 Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Whole organ cross-section chemical imaging using label-free mega-mosaic FTIR microscopy†
Paul Bassana, Ashwin Sachdevaa, Jonathan H. Shanksb, Mick D. Brownc, Noel W. Clarkec and Peter Gardner*a
aManchester Institute of Biotechnology, University of Manchester, 131 Princess Street, Manchester, M1 7DN, UK. E-mail: peter.gardner@manchester.ac.uk
bDepartment of Histopathology, The Christie NHS Foundation Trust, Manchester, UK M20 4BX, UK
cGenito Urinary Cancer Research Group, Institute of Cancer Sciences, Paterson Institute for Cancer Research, The University of Manchester, Manchester Academic Health Science Centre, Department of Urology, The Christie NHS Foundation Trust, Manchester, UK M20 4BX, UK
First published on 2nd October 2013
Abstract
FTIR chemical imaging has been demonstrated as a promising technique to construct automated systems to complement histopathological evaluation of biomedical tissue samples. The rapid chemical imaging of large areas of tissue has previously been a limiting factor in this application. Consequently, smaller areas of tissue have previously had to be sampled, possibly introducing sampling bias and potentially missing diagnostically important areas. In this report a high spatial resolution chemical image of a whole prostate cross section is shown comprising 66 million pixels. Each pixel represents an area 5.5 × 5.5 μm2 of tissue and contains a full infrared spectrum providing a chemical fingerprint. The data acquisition time was 14 hours, thus showing that a clinical time frame of hours rather than days has been achieved.
Introduction
Vibrational spectroscopic chemical imaging has become a popular technique in recent years for the study of biomedical samples, namely biopsy tissue, where it holds significant promise to complement histopathology practice.1–4 The ability to classify a biopsy specimen as healthy or diseased/malignant based purely upon its Raman or IR spectral chemical fingerprint lends itself to building automated systems for high-throughput screening, where human error is minimised. Moreover, the knowledge that can be gained from the chemistry is equally attractive. Fourier transform infrared (FTIR) chemical imaging has been successfully employed to image prostate tissue microarrays with the ability to distinguish cell-types within prostate tissue based purely on the chemical fingerprint of the IR spectrum.4 The potential to predict the presence of prostatic adenocarcinoma,4 Gleason grade and stage2,5,6 of prostate tissue has been demonstrated in a number of studies. FTIR chemical imaging of tissue sections is advantageous as it is label-free and can be performed on tissue embedded in paraffin, i.e. without chemical dewaxing1,7–9 which prevents further alteration of the chemistry of the specimen, and negates the need for expensive computational signal processing to correct scattering effects.10–12The majority of chemical imaging of biomedical samples such as tissue specimens involves individually sampling different areas of the tissue as the entire tissue sample may be too large to measure in one go introducing the problem of sampling bias. For example, if there are several sites of interest but not enough time or resources to measure them all, representative individual areas would need to be identified. This may inadvertently lead to operator bias favouring specific histological regions, leading to the potential loss of clinically relevant information in associated histologically “normal” regions. An alternative approach to large area imaging is to degrade spatial resolution by (a) pixel binning, e.g. 4 × 4 resulting in a pixel size of 22 × 22 μm2, or (b) using lower magnification optics, such as the Agilent 4 × objective for FTIR imaging. These methods may help to image larger areas. However, in both scenarios information is lost meaning that finer features such as a small cluster of cellular subtypes may not be resolvable from surrounding tissue, leading to the loss of potentially vital diagnostic features.
Prostate cancer is the most commonly diagnosed malignancy in men within the UK with over 41![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 new cases diagnosed in each year and approximately 11
000 new cases diagnosed in each year and approximately 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 associated deaths in 2010 (Cancer Research UK figures). Treatment is predicated on accurate grading and staging of the disease and in this area there is significant clinical uncertainty. Risk profiling, of which histological Gleason grading is a major component, determines treatment decision making and it is known that there are significant errors both in relation to inter-observer under and over-grading13 and to sampling error. This contributes to the overtreatment of disease with a lower risk profile14 and potential under-treatment of disease with a more aggressive biopotential. Advanced metastatic prostate cancer is currently incurable and it is therefore essential for the correct grading and staging of diseases. Patients initially diagnosed with localised prostate cancer (T1–T2 disease), i.e. the tumour has not spread beyond the prostate capsule may be offered a radical prostatectomy (RP). The surgically removed prostate specimen then goes through the standard histopathology process of formalin fixation and paraffin embedding (FFPE) prior to histological analysis by a histopathologist to confirm the primary diagnosis and to ascertain further clinical information enabling risk profiling, thereby informing further treatment decision making. This task is not trivial and requires careful analysis, which is time consuming and is subject to inter-operator variability. Therefore, an automated method of analysing the whole tissue sections, based on defined chemical spectral biomarkers rather than histological architecture is desirable. In particular, the ability to chemically image large specimens combined with recent advances in spectral histopathology demonstrates how vibrational spectroscopy is maturing rapidly and has significant potential to become clinically feasible.
000 associated deaths in 2010 (Cancer Research UK figures). Treatment is predicated on accurate grading and staging of the disease and in this area there is significant clinical uncertainty. Risk profiling, of which histological Gleason grading is a major component, determines treatment decision making and it is known that there are significant errors both in relation to inter-observer under and over-grading13 and to sampling error. This contributes to the overtreatment of disease with a lower risk profile14 and potential under-treatment of disease with a more aggressive biopotential. Advanced metastatic prostate cancer is currently incurable and it is therefore essential for the correct grading and staging of diseases. Patients initially diagnosed with localised prostate cancer (T1–T2 disease), i.e. the tumour has not spread beyond the prostate capsule may be offered a radical prostatectomy (RP). The surgically removed prostate specimen then goes through the standard histopathology process of formalin fixation and paraffin embedding (FFPE) prior to histological analysis by a histopathologist to confirm the primary diagnosis and to ascertain further clinical information enabling risk profiling, thereby informing further treatment decision making. This task is not trivial and requires careful analysis, which is time consuming and is subject to inter-operator variability. Therefore, an automated method of analysing the whole tissue sections, based on defined chemical spectral biomarkers rather than histological architecture is desirable. In particular, the ability to chemically image large specimens combined with recent advances in spectral histopathology demonstrates how vibrational spectroscopy is maturing rapidly and has significant potential to become clinically feasible.
Here we show here that the problem of sampling small areas of the clinical specimen or lowering degrading spatial resolution to image the whole specimen, can be negated by measuring the entire sample using rapid FTIR chemical imaging at a high spatial resolution. We show a 66 million pixel chemical image of a whole prostate cross section of size ca. 4 × 5 cm2, where each pixel covers 5.5 × 5.5 μm2 of tissue.
Materials and methods
FTIR chemical imaging was performed using a Varian 670-IR coupled with a Varian 620-IR microscope (Agilent Technologies – Santa Clara, CA, USA). The system was equipped with a liquid nitrogen cooled mercury–cadmium–telluride (MCT) detector comprising 128 × 128 elements producing a focal plane array. The resulting image from a single tile produces 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 384 spectra. A mosaic method was used to image larger areas, employing a motorised stage to automate image collection. The spectral resolution was 16 cm−1 and the number co-added scans for the background and sample were 16 and 4 respectively resulting in a 14 hour collection time. A study by Bhargava15 showed that accurate histological classification of prostate tissue cell-types could be performed with a spectral resolution as low as 64 cm−1. The number of scans was optimised such that it was lower than the buffer rate of the FPA, which reduces dead time and increases the efficiency of the duty cycle. This collection time will reduce hugely in the coming years accompanied by an increase in spectral resolution as suggested by a recent review on imaging.16
384 spectra. A mosaic method was used to image larger areas, employing a motorised stage to automate image collection. The spectral resolution was 16 cm−1 and the number co-added scans for the background and sample were 16 and 4 respectively resulting in a 14 hour collection time. A study by Bhargava15 showed that accurate histological classification of prostate tissue cell-types could be performed with a spectral resolution as low as 64 cm−1. The number of scans was optimised such that it was lower than the buffer rate of the FPA, which reduces dead time and increases the efficiency of the duty cycle. This collection time will reduce hugely in the coming years accompanied by an increase in spectral resolution as suggested by a recent review on imaging.16A radical prostatectomy sample of Gleason score 7 was obtained following informed consent and ethical approval (Trent Multi-centre Research Ethics Committee 01/4/061). A trans-axial whole mount section was taken from the centre of the paraffin embedded prostate and two tissue sections were fixed, one onto a large glass slide, the second onto a low-e slide (MirrIR – Kevley Technologies, Ohio, USA) which is suitable for transflection mode FTIR imaging.
Results and discussion
Fig. 1 shows a 5 μm tissue cross-section from a whole prostate with (a) the unstained FTIR based chemical image, and (b) the serial section stained with haematoxylin and eosin (H&E) commonly employed in histopathology laboratories to create contrasts in the tissue by differentially staining different cell types and connective tissue. The vertical and elongated structure in the centre of the prostate is the prostatic urethra, which is surrounded by connective tissue and glands into which prostatic fluid is secreted. Fig. 1a shows a 66 million-pixel false colour chemical image of the absorption from the carbonyl vibration arising predominantly from proteins occurring at 1655 cm−1. The image comprises 71 × 57 (4047) individual FTIR image tiles each of which is a 128 × 128 hyper-spectral chemical image.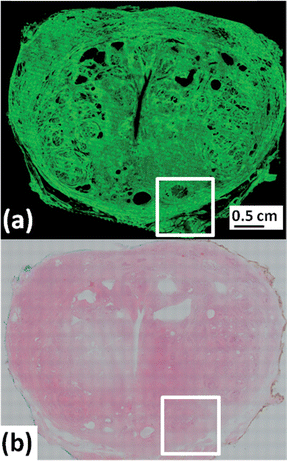 | ||
| Fig. 1 Whole prostate cross section (a) 66 million pixel chemical image of absorbance at 1655 cm−1 (amide I). (b) H&E micrograph of serial section to (a). | ||
FTIR chemical imaging and appropriate bioinformatics of the spectra allow for automated and high-throughput systems to complement the histopathologist. The analysis of the automated system is not reliant upon human interpretation, and in addition to the tissue architecture, the chemical fingerprint of the specimen is available in the form of an IR spectrum. We, and others have shown that diseased tissue regions have a spectral signature different to that of non-cancerous tissue of the same organ.2,5,6,17 Independent studies have reported this concept for different cancers in both males (e.g. prostate4) and females (e.g. cervical18). This technique is not limited to cancers and is applicable to any specimen type where spatial architecture and distribution of chemicals is useful in diagnosis.
To highlight the spatial resolution of the chemical image in Fig. 1a, the white square has been expanded and shown in Fig. 2(a–c)i. The image shows a cluster of prostatic glands (epithelia) surrounded by supporting stroma. Fig. 2a shows the outer capsule of the prostate, with the chemical image shown in Fig. 2ai depicting its fibrous nature. Capturing the whole section at a high resolution allows the operator to zoom in on regions of interest, such as the epithelial gland seen in Fig. 2(a)i which has been expanded in Fig. 2b and c. Fig. 2c is the size of a single 128 × 128 FTIR chemical image where each pixel covers an area of 5.5 × 5.5 μm2 of tissue and contains a full infrared spectrum. In principle such maps can be made from any absorption band in the infrared spectrum. In the supplementary information available online, we have rendered a chemical image of the phosphate absorption occurring at 1080 cm−1, which interestingly, has a different spatial distribution in the prostate compared with the protein chemical image of the absorbance at 1655 cm−1 shown in Fig. 1a.
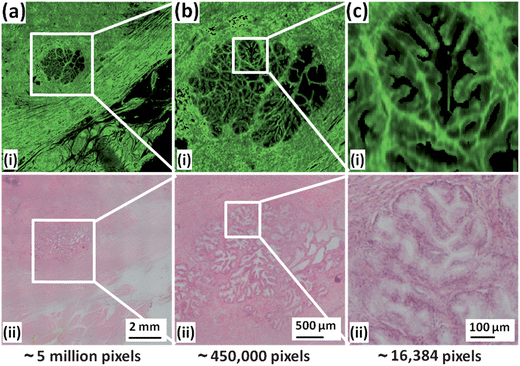 | ||
Fig. 2 Expanded regions of chemical image and H&E micrograph from Fig. 1. (a) Expansion of white square in Fig. 1. (b) Expansion of region highlighted in (a). (c) Expansion of region highlighted in (b) corresponding to a single 128 × 128 tile comprising 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 384 spectra. 384 spectra. | ||
This report shows how an entire organ such as the prostate can be imaged chemically in a clinical time frame of hours rather than days. Transflection mode FTIR imaging was used and recent studies have shown that spectra measured this way are distorted due to electric-field standing waves, meaning that quantitative data analysis is rendered unreliable.19–22 Although we have used this sub-optimal slide, it serves adequately for the qualitative demonstration and the results are directly applicable to the preferable transmission modality.
Combined with the advances by other researchers,4 an automated system could be constructed whereby the chemical image, prostate in this case, can be classified initially into its constituent cell types (the histology), after which the epithelial cells can be classified as cancerous or non-cancerous. Gazi et. al.5 have shown that it is possible to grade prostate cancers using just the IR spectrum. The vision of creating an FTIR grading system assessing tumour biopotential which is based on assistive technology is therefore closer to becoming a reality and this would potentially work alongside standard histopathology to improve diagnostic accuracy and to guide clinicians in deciding their clinical treatment planning of complex medical problems such as cancer.
Acknowledgements
We would like to thank the Engineering and Physical Sciences Research Council (EPSRC) for funding.References
- E. Ly, O. Piot, A. Durlach, P. Bernard and M. Manfait, Analyst, 2009, 134, 1208–1214 RSC.
- M. J. Baker, E. Gazi, M. D. Brown, J. H. Shanks, N. W. Clarke and P. Gardner, J. Biophotonics, 2009, 2, 104–113 CrossRef CAS PubMed.
- R. Bhargava, D. C. Fernandez, S. M. Hewitt and I. W. Levin, Biochim. Biophys. Acta, Biomembr., 2006, 1758, 830–845 CrossRef CAS PubMed.
- D. C. Fernandez, R. Bhargava, S. M. Hewitt and I. W. Levin, Nat. Biotechnol., 2005, 23, 469–474 CrossRef CAS PubMed.
- E. Gazi, M. Baker, J. Dwyer, N. P. Lockyer, P. Gardner, J. H. Shanks, R. S. Reeve, C. A. Hart, N. W. Clarke and M. D. Brown, Eur. Urol., 2006, 50, 750–761 CrossRef PubMed.
- E. Gazi, J. Dwyer, P. Gardner, A. Ghanbari-Siahkali, A. P. Wade, J. Miyan, N. P. Lockyer, J. C. Vickerman, N. W. Clarke, J. H. Shanks, L. J. Scott, C. A. Hart and M. Brown, J. Pathol., 2003, 201, 99–108 CrossRef CAS PubMed.
- R. Wolthuis, A. Travo, C. Nicolet, A. Neuville, M. P. Gaub, D. Guenot, E. Ly, M. Manfait, P. Jeannesson and O. Piott, Anal. Chem., 2008, 80, 8461–8469 CrossRef CAS PubMed.
- E. Ly, O. Piot, R. Wolthuis, A. Durlach, P. Bernard and M. Manfait, Analyst, 2008, 133, 197–205 RSC.
- A. Tfayli, O. Piot, A. Durlach, P. Bernard and M. Manfait, Biochim. Biophys. Acta, 2005, 1724, 262–269 CrossRef CAS PubMed.
- P. Bassan, A. Sachdeva, A. Kohler, C. Hughes, A. Henderson, J. Boyle, J. H. Shanks, M. Brown, N. W. Clarke and P. Gardner, Analyst, 2012, 137, 1370–1377 RSC.
- P. Bassan, A. Kohler, H. Martens, J. Lee, H. J. Byrne, P. Dumas, E. Gazi, M. Brown, N. Clarke and P. Gardner, Analyst, 2010, 135, 268–277 RSC.
- P. Bassan, H. J. Byrne, F. Bonnier, J. Lee, P. Dumas and P. Gardner, Analyst, 2009, 134, 1586–1593 RSC.
- W. C. Allsbrook, K. A. Mangold, M. H. Johnson, R. B. Lane, C. G. Lane, M. B. Amin, D. G. Bostwick, P. A. Humphrey, E. C. Jones, V. E. Reuter, W. Sakr, I. A. Sesterhenn, P. Troncoso, T. M. Wheeler and J. I. Epstein, Hum. Pathol., 2001, 32, 74–80 CrossRef PubMed.
- M. R. Cooperberg, J. M. Broering, M. S. Litwin, D. P. Lubeck, S. S. Mehta, J. M. Henning, P. R. Carroll and C. Investigators, J. Neurol., 2004, 171, 1393–1401 Search PubMed.
- R. Bhargava, Anal. Bioanal. Chem., 2007, 389, 1155–1169 CrossRef CAS PubMed.
- R. Bhargava, Appl. Spectrosc., 2012, 66, 1091–1120 CrossRef CAS PubMed.
- M. J. Baker, E. Gazi, M. D. Brown, J. H. Shanks, P. Gardner and N. W. Clarke, Br. J. Cancer, 2008, 99, 1859–1866 CrossRef CAS PubMed.
- B. R. Wood, K. R. Bambery, L. M. Miller, M. Quinn, L. Chiriboga, M. Diem and D. McNaughton, Biomedical Applications of Micro- and Nanoengineering II, 2005, 5651, 78–84 CrossRef PubMed.
- B. J. Davis, P. S. Carney and R. Bhargava, Anal. Chem., 2010, 82, 3487–3499 CrossRef CAS PubMed.
- B. J. Davis, P. S. Carney and R. Bhargava, Anal. Chem., 2010, 82, 3474–3486 CrossRef CAS PubMed.
- P. Bassan, J. Lee, A. Sachdeva, J. Pissardini, K. M. Dorling, J. S. Fletcher, A. Henderson and P. Gardner, Analyst, 2013, 138, 144–157 RSC.
- J. Filik, M. D. Frogley, J. K. Pijanka, K. Wehbe and G. Cinque, Analyst, 2012, 137, 853–861 RSC.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c3an01674a |
| This journal is © The Royal Society of Chemistry 2013 |
