Diagnostic segregation of human brain tumours using Fourier-transform infrared and/or Raman spectroscopy coupled with discriminant analysis†
Ketan
Gajjar
ab,
Lara D.
Heppenstall
a,
Weiyi
Pang
a,
Katherine M.
Ashton
b,
Júlio
Trevisan
a,
Imran I.
Patel
a,
Valon
Llabjani
a,
Helen F.
Stringfellow
b,
Pierre L.
Martin-Hirsch
ab,
Timothy
Dawson
b and
Francis L.
Martin
*a
aCentre for Biophotonics, Lancaster Environment Centre, Lancaster University, Lancaster, LA1 4YQ, UK. E-mail: f.martin@lancaster.ac.uk; Tel: +44 (0)1524 510206
bLancashire Teaching Hospitals NHS Trust, Royal Preston Hospital, Sharoe Green Lane North, Preston, Lancashire, UK
First published on 6th September 2012
Abstract
The most common initial treatment received by patients with a brain tumour is surgical removal of the growth. Precise histopathological diagnosis of brain tumours is to some extent subjective. Furthermore, currently available diagnostic imaging techniques to delineate the excision border during cytoreductive surgery lack the required spatial precision to aid surgeons. We set out to determine whether infrared (IR) and/or Raman spectroscopy combined with multivariate analysis could be applied to discriminate between normal brain tissue and different tumour types (meningioma, glioma and brain metastasis) based on the unique spectral “fingerprints” of their biochemical composition. Formalin-fixed paraffin-embedded tissue blocks of normal brain and different brain tumours were de-waxed, mounted on low-E slides and desiccated before being analyzed using attenuated total reflection Fourier-transform IR (ATR-FTIR) and Raman spectroscopy. ATR-FTIR spectroscopy showed a clear segregation between normal and different tumour subtypes. Discrimination of tumour classes was also apparent with Raman spectroscopy. Further analysis of spectral data revealed changes in brain biochemical structure associated with different tumours. Decreased tentatively-assigned lipid-to-protein ratio was associated with increased tumour progression. Alteration in cholesterol esters-to-phenylalanine ratio was evident in grade IV glioma and metastatic tumours. The current study indicates that IR and/or Raman spectroscopy have the potential to provide a novel diagnostic approach in the accurate diagnosis of brain tumours and have potential for application in intra-operative diagnosis.
Introduction
Brain tumours account for about 1.6% of cancers in England and Wales.1 In 2008, there were 9337 people diagnosed with brain and other central nervous system (CNS) tumours in the UK, causing 3674 deaths in the same year.2 Lifetime risk of developing a malignant brain or other CNS tumour is estimated 1 in 130 for men and 1 in 173 for women.3 The terminology “brain tumours” refers to a mixed group of neoplasms originating from intracranial tissues and the meninges, with degrees of malignancy ranging from benign to aggressive. Their classification is not without problems due to the fact that so-called “benign” tumours can also be lethal as a result of their site, ability to infiltrate locally, and their propensity to become malignant. The WHO classification is the most accepted; here tumours are divided into various types based on cellular origin and histological appearance, and graded according to their aggressiveness from grade I to grade IV.4 High-grade tumours are grades III and IV whilst low-grade tumours are grades I and II.4The aetiology of brain tumours remains largely obscure. The only two aetiological factors thus far proven are familial syndromes (Li-Fraumeni syndrome, neurofibromatosis, Von Hippel-Lindau syndrome, Turcot syndrome, and Gorlin syndrome) and ionizing radiation.5 Studies into potential neurocarcinogens like electromagnetic field exposure, cell phone use, tobacco, and environmental causes have failed to prove any causative link.6 Autoimmune disorders, asthma, and allergies appear to have a protective role in gliomas.7
Brain tumours, particularly high-grade (grades III and IV), have poor prognosis and patient survival is associated with age and histological type.7 In 1999, the 5-year survival rate for brain tumours in the US for all ages and types was reported as 20% (95% CI, 18–22%), without significant improvement in the preceding 30 years.8 Patients with glioblastoma multiforme (GBM) consistently have the poorest survival. Benign meningioma patients have good overall survival rates, in the range of 81% at 2 years and 69% at 5 years, but for malignant meningioma the 5-year survival rate drops to 54.6%.9 Other factors that predict overall and progression-free survival include the tumour location and the extent of resection.10
In UK, about 25% of brain tumours in adults are meningiomas.2 The vast majority (≈80%) are grade I and are commonly treated with surgical resection alone. There is a fairly well-defined relationship between completeness of resection and likelihood of recurrence of meningioma.11 About half of all primary brain tumours are gliomas, which are further classified into common subtypes such as low-grade astrocytoma (grade I and II), anaplastic astrocytoma (grade III) and glioblastoma multiforme (grade IV). This grading can be very subjective and particular tumours often do not fit neatly into any given grade.12
Metastatic brain tumours are the most frequently occurring intracranial neoplasms in adults with the US annual incidence being ≈200![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cases.13 The majority of metastases originate from primary cancers in lung (40–50%) or breast (15–25%), or from melanoma (5–20%),14 while in about 15% of patients the primary site remains unidentified.15 Treatment for metastatic brain tumours is centred around surgical and/or radiation therapy. Surgery is a viable option for patients with only one or a small number of lesions located in accessible regions of the brain and can result in rapid relief of symptoms.14,16
000 cases.13 The majority of metastases originate from primary cancers in lung (40–50%) or breast (15–25%), or from melanoma (5–20%),14 while in about 15% of patients the primary site remains unidentified.15 Treatment for metastatic brain tumours is centred around surgical and/or radiation therapy. Surgery is a viable option for patients with only one or a small number of lesions located in accessible regions of the brain and can result in rapid relief of symptoms.14,16
Currently, there are two important challenges in the management of brain tumours. The first is accurate diagnosis with determination of grade to guide treatment and predict survival whilst the second is precision in defining intra-operative surgical margins. The current approach for diagnosis and histological grading is to obtain tumour sections by biopsy or cytoreductive surgery, stain with haematoxylin and eosin (H&E) as well as applying an array of immunohistochemical neuronal marker proteins. Immunohistochemical detection of isocitrate dehydrogenase (IDH) expression is considered a useful marker in gliomas.17 However, these methods have limitations, which include subjective interpretation.18 With the recognition of an increasing number of brain tumour phenotypes as well as newly evolving variants, there appears to be a need for the development of more robust and accurate diagnostic tools.
During surgery, complete removal of the tumour is one of the most important factors for prediction of recurrence-free survival.19 Various intra-operative diagnostic imaging techniques are available to delineate the excision border; however, no single technique has the spatial resolution to the level required by surgeon-working precision. Despite the use of neuronavigational guidance tools, the precise resection of the brain tumour is hampered due to “brain shift” in stereotactic surgery.20 Brain shift refers to intra-operative brain deformation as a result of changes in tumour volume, cerebrospinal fluid drainage, intracranial pressure or the use of brain retractors that render preoperative neuronavigation registration inaccurate.21 Dedicated high-field intra-operative MRI (iMRI) systems show promising results but the major limitation is cost.22 Stummer and colleagues developed a tumour-specific fluorescent marker, 5-aminolevulinic acid (5-ALA) that allows more accurate discrimination of infiltrating tumour from normal brain parenchyma.23 The limitation of this technique is the limited penetration depth of blue light (mm). Also, non-enhancing tumours do not fluoresce well and the view is often obscured by blood products.24 Another method to delineate tumour margins is to obtain an intra-operative smear or biopsy which provides cell-level information. This is of limited usefulness as it is purely an ex vivo technique and even with rapid staining protocols, at least 20 min are needed to deliver a diagnosis. The results are also only as good as the chosen area of smear or biopsy by the operating surgeon.
Vibrational spectroscopy has potential as a bio-analytical tool for diagnosing cancer because it can probe the chemical composition and molecular structure of normal and pathological tissue.25 Attenuated total reflection Fourier-transform infrared (ATR-FTIR) spectroscopy determines the fingerprint structure of several molecules including proteins, carbohydrates, DNA/RNA or fatty acids. Infrared (IR) spectroscopic imaging measures a large number of spectral profiles from particular tissue subtypes; then computational algorithms identify potentially relevant spectral markers and facilitate classification.26 Such methods offer a means to identify robust diagnostic spectral patterns, even with substantial intra-class variability.27 Examples of such methods are principal component analysis (PCA), linear discriminant analysis (LDA) and fuzzy cluster analysis, each of which have been employed for the analysis of IR/Raman spectra derived from biological tissues.28
ATR-FTIR spectroscopy provides spectra from intact cells recorded within a few seconds and spectral images of tissue sections within minutes. However, it is an ex vivo technique and requires a dry specimen. Advantages of Raman spectroscopy include intra-operative, in vivo diagnosis with non-destructive, real-time analyses. Unlike IR spectroscopy, water does not influence Raman spectra. Raman spectroscopy has the potential to delineate tumour margins and identify tumour remnants while preserving normal tissue.29,30 Fibre-optic probes give spatial flexibility and spatial resolution may be chosen according to surgical need.30
In this study, we hypothesized that interrogation of brain tissues with IR and/or Raman spectroscopy will allow diagnostic segregation of tumours. Given that protein and DNA conformational changes occur in most pre-cancer or cancer lesions, spectroscopy techniques allow their detection. Following spectroscopic analyses, spectral data were analyzed using LDA. When appropriate, PCA was used to reduce the dataset dimensions before LDA was employed to reveal clustering. Scores plots generated following LDA were used to discriminate tumour subtypes, where closeness between clusters reveals spectral similarities and segregation indicates dissimilar classes. The cluster vector approach allowed determination of any identifiable spectral biomarkers segregating tumour subtypes. Additionally, we tested whether spectroscopic identification of subtypes of glioma was more successful than classification based on immunohistochemical biomarkers like isocitrate dehydrogenase-1 (IDH1) and p53.
Patients and methods
Study participants
With ethical approval (REC # 09/H0304/88) we obtained formalin-fixed paraffin-embedded tissue blocks (n = 52) from the Brain Tumour NorthWest (BTNW) biobank. Age, gender, histological type, WHO grade and primary site of metastatic tumours for all samples are summarized in Table 1. Tissue blocks consisted of normal brain (n = 7 patients), meningioma (n = 15), glioma (n = 15) and metastatic brain tumours (n = 15). Gliomas were further subdivided into three pathological subtypes with each category having five tissue blocks: low-grade astrocytoma (LA), anaplastic astrocytoma (AA) and glioblastoma multiforme (GBM).| BTNW code | Age (years) | Sex | Histology | WHO grade/primary site of origin of metastatic tumour | ATR-FTIR spectroscopy (number of spectra) | Raman spectroscopy (number of spectra) |
|---|---|---|---|---|---|---|
| 3 | 24 | F | Glioblastoma multiforme | Grade 4 | 20 | 50 |
| 4 | 63 | M | Glioblastoma multiforme | Grade 4 | 20 | 50 |
| 5 | 47 | F | Glioblastoma multiforme | Grade 4 | 20 | 50 |
| 12 | 64 | M | Glioblastoma multiforme | Grade 4 | 20 | 50 |
| 2 | 68 | F | Glioblastoma multiforme | Grade 4 | 20 | 50 |
| 38 | 48 | M | Anaplastic astrocytoma | Grade 3 | 20 | 50 |
| 173 | 50 | M | Anaplastic astrocytoma | Grade 3 | 20 | 50 |
| 458 | 71 | F | Anaplastic astrocytoma | Grade 3 | 20 | 50 |
| 515 | 65 | M | Anaplastic astrocytoma | Grade 3 | 20 | 50 |
| 126 | 66 | M | Anaplastic astrocytoma | Grade 3 | 20 | 50 |
| 20 | 26 | M | Low-grade astrocytoma | Grade 2 | 20 | 50 |
| 203 | 42 | F | Low-grade astrocytoma | Grade 2 | 20 | 50 |
| 365 | 42 | F | Low-grade astrocytoma | Grade 2 | 20 | 50 |
| 422 | 25 | M | Low-grade astrocytoma | Grade 2 | 20 | 50 |
| 680 | 59 | M | Low-grade astrocytoma | Grade 2 | 20 | 50 |
| 1 | 39 | F | Meningioma | Grade 1 | 20 | 50 |
| 36 | 59 | M | Meningioma | Grade 1 | 20 | 50 |
| 84 | 57 | M | Meningioma | Grade 1 | 20 | 50 |
| 88 | 57 | F | Meningioma | Grade 1 | 20 | 50 |
| 91 | 56 | F | Meningioma | Grade 1 | 20 | 50 |
| 95 | 71 | F | Meningioma | Grade 1 | 20 | 50 |
| 99 | 55 | F | Meningioma | Grade 1 | 20 | 50 |
| 139 | 56 | F | Meningioma | Grade 1 | 20 | 50 |
| 143 | 65 | F | Meningioma | Grade 2 | 20 | 50 |
| 145 | 31 | F | Meningioma | Grade 1 | 20 | 50 |
| 148 | 73 | F | Meningioma | Grade 1 | 20 | 50 |
| 151 | 47 | M | Meningioma | Grade 1 | 20 | 50 |
| 262 | 75 | F | Meningioma | Grade 1 | 20 | 50 |
| 291 | 52 | F | Meningioma | Grade 1 | 20 | 50 |
| 297 | 80 | M | Meningioma | Grade 2 | 20 | 50 |
| 34 | 83 | F | Metastasis | Unknown primary site | 20 | 50 |
| 78 | 52 | F | Metastasis | Non-small cell lung cancer | 20 | 50 |
| 137 | 63 | M | Metastasis | Colon cancer | 20 | 50 |
| 164 | 54 | M | Metastasis | Oesophageal cancer | 20 | 50 |
| 181 | 79 | F | Metastasis | Unknown primary site | 20 | 50 |
| 182 | 70 | F | Metastasis | Non-small cell lung cancer | 20 | 50 |
| 215 | 34 | M | Metastasis | Lung cancer | 20 | 50 |
| 253 | 64 | F | Metastasis | Ovarian/breast cancer | 20 | 50 |
| 295 | 72 | F | Metastasis | Non-small cell lung cancer | 20 | 50 |
| 313 | 58 | M | Metastasis | Squamous cell carcinoma | 20 | 50 |
| 379 | 72 | M | Metastasis | Unknown primary site | 20 | 50 |
| 409 | 84 | F | Metastasis | Bowel cancer | 20 | 50 |
| 119 | 44 | F | Metastasis | Breast cancer | 20 | 50 |
| 271 | 81 | F | Metastasis | Previous squamous cell carcinoma | 20 | 50 |
| 274 | 67 | M | Metastasis | Colon cancer | 20 | 50 |
| 7 | 66 | M | Normal brain | N/A | 20 | 50 |
| 83 | 58 | M | Normal brain | N/A | 20 | 50 |
| 132 | 64 | F | Normal brain | N/A | 20 | 50 |
| 136 | 51 | F | Normal brain | N/A | 20 | 50 |
| 625 | 51 | F | Normal brain | N/A | 20 | 50 |
| 678 | 48 | F | Normal brain | N/A | 20 | 50 |
| 713 | 39 | F | Normal brain | N/A | 20 | 50 |
| Total | 52 (45 tumours, 7 normal) | 1040 | 2600 |
Tissue preparation for spectroscopy
Microtomed 10-μm-thick tissue sections were obtained from paraffin-embedded blocks and mounted on low-E IR reflective (Kevley Technologies, Chesterland, OH, USA) slides for ATR-FTIR spectroscopy and Raman spectroscopy. Tissue sections were de-waxed by immersion in three sequential baths of fresh xylene (5 min), then washed and cleared in acetone (5 min) and left to air-dry. Tissue sections were then placed in a desiccator for storage until analysis. Parallel 4-μm tissue sections were obtained and stained with H&E followed by confirmation of histology for all the specimens by a neuropathologist. Further 4-μm tissue sections were obtained from glioma blocks to carry out immunohistological staining with IDH1 or p53. Tissue sections were interrogated by the biospectroscopy techniques (i.e., ATR-FTIR or Raman spectroscopy) in a randomized fashion to avoid bias.ATR-FTIR spectroscopy
IR spectra were obtained using a Bruker Vector 27 FTIR spectrometer with a Helios ATR attachment containing a diamond crystal (≈250 μm × 250 μm sampling area) (Bruker Optics Ltd., Coventry, UK). Spectra were acquired from 20 different locations across each specimen (n = 52) with a new background taken after acquisition of every 10 spectra. The ATR crystal was cleaned with distilled water and dried with dry tissue paper before the acquisition of spectral background and for each new sample. Spectra (8 cm−1 spectral resolution giving 4 cm−1 data spacing, co-added for 32 scans) were converted into absorbance by Bruker OPUS software. Raw spectra were cut between 1800 and 900 cm−1 (235 data points) and were then baseline corrected and normalized to the Amide I peak (Bruker OPUS software).Raman spectroscopy
Raman spectra were acquired from each sample using an InVia Renishaw Raman spectrometer (Renishaw plc, Gloucestershire, UK). The spectrometer contained a laser diode of 785 nm operating at 35 mW (at sample) with a Rayleigh holographic edge filter. The spectrometer's entrance slit of 50 μm combined with a 1200 lines per mm (1 cm−1 spatial resolution) diffraction grating allowed dispersion of Raman signals onto a Master Renishaw Pelletier cooled charged couple detector (CCD). A white light camera mounted on the microscope allowed the use of dark-field images to visualize locations for spectral acquisition. Spectra were acquired using a Leica ×50 objective lens (numerical aperture 0.75, ≈1 μm spatial resolution), 25 s of 100% laser power exposure and four repeat acquisitions. The Renishaw system was calibrated with a Renishaw silicon calibration source for wavenumber shifts. A total of 2600 spectra were obtained from independent locations with 50 spectra acquired from each sample. Renishaw Wire 3.1 software was used to remove cosmic rays, to average every five spectra and then cut them between the spectral ranges 1750–800 cm−1 (597 data points). Subtraction of background fluorescence was carried out using a fifth-order polynomial fit (MATLAB software), and spectra were wavelet de-noised before being vector normalized.31 Residual contaminating paraffin peaks were excluded from computational analyses.32,33 Raman spectroscopy was always carried out on tissue sections post-analyses using ATR-FTIR spectroscopy and in independent regions so as to minimize any confounding influences of tissue compression by the ATR crystal.Immunohistochemistry
Immunohistochemical staining with IDH1 was performed with mouse monoclonal anti-human IDH1-R132H antibody (Dianova, Hamburg, Germany; dilution 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100). Staining was performed following de-waxing and rehydration. Endogenous peroxidase was blocked by immersing the tissue sections in 4% H2O2 in methanol for 15 min. High-temperature antigen retrieval was performed by heating the tissue sections in citrate buffer (pH 6.0) for 2 min, under pressure and at full power (800 W) in a microwave oven. The antiserum [anti-IDH1-R132H (clone H09)] was diluted 1
100). Staining was performed following de-waxing and rehydration. Endogenous peroxidase was blocked by immersing the tissue sections in 4% H2O2 in methanol for 15 min. High-temperature antigen retrieval was performed by heating the tissue sections in citrate buffer (pH 6.0) for 2 min, under pressure and at full power (800 W) in a microwave oven. The antiserum [anti-IDH1-R132H (clone H09)] was diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200 in 0.2% bovine serum albumin in Tris-buffered saline (pH 7.6) (BSAT). The anti-p53 (DakoCytomation, DO-7) serum was diluted 1
200 in 0.2% bovine serum albumin in Tris-buffered saline (pH 7.6) (BSAT). The anti-p53 (DakoCytomation, DO-7) serum was diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20 (anti-p53) in BSAT. Tissue sections were incubated with primary antibody for 30 min at room temperature (anti-p53) or overnight at 4°C (anti-human IDH1-R132H). Following application of 3,3′-diaminobenzidine (DAB) chromogen in 0.05 M Tris–HCl buffer (pH 7.4) with 0.1% H2O2, slides were stained (15 s) with Harris' haematoxylin, rinsed with tap water, blued in warm tap water (15 s) and rinsed again.34 Preparations were dehydrated with graded alcohol solutions through to xylene and mounted with cover-slips using Styrolite mounting medium (VWR International, Poole, UK). The expression of IDH1 was determined by assessing the proportion of positively-stained tumour cells semi-quantitatively.35 The p53 staining was categorized as the following: 0 = no detectable staining over background; 1 = staining <10% of cells; 2 = 10 to 25% of the cells; 3 = 25 to 50% of cells; 4 = 50 to 75% of cells; and, 5 = staining >75% of cells.36
20 (anti-p53) in BSAT. Tissue sections were incubated with primary antibody for 30 min at room temperature (anti-p53) or overnight at 4°C (anti-human IDH1-R132H). Following application of 3,3′-diaminobenzidine (DAB) chromogen in 0.05 M Tris–HCl buffer (pH 7.4) with 0.1% H2O2, slides were stained (15 s) with Harris' haematoxylin, rinsed with tap water, blued in warm tap water (15 s) and rinsed again.34 Preparations were dehydrated with graded alcohol solutions through to xylene and mounted with cover-slips using Styrolite mounting medium (VWR International, Poole, UK). The expression of IDH1 was determined by assessing the proportion of positively-stained tumour cells semi-quantitatively.35 The p53 staining was categorized as the following: 0 = no detectable staining over background; 1 = staining <10% of cells; 2 = 10 to 25% of the cells; 3 = 25 to 50% of cells; 4 = 50 to 75% of cells; and, 5 = staining >75% of cells.36
Computational analysis
Multivariate analysis (i.e., LDA and PCA-LDA) was applied to the spectral data using MATLAB R2010a (The Maths Works, Natick, MA, USA) with a graphical user interface toolkit for spectroscopy (http://biophotonics.lancs.ac.uk/software).31 Generally, it is recommended that the number of spectra be several times greater than the number of variables/features in the dataset.28,37 As the number of spectra (n = 1040 spectra) in the IR dataset is almost five times the number of variables (235 data points), we applied LDA, which is a supervised technique and forms linear combinations of variables dependent on differences between the classes. This generates new variables, linear combinations known as “factors”, which are weighted sums of the original wavenumber–absorbance intensities. The weights for each factor are represented by a vector called a “loadings vector”. The loadings vectors are successive orthogonal solutions to the problem, the function of which appears to maximize the between-class variance responsible for diagnostic segregation over the within-class variance (mostly associated with heterogeneity in tissue sample) of the factor.37,38 Another technique called PCA allows for the reduction of the number of variables in the spectral dataset, whose principal components (PCs) can capture more than 95% of the variance present in the original dataset. PCA can be applied before LDA (thus “PCA-LDA”) to reduce computational complexity and increase the recognition accuracy in different classes.39 Following the construction of factors from LDA or PCA-LDA technique, the factor values (i.e., factor scores) can be used as Cartesian coordinates to generate 1-, 2-, or 3-dimensional (D) scatter plots (scores plots). A scores plot allows visualization of segregation of classes whilst derived cluster vector plots determine the wavenumbers responsible for segregation.40,41 Subsequently a peak detection algorithm is applied to identify the six most prominent peaks from each cluster vector, whereby the normal brain tissue is labelled as the “control” vector.27 The location of the detected peaks is then plotted along the wavenumber line of the tumour type being compared using marker symbols whose sizes are proportional to the height of their corresponding peaks (see Fig. 4C and D).39 The statistical significance of the contribution of each linear discriminant (LD) to inter-category segregation was determined by an unpaired t-test and ANOVA analysis conducted using GraphPad Prism software.Results
Characteristics of brain tissues in H&E-stained samples
Fig. 1A shows normal brain architecture while Fig. 1B shows a meningioma, which is characterized by tumour cells arranged in whorls with hyalinised and calcified central areas that are called psammoma (sand) bodies. Fig. 1C–E show increasing grades of gliomas; the high-grade glioma such as GBM shown in Fig. 1E is characterized by high cellularity, cellular and nuclear atypia, mitosis, microvascular proliferation and necrosis. Fig. 1F shows a metastatic brain tumour arising from a primary colon cancer.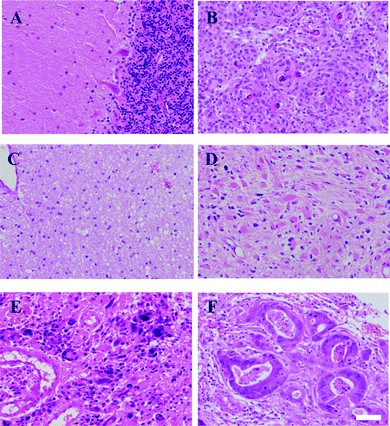 | ||
| Fig. 1 Microscopic images of normal and different tumour subtypes of the brain. Haematoxylin and eosin (H&E) staining of normal brain (Nor) tissue is shown in (A); low-grade tumours like meningioma (Men) in (B); Glioma WHO grade II or low-grade astrocytoma (LA) in (C); high-grade tumours like Glioma WHO grade III or anaplastic astrocytoma (AA) in (D); Glioma WHO grade IV or glioblastoma multiforme (GBM) in (E); and, metastatic brain tumours (Mets, primary colon cancer) in (F). Scale bar = 50 μm. | ||
IDH1 and p53 immunohistochemical analysis vs. biospectroscopy for glioma
The tissue samples from patients with gliomas (LA, AA and GBM) were stained with immunohistochemical markers, IDH1 and p53. Fig. 2A shows typical IDH1-R132H positive staining and Fig. 2B shows p53 staining of LA. Overall, positive IDH1 staining was observed in 33% (5/15) gliomas (Table 2) with three out of five LA (60%) staining positive, while one anaplastic astrocytoma (20%) and one glioblastoma (20%) were positive out of five each. For p53, positive staining was observed in 73% of the gliomas (11/15) with 80% of high-grade gliomas (AA and GBM) showing positive stain. Staining with p53 was also observed in 60% of LA. Samples that did not stain for IDH1 showed negative or minimal p53 staining.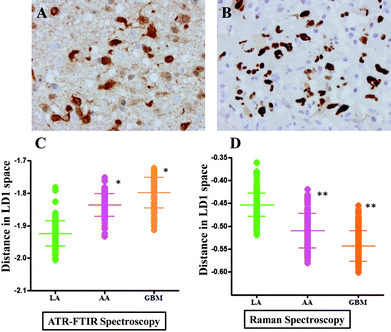 | ||
| Fig. 2 Comparative analysis of glioma tumours using immunohistochemistry (IDH1 and p53 staining) vs. vibrational spectroscopy. (A) shows characteristic staining for IDH1-R132H; and, (B) for p53 in low-grade glioma. LD1 scores plots from PCA-LDA representing spectra from LA, AA and GBM are shown in (C) for IR spectra and in (D) for Raman spectra. P-values from scores plot results using ANOVA test show that LA tumours were statistically significant from other gliomas (*, P ≤ 0.05; **, P ≤ 0.01). | ||
| IDH1 staining | Grades of glioma | ||
|---|---|---|---|
| LA (n = 5) | AA (n = 5) | GBM (n = 5) | |
| Positive (33%) | 3 | 1 | 1 |
| Negative (67%) | 2 | 4 | 4 |
| Grades of p53 staining | Grades of glioma | ||
|---|---|---|---|
| LA (n = 5) | AA (n = 5) | GBM (n = 5) | |
| Negative (27%) | 2 | 1 | 1 |
| 0–25 % | 1 | 3 | 1 |
| 26–50% | 1 | 0 | 1 |
| 51–75% | 1 | 1 | 0 |
| >75% | 0 | 0 | 2 |
IR and Raman spectra of LA, AA and GBM were analyzed using the PCA-LDA method and LD1 scores plots were obtained. As seen in Fig. 2, the LD1 scores of IR and Raman spectra for LA is significantly different from AA and GBM.
Average spectra and mean-derived spectra for IR and Raman
Fig. 3A and B shows the average spectra for normal brain tissue (n = 7) and brain tumours (n = 45) while in Fig. S1 [A and C; see ESI†] the tumours are sub-grouped according to the histological types. Overall, the IR spectra for brain tumours appear to overlap with the normal brain spectra in the biochemical-cell fingerprint region (1800 cm−1 to 900 cm−1), making it difficult to distinguish any subtle but significant differences. These similarities are attributable to dominant contributions of protein constituents. However, on closer inspection notable differences are tentatively identified in the region of 1050–1000 cm−1 (carbohydrates and collagen), 1300 cm−1 to 1150 cm−1 (Amide III and νasPO2−) and 1760 cm−1 to 1700 cm−1 (lipids, DNA/RNA and Amide I) in IR spectra, and at 950 cm−1 to 850 cm−1 (protein, tyrosine and collagen), 1265 cm−1 to 1240 cm−1 (Amide III) and 1600 cm−1 to 1530 cm−1 (amino acids and DNA/RNA) in Raman spectra.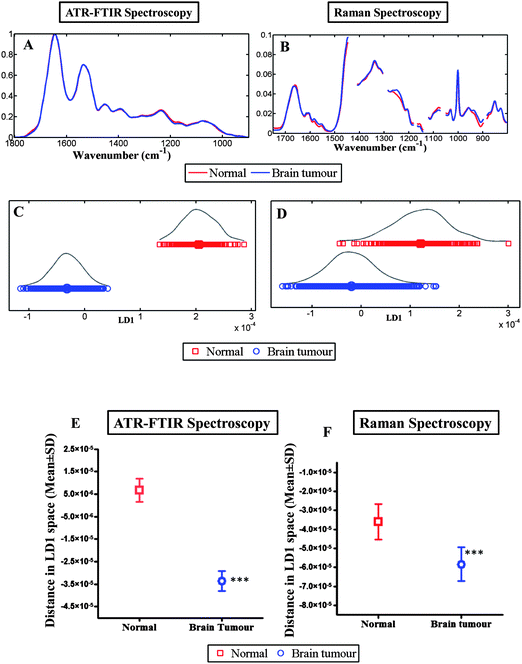 | ||
| Fig. 3 Compares the discriminating power of IR and Raman spectroscopy for normal brain tissue vs. brain tumours. (A) shows the average absorbance spectra of the biochemical-cell fingerprint regions for IR spectroscopy (1800 cm−1 to 900 cm−1); and, (B) for Raman spectroscopy (1750 cm−1 to 800 cm−1). (C and D) shows LD1 scores plot for IR and Raman spectroscopy respectively and represents the spectra from normal brain compared to brain tumours. (E and F) shows the mean ± SD of the spectral points. The difference of the spectral points for normal vs. brain tumour tissue is statistically significant (***, P ≤ 0.0001) for both techniques. | ||
The sub-grouped spectra of all tumour types show notable differences in the wavenumber regions 1100 cm−1 to 1050 cm−1 (Fig. S1A and C; see ESI†). This region corresponds to carbohydrate moieties and these differences may indicate variations in the ganglioside profile of human gliomas with increasing degree of malignancy.42,43 The 2nd-most prominent divergent spectral trend is seen in the region of 1550 cm−1 to 1400 cm−1, which tentatively is associated with protein moieties while the spectral alterations in the region of 1760 cm−1 to 1720 cm−1 may arise from the lipid constituents (e.g., phospholipids and possibly others).43
Multivariate analysis of IR and Raman spectra
Multivariate classification allows identification of diagnostic spectral patterns that remain valid for all spectra within a class, even though there may be substantial inter-class variability among spectra. On comparing spectra from normal brain to those of tumours in a 1-D scores plot using LDA (Fig. 3C), ATR-FTIR spectroscopy distinguished normal from tumour tissue without any overlap of spectral points, although a degree of overlap was evident in Raman spectra (Fig. 3D). The difference of the spectral points (mean ± SD) between normal and tumour tissue is statistically significant (P ≤ 0.0001) for both techniques. In IR spectra, the majority of the difference between normal and tumour subtypes are attributable to Amide I (1655 cm−1), Amide II (1547 and 1582 cm−1), Amide III (1304 cm−1), glycogen (1014 cm−1), carbohydrate (1173 cm−1) and νasPO2− (1234 cm−1), although there are also important contributions from protein bands (1454 and 1489 cm−1) and a lipid band (1740 cm−1). In the Raman dataset, the separation is attributed to spectral regions corresponding to CH2 deformation (1483 cm−1), tyrosine and proline (852 cm−1), ≈Amide III (1235 and 1276 cm−1), Amide I (1654 cm−1), phospholipids and glucose-I-phosphate (997 cm−1).Fig. 4A and B shows the 3-D scores plot derived after LDA of IR and Raman spectra, comparing all categories of brain tissues [i.e., Normal (Nor), meningioma (Men), low-grade astrocytoma (LA), anaplastic astrocytoma (AA), glioblastoma multiforme (GBM) and metastatic tumours (Mets)]. Good separation is observed between the normal brain tissue and different types of tumours in all LDs following ATR-FTIR spectroscopy, whereas following Raman spectroscopy a degree of overlap is observed with all tissue types except for LA in LD2 and LD3.
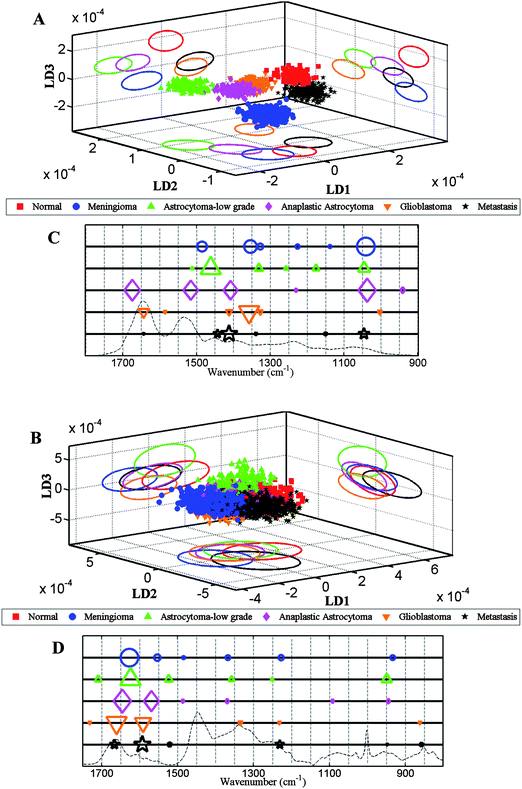 | ||
| Fig. 4 Comparison of LDA scores plots and cluster vector plots derived from the spectra acquired from normal brain and tumour tissue subtypes. (A) shows scores plot derived from IR spectra (n = 1040 spectra; 20 spectra per patient); (B) Raman spectra (n = 2600 spectra; 50 spectra per patient); and, (C and D) shows corresponding cluster vector plots (peak detection plots) with wavenumbers discriminating tumour subtypes from normal tissue. | ||
Inter-individual vs. disease differences
Despite the potential for the influence of confounding factors, Fig. S1 (B and D; see ESI†) shows that when per-individual spectra were averaged there was little evidence for marked inter-individual variability. Fig. 5 shows the inter-individual differences in the IR spectra of patients. To obtain this LD scores plot, IR spectra belonging to all patients were analyzed without providing the information about the tumour type to the LDA model. Thus, any observed clustering of the spectra is considered as “spontaneous”, indicating a common underlying biochemical signature for a particular tumour type within the cluster. Inter-individual differences in spectra allow one to distinguish small alterations in pathology from confounding factors that are a consequence of inter-individual differences. Spectra belonging to similar histological tissue types exhibit good clustering, although there is some degree of overlap between metastatic tumours, GBM and meningioma.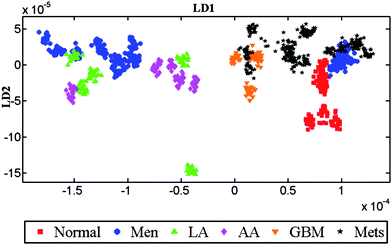 | ||
| Fig. 5 LDA scores plot of IR spectra showing the inter-class variance. To obtain this scores plot, each patient in the dataset is treated as a class without specifying the histological classes (i.e., normal or tumour). After LDA, individual patients' spectra are given a matching colour and symbol in accordance with their original tissue types. Thus, any observed clustering of patients would be spontaneous suggesting a common underlying biochemical signature. | ||
Lipid-to-protein ratio and phosphate-to-carbohydrate ratio in IR spectroscopy
The parameters for the tentatively-assigned lipid-to-protein ratio in each IR spectrum are derived from the intensity of lipid bands at 1740 cm−1 and of protein bands at 1400 cm−1. Table 3 shows the significance of difference in the lipid-to-protein ratio of various tumour types compared to normal brain tissue. The lipid-to-protein ratio of IR spectra was high in normal tissue and considerably decreased in meningiomas, high-grade gliomas and metastatic brain tumours (Fig. 6A, Table 3).| Normal vs. tumour type | P-value |
|---|---|
| Normal vs. Men | P ≤ 0.001 |
| Normal vs. LA | P > 0.05 |
| Normal vs. AA | P ≤ 0.01 |
| Normal vs. GBM | P ≤ 0.01 |
| Normal vs. Mets | P ≤ 0.001 |
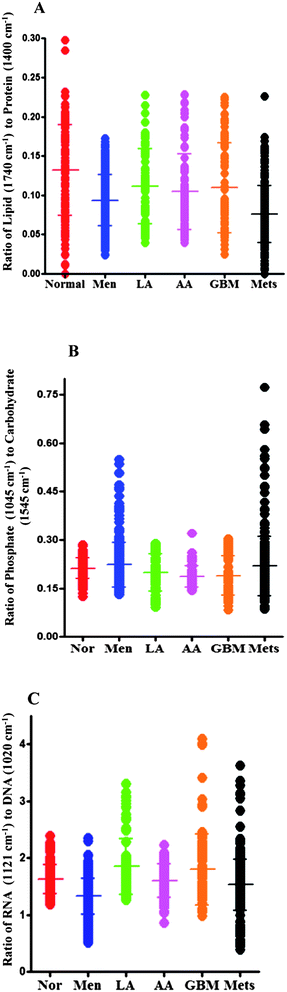 | ||
| Fig. 6 (A) Shows 1-D scores plot of lipid-to-protein ratio (based on the intensity at wavenumbers 1740 cm−1/intensity at 1400 cm−1) for normal brain tissue and different types of tumours. The transverse bars represent mean ± SD of spectrally-derived estimations for a particular tissue type. The ratio of lipid to protein is higher in normal brain tissue compared to tumours with significant difference between normal tissue and meningioma (P ≤ 0.001), high-grade gliomas (AA and GBM) (P ≤ 0.01) and metastatic tumours (P ≤ 0.001) but not with low-grade glioma (LA). (B) shows 1-D scores plots of phosphate (1045 cm−1) to carbohydrate (1545 cm−1) ratio for normal brain tissue and brain tumours. Differences in the ratio are apparent between normal tissue and high-grade gliomas (AA and GBM) (P ≤ 0.001). (C) shows ratio of RNA (1121 cm−1) to DNA (1020 cm−1) comparing the spectral points acquired by LDA for normal brain tissue and various tumour grades. The RNA-to-DNA ratio is significantly altered from normal brain tissue compared to meningioma (P ≤ 0.001) and to a lesser extent in metastatic tumours (P ≤ 0.05). | ||
Fig. 6B shows the ratio of phosphate-to-carbohydrate which is obtained by calculating the ratio of band intensities at 1045 cm−1 and 1545 cm−1 in each IR spectrum. This scale may provide information on metabolic turnover in tissues. The phosphate-to-carbohydrate ratio is reduced in high-grade gliomas (AA and GBM), but there is no significant difference in the ratio between normal tissue compared to meningioma, low-grade glioma or metastatic tumours (Table 4). The ratio of IR peak intensities located at 1121 cm−1 and 1020 cm−1 giving an RNA/DNA ratio can be used as a potential biomarker to predict the cell proliferation in the normal or malignant tissue.44,45 The RNA/DNA ratio in IR spectra was reduced significantly in meningioma (P ≤ 0.001) and to a lesser extent in metastatic tumours (P ≤ 0.05) compared to normal brain tissue (Fig. 6C, Table 5).
| Normal vs. tumour type | P-value |
|---|---|
| Normal vs. Men | P > 0.05 |
| Normal vs. LA | P > 0.05 |
| Normal vs. AA | P ≤ 0.001 |
| Normal vs. GBM | P ≤ 0.001 |
| Normal vs. Mets | P > 0.05 |
| Normal vs. tumour type | P-value |
|---|---|
| Normal vs. Men | P ≤ 0.001 |
| Normal vs. LA | P > 0.05 |
| Normal vs. AA | P > 0.05 |
| Normal vs. GBM | P > 0.05 |
| Normal vs. Mets | P > 0.05 |
Two-category discriminant analysis using PCA-LDA following IR spectroscopy
Figs. S2–S6-A (see ESI†) show 1-D scores plots comparing the IR spectra of normal brain tissue with each tumour type and Figs. S2–S6-C (see ESI†) show the corresponding loadings plots identifying wavenumbers responsible for the separation. Significant differences are observed between normal vs. individual tumour subtypes with some degree of overlap with normal in all classes. The top six distinguishing wavenumbers for each comparison with their tentative biochemical assignments are presented in Table 6. The top six tentatively-assigned wavenumbers responsible for the segregation of normal tissue from meningioma are: 1018 cm−1 (glycogen), 1173 cm−1(carbohydrate), 1543 and 1620 cm−1 (Amide I), 1582 cm−1 (Amide II) and 1740 cm−1 (lipids). The wavenumbers separating low-grade astrocytoma from normal are 1103 cm−1, 1234 cm−1 (νasPO2−), 1470 cm−1 (lipids), 1504 cm−1 (Amide II) and 1628/1686 cm−1 (Amide I). Comparison of normal tissue with anaplastic astrocytoma shows good separation and the wavenumbers responsible for segregation are 1018 cm−1 (glycogen), 1234 cm−1 (νasPO2−), 1489 cm−1 (CH bending vibration), 1551 cm−1 (Amide II), 1628 cm−1 (Amide I) and 1701 cm−1 (lipid). The tentative assignments of wavenumbers for AA appear similar to those found in LA, possibly suggesting a similar pathological process in both tumour types affecting the identical molecules but with a varying degree of biochemical alterations. The degree of alterations responsible for diagnostic segregation is reflected by the variation in the intensity of the IR and Raman spectra. Normal vs. GBM has 1107 cm−1 (glycogen), 1393 cm−1 (COO– symmetric stretching), 1474 cm−1 (proteins), 1531 cm−1 (Amide II), 1585/1659 cm−1 (Amide I) as segregating wavenumbers. Normal tissue is separated from metastatic tumours by following top six wavenumbers: 1173 cm−1 (carbohydrate), 1489 cm−1 (proteins), 1543 cm−1 (Amide II), 1632/1659 cm−1 (Amide I) and 1740 cm−1 (lipids).| Comparisons | Top six discriminating wavenumbers with tentative biochemical assignments | |||
|---|---|---|---|---|
| ATR FTIR spectroscopy | Raman spectroscopy | |||
| Normal vs. meningioma | 1018 | Glycogen | ≈911 | C–C stretching of proline ring/glucose/lactic acid |
| 1173 | Carbohydrate | ≈964 | Lipids, proteins (CH3 deformations) | |
| 1543 | Amide I | ≈1237 | Amide III | |
| 1582 | Amide II | 1276 | Amide III (α-helix) | |
| 1620 | Amide I | 1485 | Lipids and proteins (CH2 deformation), purine ring(guanine) | |
| 1740 | Lipids | 1655 | Amide I/lipids | |
| Normal vs. LA | 1103 | ν sPO2− | 800 | Undefined |
| 1234 | ν asPO2− | 903 | Undefined | |
| 1470 | CH2 bending of the methylene chains in lipids | ≈999 | Glucose-I-phosphate and symmetric ring breathing mode of phenylalanine | |
| 1504 | Amide II | 1306 | Lipids, collagen, protein amide III, DNA purine bases, phenylalanine | |
| 1628 | Amide I | 1446 | Proteins and lipids (CH2 bending mode of proteins and lipids) | |
| 1686 | Amide I | 1670 | Cholesterol esters, Amide 1 | |
| Normal vs. AA | 1018 | Glycogen | 810 | Undefined |
| 1234 | Asymmetric phosphate | 853 | Tyrosine, proline, glycogen | |
| 1489 | In-plane CH bending vibration | 911 | C–C stretching of proline ring/glucose/lactic acid | |
| 1551 | Amide II | 1004 | Lipids and proteins, phenylalanine | |
| 1628 | Amide I | ≈1455 | Protein δ (CH2/CH3) | |
| 1701 | Lipid | 1670 | Cholesterol esters, Amide 1 | |
| Normal vs. GBM | 1107 | Glycogen | ≈849 | Tyrosine and proline, glycogen |
| 1393 | COO– symmetric stretching | 904 | Undefined | |
| 1474 | Proteins? | ≈917 | C–C stretching, glycogen, lactic acid | |
| 1531 | Amide II, Modified guanine? | 1001 | Phenylalanine (symmetric ring breathing mode) | |
| 1585 | Amide I | ≈1473 | CH2 deformation | |
| 1659 | Amide I | 1673 | Lipids, Amide I | |
| Normal vs. metastasis | 1173 | Carbohydrate | ≈997 | Phospholipids, glucose-I-phosphate |
| 1489 | In-plane CH bending vibration | 1077 | Lipids (C–C vibrations) | |
| 1543 | Amide II | 1241 | Amide III | |
| 1632 | Amide I | 1446 | Proteins and lipids (CH2 bending mode of proteins and lipids) | |
| 1659 | Amide I | ≈1460 | Cytosine | |
| 1740 | C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching (lipids) O stretching (lipids) |
1654 | Amide I (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching mode of proteins, α-helix conformation)/C O stretching mode of proteins, α-helix conformation)/C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C lipid stretch C lipid stretch |
|
Two-category discriminant analysis using PCA-LDA following Raman spectroscopy
Figs. S2–S6-B (see ESI†) show 1-D scores plots comparing the Raman spectra of normal brain tissue with each tumour subtype and Figs. S2–S6-D (see ESI†) show the corresponding loadings plots identifying the wavenumbers responsible for the separation. The wavenumbers separating normal tissue from meningioma are 911 cm−1 (C–C stretching of proline ring/glucose/lactic acid), ≈964 cm−1 and 1485 cm−1 (lipids, proteins), ≈1237 cm−1 (Amide III), 1276 cm−1 (Amide III α-helix) and 1655 cm−1 (Amide I/lipids), while wavenumbers 999 cm−1 (glucose-I-phosphate and symmetric ring breathing mode of phenylalanine), 1306 cm−1 (lipids, collagen, Amide III, DNA purine bases, phenylalanine), 1446 cm−1 (proteins and lipids) and 1670 cm−1(cholesterol esters) separated normal tissue from low-grade astrocytoma. Segregation of anaplastic astrocytoma from normal tissue is due to the following wavenumbers: 853 cm−1 (tyrosine and proline), 911 cm−1 (C–C stretching of proline ring/glucose/lactic acid), 1004 cm−1 (phenylalanine, lipids and proteins), ≈1455 cm−1 (proteins) and 1670 cm−1 (cholesterol esters).30 GBM are distinguished from normal tissue by wavenumbers ≈849 cm−1 (tyrosine and proline), 1001 cm−1 (phenylalanine), 1473 cm−1 (CH2 deformation) and 1673 cm−1 (lipids, Amide I). Wavenumbers separating normal from metastatic tumours are 997 cm−1 (phospholipids, glucose-I-phosphate), 1241 cm−1 (Amide III), ≈1460 cm−1 (cytosine), 1654 cm−1 (Amide I), 1077 cm−1 and 1446 cm−1(lipids, proteins).Phospholipids-to-nucleic acid/proteins ratio and cholesterol esters-to-phenylalanine ratio in Raman spectroscopy
The ratio of phospholipids-to-nucleic acids/proteins was obtained by calculating the Raman peak intensities at 1745 cm−1 (phospholipids) and 1335 cm−1 (nucleic acids/proteins). This ratio was evaluated for normal brain and tumour subtypes (Fig. S7; see ESI†). The importance of this ratio lies in the fact that it appears to correlate with histopathological studies grading the malignancy by the nucleic acid-to-cytoplasmic ratio.46–49 The ratio of phospholipid-to-nucleic acid in Raman spectra was reduced in meningioma, high-grade gliomas and metastatic tumours.When the ratio of cholesterol esters (1670 cm−1) to phenylalanine (1001 cm−1) for normal brain tumour was compared with tumour subtypes, we observed an increase in the ratio for meningioma, whereas the ratio was reduced for LA, GBM and metastatic tumours. The ratio difference was most noticeable between meningioma and low-grade astrocytoma with reduction in the mean ± SD of spectral intensity ratio (Fig. S8; see ESI†). Additionally, the ratio of intensities at wavenumbers 1654 cm−1 (Amide 1 α-helix) to 1446 cm−1 (CH2 bending mode of proteins and lipids) in Raman spectra showed significant reduction from normal brain tissue to Men, LA, AA and metastatic tumours (Fig. S9; see ESI†). This ratio has been used to differentiate normal and cancerous tissues at different sites including brain, breast and gynaecological tissues.50–52
Discussion
This exploratory study demonstrates that ATR-FTIR spectroscopy and Raman spectroscopy have the potential to distinguish between normal brain and tumour tissue (Fig. 3E and F), as well as between various subtypes thereof. The most conspicuous differences are between meningioma and normal tissue. This finding is significant as meningiomas are amenable to surgical resection with good survival rates; resection with clear margins will reduce the risk of disease recurrence. LA, AA and GBM are also seen to separate from one another by both IR and Raman spectroscopic methods (Fig. 2C and D). Wavenumber assignment allows the identification of molecular markers that can be utilized to separate normal brain tissue from tumours and multivariate analysis with LDA or PCA-LDA allows further interrogation of the spectral data comparing normal brain tissue with various tumours. The advantage of using LDA is that it seeks directions in the original dataset (wavenumber variables) where the ratio of between-class variance compared to within-class variance is maximal. The disadvantage of this method is potential over-fitting of the spectra. This may be avoided by having a large spectral dataset with at least four to five times the number of spectra compared to the number of variables.As seen in the 3-D scores plot in Fig. 4(A and B), ATR-FTIR spectroscopy is able to distinguish meningioma from various grades of gliomas as well as from metastatic tumours, while Raman spectroscopy was able to distinguish low-grade glioma from high-grade gliomas and from normal brain tissue. From a clinical as well as histological perspective, low-grade gliomas are poorly demarcated and the ability of vibrational spectroscopy to distinguish their extent during surgery could be of potential advantage. Moreover, it is often difficult to determine where the tumour ends and normal tissue begins either by imaging, direct observation during surgery or gross pathological examination.53
LD1 scores plots following PCA-LDA of normal brain compared to individual tumour subtypes for IR (Figs. 2–6, panel A) and Raman spectra (Figs. 2–6, panel B) showed separation of tissue classes with a degree of overlap between them. These between-class similarities and differences could be attributable to the inherent heterogeneity of tumour tissues. Brain tumours may contain various grades of neoplastic tissue, stromal elements, haemorrhage and necrotic tissue, making the spatially-resolved IR spectral data derived from μm-size tissue sample sections non-representative of the tumour class. Despite this limitation important molecular markers were identified by wavenumber assignments resulting in segregation of normal tissue from different tumour types. In derived PCA-LDA scores plots (Fig. 5), marked within-class variation (i.e., heterogeneity) was noted; even so, good discrimination between different grades of glioma was also observed (i.e., towards between-category discriminating biomarkers). This is explained by the fact that the gene and protein expression of morphologically-similar astrocytoma tissues can vary depending on the patients' tumour grade.
The transition of normal brain tissue to neoplastic tissue is connected with qualitative and quantitative changes of lipids. Lipid-to-protein ratio is of particular interest due to its ability to distinguish normal brain tissue and tumour tissue. Tumour tissue shows marked decreases in the bands associated with lipids and subtle changes in the main protein bands. Earlier studies have shown that the magnitude of the lipid-to-protein ratio correlates with the progression of malignancy in gliomas.54 As seen in Fig. 6A, the lipid-to-protein ratio decreases significantly going from normal to glioma tissue as well as in metastatic brain tumours. However the decrease in the band intensity does not appear to reflect the worsening grades of glioma in our study. A plausible explanation for this could be that although astrocytic gliomas are classified in grade II–IV, it is not an absolute classification. A single tissue section of any grade of glioma may still encompass regions characteristic of all four grades of malignancy.43 We also observed reductions in the phosphate-to-carbohydrate ratio in grade III and grade IV gliomas compared to normal brain tissue (Fig. 6B, Table 3), possibly suggesting similar underlying biochemical alterations in grade III and grade IV gliomas. Certainly it is arguable from the above ratio observations that glioma grade III (anaplastic astrocytoma) is not in the middle of the biological spectrum but closer to the highly malignant glioma grade IV (glioblastoma). The ratio of phospholipids to proteins in Raman spectra (Fig. S7; see ESI†) for normal brain and different tumour subtypes has shown a decreasing trend with progression of gliomas. As well as the reduction in lipids with increasing grade of malignancy, relative increases in nucleic acids and proteins in tumour tissues may contribute to the reduction in the ratio.
The concentration of minor lipid cholesterol esters can increase up to 100 times in gliomas compared to the trace amounts found in normal brain tissue.55 In a recent study utilizing Raman spectroscopy for grading of astrocytoma, phenylalanine bands appeared to give important contributions discriminating high-grade gliomas (AA and GBM) compared to normal tissue.30 Reduced band intensities for phenylalanine had been reported in dysplastic tissue compared to normal tissue in previous studies.48,56 Koljenović et al. discriminated vital from necrotic glioblastoma tissues by Raman spectroscopy;57 they demonstrated that necrotic tissue contains higher levels of cholesterol than vital tumour tissue. Yamada et al. came to the same conclusions by comparing necrotic and vital carcinoma tissues.58 In our study, the most prominent contribution for distinction between normal brain tissue and gliomas is at 1670–1674 cm−1 in Raman spectra, corresponding to cholesterol and cholesterol esters.30,59–61 The second prominent band resulting into segregation of normal tissue from gliomas is at 1001 cm−1 corresponding to phenylalanine. The ratio of cholesterol esters to phenylalanine (Fig. S8; see ESI†) has the potential to be used as a marker to differentiate between meningioma and low-grade astrocytoma. In Raman spectra, the wavenumber contribution at 850 cm−1 (tyrosine and proline) is able to discriminate normal tissue from glioblastoma in our study. Similar observations were made in a recent study in which the bands at 850 cm−1 in Raman spectroscopy were considered to give evidence of high-grade tumours.30
Isocitrate dehydrogenase 1 (IDH1) mutations have recently been identified as early and frequent genetic alterations in astrocytomas and secondary glioblastomas, whereas primary glioblastomas very rarely contain IDH1 mutations. IDH1 expression is emerging as an important biomarker for gliomas with about 80% low-grade gliomas staining positive for IDH1-R132H mutation.17 In addition, several studies have demonstrated that an IDH1 mutation is associated with good prognosis and can be utilized as marker of prognosis in gliomas.17,35 In our cohort of gliomas (Table 2), IDH1 staining was positive for 60% of LA with an overall positive rate of 33% for all gliomas. A recent study, using the same antibody, has found similar low positive results for IDH1 (23.72%).62 When the glioma samples were analyzed to identify differences in the spectral intensity for both IR and Raman spectra, we found significant (P ≤ 0.01) differences in the spectra between all three grades of gliomas (Fig. 2C and D). This finding shows the ability of biospectroscopy to distinguish different grades of gliomas. Spectral biomarkers appeared to provide more robust identification of aberrant tissue than immunohistochemical markers.
The major limitation of ATR-FTIR spectroscopy lies in the fact that it is an ex vivo technique and can only be performed on fixed tissue. Raman spectroscopy is one of the optical spectroscopy techniques currently under investigation for in vivo endoscopic applications. In contrast to the ex vivo histological analysis, the concept of in vivo Raman spectroscopy is the distinction of tissues within the tumour intra-operatively. Studies using fibre-optic Raman probes under in vivo conditions have been reported for the oesophagus,63 brain,29 pre-cancer lesions in the cervical epithelium,64 lesions of breast tissue,65 and polyps in colon.66
It would be expected that substantial modifications occur at the molecular level during the process of development of brain tumours before visible changes are apparent on histological assessment by conventional H&E staining and microscopic examination for structural changes. ATR-FTIR spectroscopy and/or Raman spectroscopy allows qualitative and quantitative analysis of basic cellular components like lipids, proteins, carbohydrates and nucleic acids within biological tissues. IR/Raman spectroscopy thus has the potential to detect early changes in tissue resulting from the development of the tumour which may not be apparent in tissue sections or cell preparations.67 The information obtained by IR/Raman spectroscopy can be combined with conventional methods like histopathological grading of smears/biopsies from tumour margins or stereotactic biopsies to diagnose and grade brain tumours. This will allow for more accurate planning and execution of surgery and/or radiation therapy resulting in the concept of personalized medicine for individualized treatment with potentially better long-term survival and cure rates.
Conclusion
In this exploratory study, we demonstrate that interrogation of human brain tissue with ATR-FTIR or Raman spectroscopy allows one to readily segregate normal tissue from brain tumours. The responsible molecular changes for this segregation were primarily alterations in lipids and proteins with a reduction in the tentatively-assigned lipid (1740 cm−1)-to-protein (1400 cm−1) ratio going from normal brain tissue to meningioma and gliomas, with a marked decrease in metastatic tumours. Certain ratios have the potential to be used as a spectral biomarker for diagnosing primary brain tumours in both IR and Raman spectroscopy. In IR spectroscopy, alterations in phosphate (1045 cm−1)-to-carbohydrate (1545 cm−1) were only seen in high-grade gliomas, whereas RNA (1121 cm−1)-to-DNA (1020 cm−1) ratio was significantly altered only in meningioma. The ratio of cholesterol esters (1670 cm−1) to phenylalanine (1001 cm−1) appears to differentiate meningioma from low-grade astrocytoma in Raman spectroscopy. IR and Raman spectroscopy were able to segregate various grades of glioma more readily compared to current staining methods like IDH1 and p53. Like IDH1, spectroscopy has the potential to be used as a prognostic marker of survival. Further validation of this approach exploiting biospectroscopy techniques with an appropriate data architecture68 utilising a new extended set of samples is required.Conflict of interest statement: none declared
None of the authors have financial interests or any other interests that might be prejudicial to the interpretation or presentation of the work contained within this manuscript.Abbreviations
| AA | Anaplastic astrocytoma |
| 5-ALA | 5-Aminolevulinic acid |
| ATR | Attenuated total reflection |
| FFPE | Formalin-fixed paraffin-embedded |
| FTIR | Fourier-transform infrared |
| GBM | Glioblastoma multiforme |
| H&E | Haematoxylin and eosin |
| IDH1 | Isocitrate dehydrogenase-1 |
| IHC | Immunohistochemistry |
| iMRI | Intraoperative MRI |
| IR | Infrared |
| LA | Low-grade astrocytoma |
| LDA | Linear discriminant analysis |
| Men | Meningioma |
| Mets | Metastatic tumours |
| Nor | Normal |
| PC | Principal component |
| PCA | Principal component analysis |
| WHO | World Health Organization. |
Acknowledgements
Rosemere Cancer Foundation supported this work. We thank theatre staff, nurses, porters, neuropathology laboratory staff, and patients of Lancashire Teaching Hospitals NHS Trust who facilitated this study.References
- M. J. Quinn, P. Babb, A. Brock, L. Kirby and J. Jones, Cancer Trends in England and Wales 1950–1999, Office of National Statistics, London, 2001 Search PubMed.
- Cancer Research UK, Brain and other CNS tumour statistics – UK, http://info.cancerresearchuk.org/cancerstats/types/brain, accessed 6 January, 2012.
- P. D. Sasieni, J. Shelton, N. Ormiston-Smith, C. S. Thomson and P. B. Silcocks, Br. J. Cancer, 2011, 105(3), 460–465 CrossRef CAS.
- D. N. Louis, H. Ohgaki, O. D. Wiestler, W. K. Cavenee, P. C. Burger, A. Jouvet, B. W. Scheithauer and P. Kleihues, Acta Neuropathol., 2007, 114(2), 97–109 CrossRef.
- B. W. Stewart and P. Kleihues, Tumours of the Nervous System, International Agency for Research on Cancer, Lyon, 2003 Search PubMed.
- J. Gu, Y. Liu, A. P. Kyritsis and M. L. Bondy, Neurotherapeutics, 2009, 6(3), 427–435 CrossRef CAS.
- M. Wrensch, Y. Minn, T. Chew, M. Bondy and M. S. Berger, Neuro-Oncology, 2002, 4(4), 278–299 Search PubMed.
- J. M. Legler, L. A. Ries, M. A. Smith, J. L. Warren, E. F. Heineman, R. S. Kaplan and M. S. Linet, J. Natl. Cancer Inst., 1999, 91(16), 1382–1390 CrossRef CAS.
- B. J. McCarthy, F. G. Davis, S. Freels, T. S. Surawicz, D. M. Damek, J. Grutsch, H. R. Menck and E. R. Laws, Jr, J. Neurosurg., 1998, 88(5), 831–839 CrossRef CAS.
- M. Nakamura, N. Konishi, S. Tsunoda, H. Nakase, T. Tsuzuki, H. Aoki, H. Sakitani, T. Inui and T. Sakaki, Oncology, 2000, 58(2), 108–116 CrossRef CAS.
- T. Kinjo, O. al-Mefty and I. Kanaan, Neurosurgery, 1993, 33(3), 394–399 CrossRef CAS.
- D. N. Louis, Annu. Rev. Pathol.: Mech. Dis., 2006, 1, 97–117 CrossRef CAS.
- I. T. Gavrilovic and J. B. Posner, J. Neuro-Oncol., 2005, 75(1), 5–14 CrossRef.
- L. J. Schouten, J. Rutten, H. A. Huveneers and A. Twijnstra, Cancer, 2002, 94(10), 2698–2705 CrossRef.
- K. S. Polyzoidis, G. Miliaras and N. Pavlidis, Cancer Treat. Rev., 2005, 31(4), 247–255 CrossRef.
- J. S. Barnholtz-Sloan, A. E. Sloan, F. G. Davis, F. D. Vigneau, P. Lai and R. E. Sawaya, J. Clin. Oncol., 2004, 22(14), 2865–2872 CrossRef.
- D. Capper, F. Sahm, C. Hartmann, R. Meyermann, A. von Deimling and J. Schittenhelm, Am. J. Surg. Pathol., 2010, 34(8), 1199–1204 CrossRef.
- W. C. Allsbrook, Jr, K. A. Mangold, M. H. Johnson, R. B. Lane, C. G. Lane and J. I. Epstein, Hum. Pathol., 2001, 32(1), 81–88 CrossRef.
- R. Stupp, M. Reni, G. Gatta, E. Mazza and C. Vecht, Crit. Rev. Oncol. Hematol., 2007, 63(1), 72–80 CrossRef.
- N. Sanai and M. S. Berger, Neurotherapeutics, 2009, 6(3), 478–486 CrossRef.
- S. N. Kalkanis, D. Kondziolka, L. E. Gaspar, S. H. Burri, A. L. Asher, C. S. Cobbs, M. Ammirati, P. D. Robinson, D. W. Andrews, J. S. Loeffler, M. McDermott, M. P. Mehta, T. Mikkelsen, J. J. Olson, N. A. Paleologos, R. A. Patchell, T. C. Ryken and M. E. Linskey, J. Neuro-Oncol., 2010, 96(1), 33–43 CrossRef.
- J. H. Sherman, K. Hoes, J. Marcus, R. J. Komotar, C. W. Brennan and P. H. Gutin, Curr. Neurol. Neurosci. Rep., 2011, 11(3), 313–319 CrossRef.
- W. Stummer, S. Stocker, S. Wagner, H. Stepp, C. Fritsch, C. Goetz, A. E. Goetz, R. Kiefmann and H. J. Reulen, Neurosurgery, 1998, 42(3), 518–525 CrossRef CAS.
- F. W. Floeth, M. Sabel, C. Ewelt, W. Stummer, J. Felsberg, G. Reifenberger, H. J. Steiger, G. Stoffels, H. H. Coenen and K. J. Langen, Eur. J. Nucl. Med. Mol. Imaging, 2011, 38(4), 731–741 CrossRef CAS.
- F. L. Martin, Nat. Methods, 2011, 8(5), 385–387 CrossRef CAS.
- D. C. Fernandez, R. Bhargava, S. M. Hewitt and I. W. Levin, Nat. Biotechnol., 2005, 23(4), 469–474 CrossRef CAS.
- J. G. Kelly, J. Trevisan, A. D. Scott, P. L. Carmichael, H. M. Pollock, P. L. Martin-Hirsch and F. L. Martin, J. Proteome Res., 2011, 10(4), 1437–1448 CrossRef CAS.
- F.L. Martin, J.G. Kelly, V. Llabjani, P.L. Martin-Hirsch, I.I. Patel, J. Trevisan, N.J. Fullwood and M.J. Walsh, Nat. Protoc., 2010, 5(11), 1748–1760 CrossRef CAS.
- M. Kirsch, G. Schackert, R. Salzer and C. Krafft, Anal. Bioanal. Chem., 2010, 398(4), 1707–1713 CrossRef CAS.
- C. Beleites, K. Geiger, M. Kirsch, S. B. Sobottka, G. Schackert and R. Salzer, Anal. Bioanal. Chem., 2011, 400(9), 2801–2816 CrossRef CAS.
- J. Trevisan, P. P. Angelov, I. I. Patel, G. M. Najand, K. T. Cheung, V. Llabjani, H. M. Pollock, S. W. Bruce, K. Pant, P. L. Carmichael, A. D. Scott and F. L. Martin, Analyst, 2010, 135(12), 3266–3272 RSC.
- E. O. Faoláin, M. B. Hunter, J. M. Byrne, P. Kelehan, H. A. Lambkin, H. J. Byrne and F. M. Lyng, J. Histochem. Cytochem., 2005, 53(1), 121–129 CrossRef.
- B. D. Beier and A. J. Berger, Analyst, 2009, 134(6), 1198–1202 RSC.
- M. N. Singh, H. F. Stringfellow, M. J. Walsh, K. M. Ashton, E. Paraskevaidis, K. R. Abdo, P. L. Martin-Hirsch, D. H. Phillips and F. L. Martin, Toxicology, 2008, 249(1), 85–90 CrossRef CAS.
- S. Takano, W. Tian, M. Matsuda, T. Yamamoto, E. Ishikawa, M. K. Kaneko, K. Yamazaki, Y. Kato and A. Matsumura, Brain Tumor Pathol., 2011, 28(2), 115–123 CrossRef CAS.
- F. S. Pardo, D. W. Hsu, R. Zeheb, J. T. Efird, P. G. Okunieff and D. M. Malkin, Br. J. Cancer, 2004, 91(9), 1678–1686 CAS.
- V. Llabjani, J. Trevisan, K. C. Jones, R. F. Shore and F. L. Martin, Environ. Sci. Technol., 2011, 45(14), 6129–6135 CrossRef CAS.
- T. Hastie, R. Tibshirani and J. H. Friedman, Elements of Statistical Learning: Data Mining, Inference, and Prediction, Springer, New York, 2009 Search PubMed.
- V. Llabjani, J. D. Crosse, A. A. Ahmadzai, I. I. Patel, W. Pang, J. Trevisan, K. C. Jones, R. F. Shore and F. L. Martin, Environ. Sci. Technol., 2011, 45(24), 10706–10712 CrossRef CAS.
- V. Llabjani, J. Trevisan, K. C. Jones, R. F. Shore and F. L. Martin, Environ. Sci. Technol., 2010, 44(10), 3992–3998 CrossRef CAS.
- M. J. German, A. Hammiche, N. Ragavan, M. J. Tobin, L. J. Cooper, S. S. Matanhelia, A. C. Hindley, C. M. Nicholson, N. J. Fullwood, H. M. Pollock and F. L. Martin, Biophys. J., 2006, 90(10), 3783–3795 CrossRef CAS.
- F. S. Parker, Applications of IR, Raman, and Resonance Raman Spectroscopy in Biochemistry, Plenum, New york, 1983 Search PubMed.
- G. Steiner, A. Shaw, L. P. Choo-Smith, M. H. Abuid, G. Schackert, S. Sobottka, W. Steller, R. Salzer and H. H. Mantsch, Biopolymers, 2003, 72(6), 464–471 CrossRef CAS.
- E. Gazi, J. Dwyer, P. Gardner, A. Ghanbari-Siahkali, A. P. Wade, J. Miyan, N. P. Lockyer, J. C. Vickerman, N. W. Clarke, J. H. Shanks, L. J. Scott, C. A. Hart and M. Brown, J. Pathol., 2003, 201(1), 99–108 CrossRef CAS.
- R. K. Sahu, S. Argov, A. Salman, M. Huleihel, N. Grossman, Z. Hammody, J. Kapelushnik and S. Mordechai, Technol. Cancer Res. Treat., 2004, 3(6), 629–638 CAS.
- S. K. Teh, W. Zheng, K. Y. Ho, M. Teh, K. G. Yeoh and Z. Huang, Br. J. Cancer, 2008, 98(2), 457–465 CrossRef CAS.
- G. Y. Lauwers and R. H. Riddell, Gut, 1999, 45(5), 784–790 CrossRef CAS.
- Z. Huang, A. McWilliams, H. Lui, D. I. McLean, S. Lam and H. Zeng, Int. J. Cancer, 2003, 107(6), 1047–1052 CrossRef CAS.
- J. R. Mourant, K. W. Short, S. Carpenter, N. Kunapareddy, L. Coburn, T. M. Powers and J. P. Freyer, J. Biomed. Opt., 2005, 10(3), 031106 CrossRef.
- A. Mizuno, H. Kitajima, K. Kawauchi, S. Muraishi and Y. Ozaki, J. Raman Spectrosc., 1994, 25, 25–29 CrossRef CAS.
- C. J. Frank, R. L. McCreery and D. C. Redd, Anal. Chem., 1995, 67(5), 777–783 CrossRef CAS.
- A. Mahadevan-Jansen and R. Richards-Kortum, J. Biomed. Opt., 1996, 1, 31–70 CrossRef.
- D. Schiff, P. D. Brown and C. Giannini, Neurology, 2007, 69(13), 1366–1373 CrossRef.
- C. Krafft, S. B. Sobottka, G. Schackert and R. Salzer, Analyst, 2004, 129(10), 921–925 RSC.
- C. Nygren, H. von Holst, J. E. Mansson and P. Fredman, Br. J. Neurosurg., 1997, 11(3), 216–220 CrossRef CAS.
- N. Stone, C. Kendall, N. Shepherd, P. Crow and H. Barr, J. Raman Spectrosc., 2002, 33, 564–573 CrossRef CAS.
- S. Koljenović, L. P. Choo-Smith, T. C. Bakker Schut, J. M. Kros, H. J. van den Berge and G. J. Puppels, Lab. Invest., 2002, 82(10), 1265–1277 Search PubMed.
- T. Yamada, N. Miyoshi, T. Ogawa, K. Akao, M. Fukuda, T. Ogasawara, Y. Kitagawa and K. Sano, Clin. Cancer Res., 2002, 8(6), 2010–2014 CAS.
- A. Beljebbar, S. Dukic, N. Amharref and M. Manfait, Anal. Bioanal. Chem., 2010, 398(1), 477–487 CrossRef CAS.
- A. Beljebbar, N. Amharref, A. Leveques, S. Dukic, L. Venteo, L. Schneider, M. Pluot and M. Manfait, Anal. Chem., 2008, 80(22), 8406–8415 CrossRef CAS.
- M. Kohler, S. Machill, R. Salzer and C. Krafft, Anal. Bioanal. Chem., 2009, 393(5), 1513–1520 CrossRef.
- P. Birner, K. Toumangelova-Uzeir, S. Natchev and M. Guentchev, Folia Neuropathol., 2011, 49(2), 88–93 Search PubMed.
- M. S. Bergholt, W. Zheng, K. Lin, K. Y. Ho, M. Teh, K. G. Yeoh, J. B. So and Z. Huang, Technol. Cancer Res. Treat., 2011, 10(2), 103–112 CAS.
- A. Mahadevan-Jansen, M. F. Mitchell, N. Ramanujam, U. Utzinger and R. Richards-Kortum, Photochem. Photobiol., 1998, 68(3), 427–431 CrossRef CAS.
- J. T. Motz, S. J. Gandhi, O. R. Scepanovic, A. S. Haka, J. R. Kramer, R. R. Dasari and M. S. Feld, J. Biomed. Opt., 2005, 10(3), 031113 CrossRef.
- A. Molckovsky, L. M. Song, M. G. Shim, N. E. Marcon and B. C. Wilson, Gastrointest. Endosc., 2003, 57(3), 396–402 CrossRef.
- I. I. Patel, J. Trevisan, P. B. Singh, C. M. Nicholson, R. K. Krishnan, S. S. Matanhelia and F. L. Martin, Anal. Bioanal. Chem., 2011, 401(3), 969–982 CrossRef CAS.
- J. Trevisan, P. P. Angelov, P. L. Carmichael, A. D. Scott and F. L. Martin, Analyst, 2012, 137(14), 3202–3215 RSC.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c2ay25544h |
| This journal is © The Royal Society of Chemistry 2013 |
