichi
Akiyama
c and
Yukio
Nagasaki
*ade
aDepartment of Materials Science, Graduate School of Pure and Applied Sciences, University of Tsukuba, Tennoudai 1-1-1, Tsukuba, Ibaraki, 305-8573, Japan. E-mail: yukio@nagalabo.jp; Fax: (+81)-29-853-5749; Tel: (+81)-29-853-5749
bDepartment of Molecular Oncology, Graduate School of Medical and Dental Sciences, Kagoshima University, 8-35-1 Sakuragaoka, Kagoshima 890-8520, Japan
cDepartment of Medical Oncology, Institute of Health Biosciences, The University of Tokushima Graduate School, 3-18-15 Kuramoto-cho, Tokushima 770-8503, Japan
dMaster’s School of Medical Sciences, Graduate School of Comprehensive Human Sciences, University of Tsukuba, Tennoudai 1-1-1 Tsukuba, Ibaraki 305-8573, Japan
eSatellite Laboratory, International Center for Materials Nanoarchitectonics (MANA), National Institute for Materials Science (NIMS), University of Tsukuba, Tennoudai 1-1-1, Tsukuba, Ibaraki, 305-8573, Japan
First published on 30th January 2013
Abstract
A pH-sensitive nanoparticle was prepared using our original amphiphilic block copolymer, poly(ethylene glycol)-b-poly(4-vinylbenzylphosphonate) (PEG-b-PVBP), which possesses phosphate groups as a side chain of its hydrophobic segment (termed here, phosphate nanoparticle (PNP)). The cationic anticancer drug, doxorubicin (DOX) was incorporated into a PNP (DOX@PNP), and its loading capacity was 320 mg g−1-PNP. Electrostatic and hydrophobic interactions in the core of the PNP might act synergistically to significantly improve its loading capacity. The cytotoxicity of the DOX@PNP was examined using the drug-sensitive human epidermoid KB carcinoma cell line (KB-3-1) and two different multi-drug resistance (MDR) KB cell lines (P-glycoprotein (P-gp) overexpressed (KB-C-2) and multidrug resistance protein 1 (MRP1) overexpressed (KB/MRP) cell lines). The DOX@PNP displayed a lower cytotoxic activity than free DOX against KB-3-1 cells. In contrast, the DOX@PNP showed a higher cytotoxic activity than free DOX against MDR cells. Of particular note, the cytotoxicity of the DOX@PNP against KB-C-2 cells was much higher than that against KB/MRP cells, suggesting that different mechanisms of drug reflux via the ATP binding cassette (ABC) transporting system play an important role in nanoparticle-assisted chemotherapy. Observation with confocal laser scanning microscopy (CLSM) indicated that the DOX@PNP was taken up by cells via the endocytosis pathway. The DOX@PNP was initially localized in the late endosome and lysosome, with the subsequent release of DOX from the DOX@PNP in response to the acidic pH of the late endosome and lysosome. Quantitative analysis using flow cytometry confirmed that the uptake of the DOX@PNP into KB-C-2 cells was much higher than that into KB/MRP cells, which was one of the reasons for the enhanced toxicity of the DOX@PNP against KB-C-2 cells compared to that against KB/MRP cells. Reflux of the liberated free DOX in the cytosol, via an endosomal membrane transporter, is considered one of the reasons for the low efficiency of DOX@PNP chemotherapy against KB/MRP cells. However, compared to the free DOX dose, a high dose of the DOX@PNP was effectively delivered to the nuclei of the KB/MRP cells. On the basis of these results, the pH-sensitive DOX@PNP is anticipated as one of the effective chemotherapeutic drugs with enhanced cytotoxicity for multiple types of MDR cancer cells.
1. Introduction
Anticancer drug-based chemotherapy is one of the most widely prescribed therapeutic methods for various cancers. However, the therapeutic effects of many anticancer drugs are limited. One of the major problems of anticancer drug-based chemotherapy is its severe side effects. Anticancer drugs kill not only cancer cells but also normal cells. Another serious problem associated with chemotherapy for cancer is multi-drug resistance (MDR) against anticancer drugs.1,2 After repeated administrations of the same anticancer drug, cancer cells overexpress the ATP-binding cassette (ABC) transporters, followed by reduction in intracellular drug concentrations. Several ABC transporters, including P-glycoprotein (P-gp)3–5 and multidrug resistance protein 1 (MRP1),6–8 have been identified in cancer cells. P-gp3–5 is a 170 kDa membrane glycoprotein that is known to be expressed in the cellular membrane. P-gp obtains energy from ATP hydrolysis, thus functioning as an ATP-dependent efflux pump that excludes anticancer drugs from cells. P-gp has a broad specificity to various anticancer drugs including anthracyclines (doxorubicin (DOX), mitoxantone, and daunorubicin), taxanes (paclitaxel and docetaxel), vinca alkaloids (vincristine and vinblastine), epipodophyllotoxins (etoposide and teniposide), and various other compounds. MRP16–8 is another member of the ABC transporter, is a 190 kDa membrane glycoprotein that is overexpressed in many non P-gp mediated MDR cancer cells. MRP1 functions as a transporter of organic anions and a variety of hydrophobic compounds, such as anthracyclines, vinca alkaloids, and anthracenedione, and particularly of glutathione conjugate drugs. MRP1 apparently works in conformity with the glutathione detoxification system, thus, its drug efflux function depends on the intracellular levels of glutathione. Additionally, MRP1 is located on the cellular membrane and cytoplasmic vesicles, and appears to transport drugs to the outside of cells.8–11 Therefore, P-gp and MRP1 possess different drug reflux mechanisms. In particular, MRP1 refluxes drugs from not only the cell membrane but also the cytoplasmic vesicles. Today, various studies have shown MDR in most cultured cancer cells.3–11 For example, the MDR cell line of human epidermoid KB carcinoma, KB-C-2 cells that overexpress P-gp, was originally isolated from parental drug-sensitive KB-3-1 cells exposed to increasing concentrations of colchicine.12 Ueda and co-workers also reported that the MRP1 overexpressing KB carcinoma cell line, KB/MRP, was isolated from KB-3-1 cells transfected with MRP cDNA.13Recently, anticancer drug-loaded nanoparticles, including polymeric micelles, have been developed to precisely and safely deliver the appropriate concentrations of anticancer drugs to cancer cells.14–17 These nanoparticles show extended bioavailability with reduced nonspecific accumulation in normal tissues and preferential accumulation in tumor tissues because of their enhanced permeability and retention (EPR) effect.18 We have previously reported that DOX-loaded and pH-sensitive nanogel works effectively against MDR cancer cell lines.19,20 The pH-triggered release of the entrapped drug and the avoidance of reflux by P-gp located in the cellular membrane are important factors for increasing the chemotherapeutic effect of drugs on MDR cancer cells. Since then, many other groups have reported the administration of pH-sensitive nanoparticle-assisted chemotherapy for MDR cancer cell lines.21–26 Quite recently, Kataoka and his coworkers reported that pH-sensitive nanoparticle therapy in MDR tumor transplanted mice in vivo.27,28 In most of the studies, however, the effective anticancer drug delivery of pH-sensitive nanoparticles against MDR cancer cells was reported only for P-gp overexpressed cancer cells. Therefore, evaluation of the cytotoxicity of pH-sensitive nanoparticles against MDR cancer cells overexpressed other efflux pumps, such as MRP1, which is also quite important to overcome the multiple types of MDR cancer cells.
Although nanoparticle-assisted drug delivery works well against MDR cancers, the delivery of the drug by nanoparticles to the tumor area is still challenging. Polyamine-based pH-responsive matrices often cause strong toxicity. When particular functions are introduced into the nanoparticle, its physicochemical characteristics change significantly, such as its biocompatibility, colloidal stability, and drug-leakage tendency. Especially, stability of the nanoparticle is the most important point for successful drug delivery. For example, disintegration of a drug-loaded nanoparticle after their dilution in the biological environment is a major drawback for their application in vivo because it can result in rapid drug release. Most of the drug-loaded polymeric micelles formed by only hydrophobic or electrostatic interactions cannot avoid this serious problem. In order to improve the stability of the delivery characteristics of nanoparticles, we have designed and developed a novel family of pH-sensitive nanoparticles by using our original amphiphilic block copolymer, PEG-b-poly(4-vinylbenzylphosphonate) (PEG-b-PVBP; termed here, phosphate nanoparticle (PNP)), which possesses phosphate groups as a side chain of its hydrophobic segment.29 These phosphonate groups were introduced as a side chain of the hydrophobic segment to introduce electrostatic interaction with cationic drugs. Our strategy was to improve the stability of the incorporated drugs in the core of the nanoparticles via both hydrophobic and electrostatic interactions to effectively deliver drugs to the target area. In this study, DOX was incorporated into the PNP (DOX@PNP) for anticancer drug delivery. The DOX@PNP displayed high stability against dilution, changes in ionic strength and presence of serum proteins.29 Furthermore, the DOX@PNP showed pH-sensitive drug release at the late endosome/lysosome mimicking an acidic pH,29 because of the protonation of the phosphonate groups of the PVBP chains (Fig. 1(a)). In this study, we report the in vitro behavior of the DOX@PNP to two MDR cancer cell lines, KB-C-2 and KB/MRP (Fig. 1(b)). This is the first report of the use of anticancer drug-loaded pH-sensitive nanoparticles to overcome MDR KB-C-2 and KB/MRP cell lines with different drug efflux mechanisms. This system may provide increased cytotoxicity for multiple types of MDR efflux pumps.
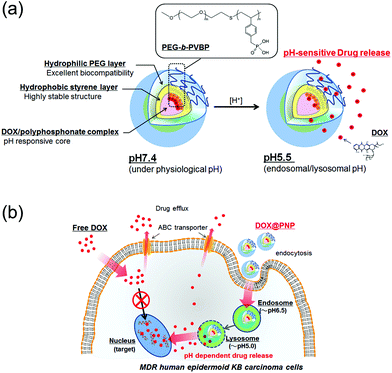 | ||
| Fig. 1 (a) Schematic illustration of the concept of pH-sensitive DOX release from the DOX@PNP and (b) a proposed model of the enhanced cellular entry and pH-sensitive DOX release of the DOX@PNP in MDR KB carcinoma cells to overcome drug efflux mechanisms. | ||
2. Experimental methods
2.1 Materials
A PEG-b-PVBP block copolymer was synthesized as described in our previous report.30 The repeating units for the PEG and PVBP blocks were 113 and 20, respectively. Methanol was purchased from Kanto Chemicals (Tokyo, Japan). Doxorubicin hydrochloride (DOX) and colchicine were purchased from Wako Pure Chemical Industries (Osaka, Japan). WST reagent (Cell Counting Kit-8) and Hoechst 33342 were purchased from Dojindo Laboratories (Kumamoto, Japan). Lysotracker Green DND26 was purchased from Molecular Probes (Eugene, OR, USA). Dulbecco's Modified Eagle's Medium (DMEM) was purchased from Sigma-Aldrich (St Louis, MO, USA). All chemicals were used without further purification.2.2 Preparation of DOX@PNPs
0.2 mL of PEG-b-PVBP solution (22 mM) in methanol was added to 0.64 mL of DOX (3.45 mM) aqueous solution, and the final volume of the mixture was adjusted to 2.4 mL with water. After the methanol was removed by evaporation, the unbound DOX was removed by ultrafiltration by using Amicon Ultra-4 centrifugal filter devices (MWCO 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 Da, Millipore) pretreated with DOX. The final concentration of DOX in the DOX@PNP dispersions was determined by measuring the absorbance at 485 nm by using a UV-2400PC spectrometer (Shimadzu, Japan). Effective mobility and z-averaged hydrodynamic diameters of the DOX@PNPs were determined by dynamic light scattering (DLS) using ZetasizerNano ZS (Malvern Instruments, U.K.) equipped with a He–Ne laser that operated at a wavelength of 635 nm. DLS measurement was performed at a detection angle of 173° at room temperature. The ζ-potential of the particles was calculated from the electrophoretic mobility value by using the Smoluchowski equation. Software provided by the manufacturer, which employs cumulants analysis and non-negatively constrained least-squares particle size distribution analysis routines, was used to analyze the size of the particles, polydispersity indices (PDI), and ζ-potential.
000 Da, Millipore) pretreated with DOX. The final concentration of DOX in the DOX@PNP dispersions was determined by measuring the absorbance at 485 nm by using a UV-2400PC spectrometer (Shimadzu, Japan). Effective mobility and z-averaged hydrodynamic diameters of the DOX@PNPs were determined by dynamic light scattering (DLS) using ZetasizerNano ZS (Malvern Instruments, U.K.) equipped with a He–Ne laser that operated at a wavelength of 635 nm. DLS measurement was performed at a detection angle of 173° at room temperature. The ζ-potential of the particles was calculated from the electrophoretic mobility value by using the Smoluchowski equation. Software provided by the manufacturer, which employs cumulants analysis and non-negatively constrained least-squares particle size distribution analysis routines, was used to analyze the size of the particles, polydispersity indices (PDI), and ζ-potential.
2.3 Cell culture
KB-3-1 cells12 and KB/MRP cells13 were cultured in DMEM containing 10% fetal bovine serum (FBS) and 1% streptomycin at 37 °C in a humidified 5% CO2 atmosphere. KB-C-2 cells12 were cultured in DMEM containing 10% FBS, 1% streptomycin and 2 mg mL−1 colchicine at 37 °C in a humidified 5% CO2 atmosphere.2.4 In vitro cytotoxicity studies
The cytotoxicity of the DOX@PNP was assessed using KB-3-1, KB-C-2, and KB/MRP cells with a standard WST assay. Briefly, the cells were seeded in 96-well microtiter plates, with 1 × 104 cells per well and allowed to adhere for 24 h prior to the assay. The cells were exposed to various doses (0–100 μg mL−1 on DOX basis) of DOX alone, DOX@PNP, and empty PNP for 24–72 h at 37 °C, followed by washing with PBS, the cellular viability was evaluated by WST assay reading the absorbance at 450 nm.2.5 Confocal microscopy of live cells
The cellular uptake of free DOX and DOX@PNP was characterized by live cell confocal laser scanning microscopy (CLSM) by using a Carl Zeiss LSM 700 confocal microscope (Peabody, MA). KB-3-1, KB-C-2, and KB/MRP cells were plated in live cell chambers and after 24 h (37 °C, 5% CO2) exposed to free DOX and DOX@PNPs for 0.5–24 h. Following this, the cells were incubated with Lysotracker Green DND26, a marker of late endosomes and lysosomes, and Hoechst 33342, a marker of nuclei, for 10 min. The concentration of DOX in the medium was 10 μg mL−1. Finally, the cells were washed 3 times with PBS and kept in complete media for confocal imaging.2.6 Cellular uptake analysis by flow cytometry
KB-3-1, KB-C-2, and KB/MRP cells were seeded in 48-well plates at a density of 1 × 104 cells per well and incubated overnight in 500 μL of DMEM for 24 h. After the medium was changed to fresh DMEM, the free DOX and DOX@PNP solutions were applied to each well. The final concentration of DOX in the medium was 10 μg mL−1 for each well. After incubation for 0.5–24 h, the medium was removed and the cells were washed 3 times with PBS. The cells were then trypsinized and collected. Finally, the cells were suspended in 1 mL of PBS. Flow cytometric analysis was carried out using the Gauva EasyCyte Mini System (Guava Technologies, CA, USA). The average pixel intensity of the nuclear DOX fluorescence in observed images was determined using Image J software. After each of the obtained images was split into the three primary colors, the average pixel intensity of the red channel images was determined.313. Results and discussion
The DOX@PNP was prepared by mixing the aqueous solution of DOX and the methanol solution of PEG-b-PVBP at pH 7.0 as described in our previous report.29 The obtained DOX@PNP was characterized according to our previous method.29 The loading capacity of DOX into the PNP was found to be 320 mg g−1 PNP, as determined by UV absorption at 485 nm. The fluorescent intensity of a DOX couple with PEG-b-PVBP quenched by ca. 80% compared to that of free DOX in an aqueous solution. This is one of the other proofs that DOX is stably internalized in the core of the nanoparticle. This extremely high loading capacity of the PNP against DOX might be because of effective and synergistic electrostatic and hydrophobic interactions in the core of the PNP. The ζ-potential of the DOX@PNP in pure water (10 mM NaCl, pH 7.4) was −3.0 mV. Fig. S1 in the ESI† shows the particle size distribution of the DOX@PNP in phosphate buffer saline (PBS; 10 mM phosphate buffer, pH 7.4, 150 mM NaCl). The DOX@PNP showed a unimodal distribution in the histogram analysis. The average particle size and PDI of the DOX@PNP were 42.06 nm and 0.186, respectively. The average particle size and PDI of the DOX@PNP did not change in PBS and serum protein solution (10 mM phosphate buffer, pH 7.4, 150 mM NaCl, 10% FBS). The DOX@PNP also showed a time- and pH-dependent drug release profile, as shown in our previous report.29 These characterization results of the DOX@PNP were in good agreement with our previous report.29 This is one of the proofs for successful preparation of DOX@PNPs.To evaluate the cytotoxicity of the DOX@PNP, WST assay with KB-3-1, KB-C-2, and KB/MRP cells was performed. The 50% inhibitory concentration (IC50) values of free DOX and DOX@PNP against KB-3-1, KB-C-2, and KB/MRP cells were evaluated from WST assay at 48 h (Fig. S2 in ESI†) and results are shown in Table S1 in ESI.† The IC50 values of free DOX against KB-C-2 and KB/MRP cells were in good agreement with the previous report.32 These results also indicate that the used KB-C-2 and KB/MRP cells possess enough MDR potential against DOX. Fig. 2 shows the results of the WST assay for free DOX, DOX@PNP, and empty PNP against the KB cell lines for 24 h. As shown in Fig. 2(a)–(c), the empty PNP showed almost no cytotoxicity for each cell line at the concentrations used in this study. In the case of KB-3-1 cells, cell viability was progressively reduced after 24 h in a dose dependent manner by free DOX and DOX@PNP (Fig. 2(a)). The DOX@PNP showed a lower cytotoxicity than free DOX, indicating that the confinement of DOX in the PNP decreased its efficiency. The cytotoxicity of the DOX@PNP became almost the same as that of free DOX after 48 h, as shown in Fig. 3(a). Therefore, once a sufficient amount of DOX was released into the cytosol, its cytotoxic effect became almost the same as that of free DOX.
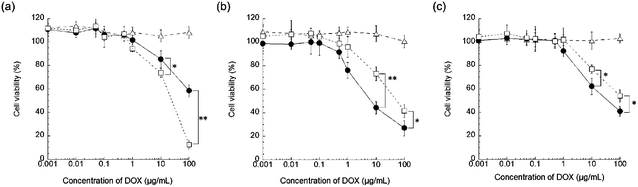 | ||
| Fig. 2 The cytotoxicity of free DOX (open squares), DOX@PNP (closed circles), and empty PNP (open triangles) against (a) KB-3-1 cells, (b) KB-C-2 cells, and (c) KB/MRP cells at 24 h. The relative viabilities of the cells are expressed as a function of the DOX concentration. The data are presented as the mean ± SD (n = 5) (*p < 0.05, **p < 0.01). | ||
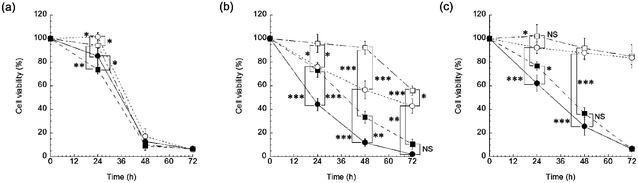 | ||
| Fig. 3 Time- and drug concentration-dependent cytotoxicity of free DOX and DOX@PNP against (a) KB-3-1 cells, (b) KB-C-2 cells, and (c) KB/MRP cells. The DOX concentrations were 1 μg mL−1 (open squares: free DOX, open circles: DOX@PNP) and 10 μg mL−1 (closed squares: free DOX, closed circles: DOX@PNP). The data are presented as the mean ± SD (n = 5) (*p < 0.05, **p < 0.01, ***p < 0.005, NS indicates not significant). | ||
In the KB-C-2 cell, which is a P-gp overexpressed cell line, the DOX@PNP showed higher cytotoxicity than that of free DOX against KB-C-2 cells, as shown in Fig. 2(b). The cytotoxicity of free DOX significantly decreased, while the DOX@PNP had a much greater effect than that of free DOX against KB-C-2 cells. It was clearly demonstrated that P-gp pumping worked effectively for the free DOX treatment, while it did not work well for the DOX@PNP. The endocytotic internalization of the DOX@PNP into the KB-C-2 cell, followed by the release of DOX into the cytosol apart from cell membrane, might effectively avoid the efflux mediated by the P-gp pump, which is known to be located in the cell membrane. Therefore, the intracellular drug concentration was increased and DOX might reach the nuclei effectively by DOX@PNP. The tendency was similar after 48 h incubation as shown in Fig. S2(b) in ESI.†Fig. 3(b) shows the time- and drug concentration-dependent cytotoxicity of the DOX@PNP against KB-C-2 cells. The DOX@PNP also displayed a higher cytotoxicity than free DOX against KB-C-2 cells up to 72 h, even in vitro.
In order to evaluate the effect of the DOX@PNP on the different efflux mechanisms of MDR cancer cells, the MRP1 overexpressed cell line KB/MRP was also employed for in vitro evaluation. The DOX@PNP displayed higher cytotoxicity than free DOX (Fig. 2(c)). The time course of cell viability is shown in Fig. 3(c). However, this result shows a higher but not a significant difference between the free DOX and the DOX@PNP compared with against KB-C-2. It is known that MRP1 is overexpressed not only in the cell membrane but also in the endoplasmic reticulum, for example, in endosomes and lysosomes. Once the nanoparticle has released its DOX into cytoplasm, MRP1 might excrete DOX from the endoplasmic reticulum, which might decrease its efficiency. However, a high dose of the DOX@PNP (10 μg mL−1) worked well compared with free DOX, indicating that a sufficient amount of liberated DOX in the cytosol was able to reach the nuclei.
To obtain further information on cellular trafficking, CLSM and flow cytometric analyses were carried out. Fig. S3 in the ESI† shows the CLSM images of the KB-3-1 cells incubated with free DOX and the DOX@PNPs. As can be seen in Fig. S3,† free DOX rapidly internalized in the cell and spread through the cytosol, while the DOX@PNP internalized slowly and became located in the late endosomes or lysosomes at the early stage (this was confirmed by co-localization of late endosomes or lysosomes and DOX at 1 h of incubation) followed by a gradual spread into the cytosol, indicating endocytotic internalization of the DOX@PNP followed by the release of DOX into the cytosol. Fig. S4 in the ESI† shows the cellular uptake determined by flow cytometric analysis of free DOX and DOX@PNP in KB-3-1 cells. These results clearly indicate that the KB-3-1 cell readily uptakes free DOX after 1 h incubation; this uptake then continues to gradually increase during further incubation. In contrast, the initial uptake of the DOX@PNP was very slow compared with free DOX, suggesting that a very low amount of free DOX liberated in the cell from the DOX@PNP might be internalized via endocytosis (because of the concentration quenching of DOX in the PNP). The fluorescence intensity of the cells treated with the DOX@PNP increased gradually up to 24 h. Fig. S5 in ESI† shows the quantitative analysis of nuclear DOX fluorescence intensity in the KB-3-1 cells. As can be seen in Fig. S5 in ESI,† the nuclear DOX concentration at 1 h was very low and gradually increased up to 24 h to attain a free DOX level, indicating slow release of DOX from the DOX@PNP to nuclei. The fact means that the DOX@PNP retained its structure and internalized probably via endocytosis followed by releasing free DOX by disintegration of the PNP.
Fig. 4 shows CLSM images of the KB-C-2 cells incubated with free DOX and DOX@PNP. As anticipated, almost no internalization was observed even after 24 h, when free DOX was employed, indicating that the P-gp pumping system worked effectively. In contrast, fluorescence based on DOX was increased in the KB-C-2 cells incubated with the DOX@PNP. The DOX was located in the late endosome or lysosome after 1 h of incubation. After 6 h of incubation, DOX fluorescence from the nuclei was observed and this became strong by 24 h of incubation. These results visually indicate that the DOX@PNP was taken up by the KB-C-2 cells via the endocytic pathway, following which liberated DOX from the DOX@PNP reached the nuclei. DOX@PNP was thus visually confirmed to avoid the P-gp efflux mechanism of KB-C-2 cells. It is already reported that fluorescence of DOX is significantly quenched in the PNP,29 indicating that the strong fluorescent signal in DOX@PNP treated KB-C-2 cells implies extremely high accumulation avoiding an efflux mechanism though it is not quantitative. Fig. 5 shows a quantitative analysis of cellular uptake in KB-C-2 cells. Contrary to the very low uptake of free DOX, a time-dependent increase in the fluorescent intensity was clearly observed for the DOX@PNP, indicating an increase in free DOX in the target cells. Fig. S6 in ESI† shows nuclear accumulation of DOX in the KB-C-2 cells. It is clear that significant high concentration of DOX in nuclei was observed in the DOX@PNP especially after 24 h, while free DOX kept a low concentration level. This result also indicated that the liberated DOX from the DOX@PNP effectively delivered to the nuclei as compared to free DOX. This tendency was similar to that for KB/MRP cells as shown in Fig. S7 in ESI.† Because of the DOX@PNP internalized into the MDR cells via endocytosis, the liberated DOX from the DOX@PNP can intercalate DNA of KB-C-2 cells to inhibit DNA replication and cell growth, as compared to free DOX. Although all of liberated DOX from the DOX@PNP might not intercalate DNA, the DOX@PNP elicited a significant amount of apoptosis caused in the KB-C-2 cells. On the basis of these results, it is concluded that the DOX was successfully released from the DOX@PNP after its internalization into the cells.
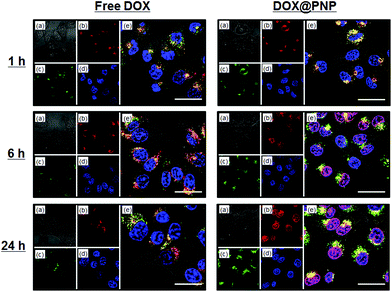 | ||
| Fig. 4 Confocal fluorescence microscopy images of KB-C-2 cells incubated with free DOX (left) and DOX@PNP (right). (a) Bright field, (b) DOX, (c) Lysotracker Green DND26, (d) Hoechst 33342, and (e) a merged image. DOX concentration = 10 μg mL−1. Scale bar = 20 μm. | ||
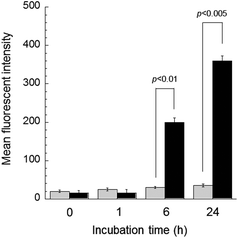 | ||
| Fig. 5 Flow cytometry analysis of the DOX uptake of KB-C-2 cells by comparison of the mean fluorescence intensity of free DOX (gray bar) and DOX@PNP (black bar). DOX concentration = 10 μg mL−1. | ||
The CLSM imaging for determining cellular trafficking was further conducted in the KB/MRP cell line. Fig. 6 shows the CLSM images of the KB/MRP cells incubated with free DOX and DOX@PNPs. Again, almost no fluorescence based on DOX was observed for the addition of free DOX, especially in the nuclei. Thus, the MRP1 system also worked effectively to free DOX in this experiment. On the other hand, fluorescence based on DOX was observed in the late endosome or lysosome after 1 h of incubation with the DOX@PNP. After 6 and 24 h, weak but definite fluorescence of DOX was observed in the nuclei. These results indicate that the DOX was successfully released from the DOX@PNP in the late endosome or lysosome and reached the nuclei of KB/MRP cells via the endocytic pathway. However, compared with KB-3-1 and KB-C-2, the DOX fluorescence of DOX@PNP treated KB/MRP cells was obviously weak. The results of the flow cytometry also showed this tendency. Fig. 7 shows the flow cytometry analysis of DOX uptake into KB/MRP cells. In the case of free DOX, the cellular uptake of DOX into the KB/MRP cells was very low. This may have been due to the effect of the reflux of MRP1. Although the fluorescence of the DOX@PNP remained quenched 1 h after incubation, it increased gradually up to 24 h. Finally, the cellular uptake of the DOX@PNP in KB/MRP cells was about 5-times higher than that of free DOX at 24 h of incubation, indicating that there was effective avoidance of the reflux mechanism, even for the MRP1 cell line. After 24 h, however, this remarkable difference of cell cytotoxicity was not observed, probably due to the strong reflux tendency of the KB/MRP cell lines (Fig. 3(c)). This effective avoidance continued up to 72 h at 10 μg mL−1 dosage of the DOX@PNP as compared with free DOX. However, in fact, DOX was effectively delivered to the nuclei of KB/MRP cells, as shown in Fig. 6.
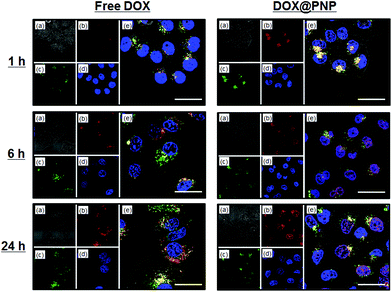 | ||
| Fig. 6 Confocal fluorescence microscopy images of KB/MRP cells incubated with free DOX (left) and DOX@PNP (right). (a) Bright field, (b) DOX, (c) Lysotracker Green DND26, (d) Hoechst 33342, and (e) a merged image. DOX concentration = 10 μg mL−1. Scale bar = 20 μm. | ||
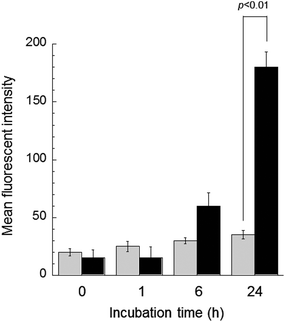 | ||
| Fig. 7 Flow cytometry analysis of DOX uptake of KB/MRP cells by comparison of the mean fluorescence intensity of free DOX (gray bar) and DOX@PNP (black bar). DOX concentration = 10 μg mL−1. | ||
4. Conclusions
In conclusion, we present here the first example of the use of anticancer drug-loaded pH-sensitive nanoparticles to target MDR human epidermoid KB-C-2 and KB/MRP carcinoma cells with different drug efflux mechanisms. The DOX-loaded pH-sensitive DOX@PNP displayed a higher level of bioavailability and more effective cytotoxicity than free DOX against MDR cancer cells. The DOX@PNP showed different cytotoxicity against P-gp overexpressed KB-C-2 and MRP1 overexpressed KB/MRP cells, due to the differences in the drug reflux mechanism of these two MDR cells. Observation with CLSM showed that the DOX@PNP in the cells was found to be initially localized within the late endosome or lysosome, with subsequent release of the DOX in response to the acidic pH of late endosome/lysosome, ultimately, diffusion via the cytoplasm into the cell nuclei. These results indicate that the DOX@PNP was taken up by cells via the endocytic pathway and showed pH-sensitive drug release. Therefore, the DOX@PNP effectively bypassed the efflux mechanism and reached the nuclei of MDR cancer cells. Additionally, based on the flow cytometry measurements, the DOX@PNPs showed a higher cellular uptake level than free DOX against the MDR KB cell lines. From these results, the pH-sensitive DOX@PNP appears to be an effective chemotherapy with an enhanced therapeutic efficacy for multiple types of MDR cancer cells.Acknowledgements
We would like to thank Prof. Kazumitsu Ueda (Kyoto University, Japan) for providing the KB/MRP cells. This research was partially supported by a Grant-in-Aid for Scientific Research on Innovative Areas “Molecular Soft Interface Science” (No. 20106011), from the Ministry of Education, Science, Sports and Culture of Japan. M.K. is grateful for research fellowships from the Japan Society for the Promotion of Science (JSPS) for young scientists.Notes and references
- M. M. Gottesman, T. Fojo and S. E. Bates, Nat. Rev. Cancer, 2002, 2, 48 CrossRef CAS.
- G. Szakács, J. K. Paterson, J. A. Ludwig, C. Booth-Genthe and M. M. Gottesman, Nat. Rev. Drug Discovery, 2006, 5, 219 CrossRef.
- M. M. Gottesman and I. Pastan, J. Biol. Chem., 1988, 263, 12163 CAS.
- J. A. Endicott and V. Ling, Annu. Rev. Biochem., 1989, 58, 137 CrossRef CAS.
- S. V. Ambudkar, C. Kimchi-Sarfaty, Z. E. Sauna and M. M. Gottesman, Oncogene, 2003, 22, 7468 CrossRef CAS.
- S. P. C. Cole, G. Bhardwaj, J. H. Gerlach, J. E. Mackie, C. E. Grant, K. C. Almquist, A. J. Stewart, E. U. Kurz, A. M. V. Duncan and R. G. Deeley, Science, 1992, 258, 1650 CAS.
- D. R. Hipfner, R. G. Deeley and S. P. C. Cole, Biochim. Biophys. Acta, 1999, 1461, 359 CrossRef CAS.
- P. Borst, R. Evers, M. Kool and J. Wijnholds, J. Natl. Cancer Inst., 2000, 92, 1295 CrossRef CAS.
- L. A. Doyle, W. Yang, L. V. Abruzzo, T. Krogmann, Y. Gao, A. K. Rishi and D. D. Ross, Proc. Natl. Acad. Sci. U. S. A., 1998, 95, 15665 CrossRef CAS.
- M. M. Gottesman and I. Pastan, Annu. Rev. Biochim., 1993, 62, 385 CrossRef CAS.
- K. T. Oh, H. J. Baik, A. H. Lee, Y. T. Oh, Y. S. Youn and E. S. Lee, Int. J. Mol. Sci., 2009, 10, 3776 CrossRef CAS.
- S. Akiyama, A. Fojo, J. A. Hanover, I. Pastan and M. M. Gottesman, Somat. Cell. Mol. Genet., 1985, 11, 117 CrossRef CAS.
- Y. Taguchi, A. Yoshida, Y. Takada, T. Komano and K. Ueda, FEBS Lett., 1997, 401, 11 CrossRef CAS.
- K. Kataoka, A. Harada and Y. Nagasaki, Adv. Drug Delivery Rev., 2001, 47, 113 CrossRef CAS.
- A. V. Kabanov, E. V. Batrakova and V. Y. Alakhov, J. Controlled Release, 2002, 82, 189 CrossRef CAS.
- N. Rapoport, Prog. Polym. Sci., 2007, 32, 962 CrossRef CAS.
- Y. Bae and K. Kataoka, Adv. Drug Delivery Rev., 2009, 61, 768 CrossRef CAS.
- Y. Matsumura and H. Maeda, Cancer Res., 1986, 46, 6387 CAS.
- M. Oishi, H. Hayashi, M. Iijima and Y. Nagasaki, J. Mater. Chem., 2007, 17, 3720 RSC.
- M. Oishi and Y. Nagasaki, React. Funct. Polym., 2007, 67, 1311 CrossRef CAS.
- L. S. Jabr-Milane, L. E. van Vlerken, S. Yadav and M. M. Amiji, Cancer Treat. Rev., 2008, 34, 592 CrossRef CAS.
- E. S. Lee, Z. Gao, D. Kim, K. Park, I. C. Kwon and Y. H. Bae, J. Controlled Release, 2008, 129, 228 CrossRef CAS.
- E. S. Lee, Z. Gao and Y. H. Bae, J. Controlled Release, 2008, 132, 164 CrossRef CAS.
- M. Saad, O. B. Garbuzenko and T. Minko, Nanomedicine (Lond), 2008, 3, 761 CrossRef CAS.
- M. K. Danquah, X. A. Zhang and R. I. Mahato, Adv. Drug Delivery Rev., 2011, 63, 623 CrossRef.
- P. Prasad, J. Cheng, A. Shuhendler, A. M. Rauth and X. Y. Wu, Drug Delivery Transl. Res., 2012, 2, 95 CrossRef CAS.
- M. Murakami, H. Cabral, Y. Matsumoto, S. Wu, M. R. Kano, T. Yamori, N. Nishiyama and K. Kataoka, Sci. Transl. Med., 2011, 3, 64ra2 CrossRef CAS.
- H. Cabral, Y. Matsumoto, K. Mizuno, Q. Chen, M. Murakami, M. Kimura, Y. Terada, M. R. Kano, K. Miyazono, M. Uesaka, N. Nishiyama and K. Kataoka, Nat. Nanotechnol., 2011, 6, 815 CrossRef CAS.
- M. Kamimura, J. O. Kim, A. V. Kabanov, T. K. Bronich and Y. Nagasaki, J. Controlled Release, 2012, 160, 486 CrossRef CAS.
- M. Kamimura, N. Kanayama, K. Tokuzen, K. Soga and Y. Nagasaki, Nanoscale, 2011, 3, 3705 RSC.
- H. Meng, M. Liong, T. Xia, Z. Li, Z. Ji, J. I. Zink and A. E. Nel, ACS Nano, 2010, 4, 4539 CrossRef CAS.
- H. Okumura, Z. Chen, M. Sakou, T. Sumizawa, T. Furukawa, M. Komatsu, R. Ikeda, H. Suzuki, K. Hirota, T. Aikou and S. Akiyama, Mol. Pharmacol., 2000, 58, 1563 CAS.
Footnotes |
| † Electronic supplementary information (ESI) available: Particle size distribution of DOX@PNP, cell cytotoxicity of KB cells at 48 h, CLMS images of the KB-3-1 cells, flow cytometry analysis of KB-3-1 cells, and nuclear DOX fluorescence of KB cells are described. See DOI: 10.1039/c2bm00156j |
| ‡ Present address: Department of Pharmaceutical Sciences and Center for Drug Delivery and Nanomedicine, College of Pharmacy, University of Nebraska Medical Center, Omaha, NE 68198-5830, USA. |
| This journal is © The Royal Society of Chemistry 2013 |
