 Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Illuminating disease and enlightening biomedicine: Raman spectroscopy as a diagnostic tool
David I. Ellis*a, David P. Cowchera, Lorna Ashtona, Steve O'Hagana and Royston Goodacreab
aSchool of Chemistry, Manchester Institute of Biotechnology, The University of Manchester, 131 Princess Street, Manchester M1 7ND, UK. E-mail: D.Ellis@manchester.ac.uk
bManchester Centre for Integrative Systems Biology, Manchester Institute of Biotechnology, University of Manchester, 131 Princess Street, Manchester M1 7DN, UK
First published on 31st May 2013
Abstract
The discovery of the Raman effect in 1928 not only aided fundamental understanding about the quantum nature of light and matter but also opened up a completely novel area of optics and spectroscopic research that is accelerating at a greater rate during the last decade than at any time since its inception. This introductory overview focuses on some of the most recent developments within this exciting field and how this has enabled and enhanced disease diagnosis and biomedical applications. We highlight a small number of stimulating high-impact studies in imaging, endoscopy, stem cell research, and other recent developments such as spatially offset Raman scattering amongst others. We hope this stimulates further interest in this already exciting field, by ‘illuminating’ some of the current research being undertaken by the latest in a very long line of dedicated experimentalists interested in the properties and potential beneficial applications of light.
 David I. Ellis | David Ellis was educated on the Welsh coast at the University of Wales, Aberystwyth, obtaining a BSc in Environmental Science and a PhD in Analytical Biotechnology/Microbiology. His research involving the rapid and quantitative detection of foodborne bacteria using FT-IR and machine learning has been widely publicised, featuring on BBC TV and radio, the Science Museum in London, as well as the national and international press (e.g. WIRED). He now works at the Manchester Institute of Biotechnology (MIB), as a Senior Experimental Officer undertaking research and managing the labs of Roy Goodacre (http://biospec.net) and Doug Kell (http://dbkgroup.org), in the School of Chemistry, University of Manchester, UK. |
 David P. Cowcher | David Cowcher obtained an undergraduate Masters degree in Chemistry and Forensic Science in 2009 from the University of Manchester, which included a year working as an analytical chemist for GlaxoSmithKline. He is currently studying for a PhD with Roy Goodacre at the Manchester Institute of Biotechnology, University of Manchester, on the development of enhanced Raman scattering methods for biochemical analysis. |
 Lorna Ashton | Lorna Ashton studied for her BSc with the Open University before joining the University of Manchester where she obtained her PhD in Biomolecular Sciences and Raman Optical Activity. She has since worked as a post doctoral research associate specialising in Raman Spectroscopy and two-dimensional correlation analysis for the characterisation of conformational transitions in biomolecules. Lorna is currently working as part of a BBSRC funded project for rapid evolution of enzymes and synthetic microorganisms developing Raman spectroscopy as a high-throughput analytical technique for industrial bioprocesses. |
 Steve O'Hagan | Steve O'Hagan gained BSc and MSc degrees in Chemistry at the University of Manchester. The MSc research was to characterise ‘Molecular Beam Sources’ using mass spectrometry and TOF kinetic energy measurements; computer simulations were also used. Steve gained a PhD in Chemistry at the University of Warwick; several mass spectrometry and chemometric techniques were employed to analyse engine oils and additives. After several years working for commercial laboratories, he joined the University of Manchester as a Computer Officer, working with Doug Kell's and Roy Goodacre's research groups. Interests include: genetic programming; laboratory automation; analytical laboratory data analysis; chemometrics; as well as scientific visualization. |
 Royston Goodacre | Roy Goodacre is a PhD graduate from the University of Bristol (UK) where he studied mass spectrometry of microbial systems. After a postdoc, Wellcome Trust fellowship and lectureship in the University of Wales, Aberystwyth, he is now Professor of Biological Chemistry at the University of Manchester (UK). His group's main areas of research (http://www.biospec.net/) are broadly within analytical biotechnology, metabolomics and systems biology. His expertise involves many forms of Raman spectroscopy (including deep UV resonance Raman and SERS), FT-IR spectroscopy, and mass spectrometry, as well as advanced chemometrics, machine learning and evolutionary computational methods. He is Editor-in-Chief of the journal Metabolomics, on the Editorial Advisory Boards of Analyst and Journal of Analytical and Applied Pyrolysis, a founding director of the Metabolomics Society and a director of the Metabolic Profiling Forum. |
Introduction
The properties of light have been of interest to experimentalists for millennia. From the publication of Ibn al-Haytham's seven volume treatise the ‘Book of Optics’ (Kitāb al-Manāẓir) in the early 11th century (leading to him being regarded as the father of modern optics1), through to one of the very first publications in a scientific journal, Isaac Newton's paper on the theory of the properties of light2 itself followed some years later by his famous book, Opticks, in 1704.3 Whilst separated by seven centuries, these two polymaths and their respective bodies of work shared similarities, perhaps the most important being that both were the result of systematic and methodical experimentation. In the century following the publication of Newton's Opticks, as early as the mid-1800s, the interaction of light within tissue was being used by physicians to assist in disease recognition.4It was during a sea voyage from India to England in 1921 that the brilliant Indian physicist C.V. Raman, another renowned experimentalist, undertook some on-board experiments which were later submitted to Nature in a letter called the ‘The Colour of the Sea’.5 Unable to accept Lord Rayleigh's6 explanation that the colour of the sea was just a reflection of the colour of the sky,7 Raman's experiments showed that the colour of the sea was in fact a direct result of the molecular scattering of light and independent of absorption or the reflection of light from the sky.8 This was very closely followed by another letter to Nature concerning the molecular scattering of light in liquids and solids.9 These experiments opened up a deep interest in C.V. Raman and a new field of research on his return to Kolkata (Calcutta) on the scattering of light,5 as well as the publication of another article on the molecular diffraction and quantum structure of light in the following year.10
Raman and collaborators such as K.S. Krishnan began a series of seminal experiments concerning the scattering of light in a large number of liquids, as well as theories about the potential applications of their experiments, which culminated in their discovery of the inelastic scattering effect named after Raman in 1928 on 28 February,11 and his award of the Nobel Prize for Physics in 1930. This discovery was also independently observed by Landsberg and Mandelstam later in 1928 (see Fig. 1 for a timeline of events in Raman spectroscopy). There was a great deal of interest in the Raman effect and this not only aided the fundamental understanding about the quantum nature of light, and its interaction with matter at the molecular level, but also opened up a completely novel area of optics and spectroscopic research that is, particularly in terms of biological and biomedical applications,12,13 accelerating at a greater rate during the last decade than at any time since its inception.
 | ||
| Fig. 1 A timeline of events and discoveries in the history of Raman spectroscopy, Nobel Prizes marked in red (photo of Sir C.V. Raman in the sculpture park of Nehru Science Centre, Mumbai, India, by Prof. Paul O'Brien FRS, School of Chemistry, University of Manchester). | ||
Whereas infrared (IR) spectroscopies measure the absorption of energy, Raman spectroscopy measures the exchange of energy with electromagnetic (EM) radiation of a particular wavelength, usually provided by a monochromatic light source such as a laser in the visible to near-IR portion of the EM; although it is also possible to conduct experiments in the near- and deep-UV. From the exchange in EM energy a measurable Raman shift in the wavelength of incident laser light is observed, this is also referred to as the inelastic light scattering effect.14–16 It is usually the Stokes shift which is measured, as this has a higher probability of occurring than anti-Stokes.17 It is important to note that this shift is complementary to IR absorption and a spectroscopic ‘fingerprint’13,17,18 of the same sample can be analysed and constructed by both vibrational spectroscopies. Whilst mid-IR spectroscopy is known to be intensely sensitive to and a high absorber of water, this is generally not the case with Raman spectroscopy as water is a weak scatterer.13 For biomedical and routine clinical applications (and with low laser output power at the point of contact), this allows for the direct collection in vivo of Raman spectra. The utility of Raman spectroscopy has been demonstrated for a diverse and wide range of potential biological and biomedical applications, such as bacterial identification,19,20 chemical hazards and illicit substance detection,21,22 as well as food and product authentication,23,24 with a great deal of interest and research into its potential for disease diagnosis and use in biomedical applications seen during the last decade (Fig. 2).
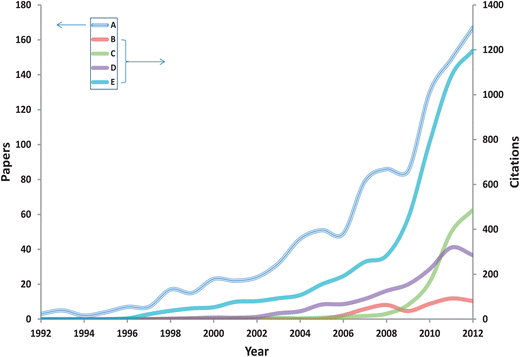 | ||
| Fig. 2 Results of bibliometric analysis of the number of publications (A) and citations (B/C/D/E) per year listed on ISI Thomson Web of Science® (http://wos.mimas.ac.uk/) for the period 1992–2012, using the search terms (A) Raman AND disease, (B) spatially offset Raman AND disease, (C) SERS AND disease, (D) Raman endoscopy, (E) Raman imaging AND disease. | ||
Some of the previously documented limitations of Raman spectroscopy for biomedical applications have for example included issues such as weak scattering signals, subsequently long spectral acquisition times, fluorescence from biological samples, and interference from silica within fibre-optics. However, within this exciting and highly research active era of biomedical Raman spectroscopy, and with the hard work and application of experimentalists (in the long tradition of those already mentioned above), these so-called limitations are being overcome by groups of scientists and engineers around the globe who are not content to remain within the bounds of current knowledge, or the confines of commercially available optics for that matter, and who are constantly pushing the field forward. This introductory overview focuses on some of the recent developments within this exciting field, highlighting a small number of high-impact studies in imaging, endoscopy, stem cell research, and other recent developments such as spatially offset Raman scattering (SORS), coherent anti-Stokes Raman scattering (CARS) and stimulated Raman scattering (SRS), amongst others.
Imaging
Biomedical imaging is an extremely useful and important field which allows for the collection and processing of highly complex biochemical and physiological data, and the creation, manipulation, and in-depth analysis of three-dimensional colourised images to aid molecular sensing,25 drug transport,26 characterization of cells27 and tissues,28 and, perhaps the ultimate goal, the rapid diagnosis of disease (vide infra). Label-free optical imaging, particularly in vivo, would be highly advantageous as dyes or fluorescent labels, which are needed as contrast agents, can be toxic or perturbative to the cell/tissue. Optical techniques have also been said to have the potential to be complementary to existing techniques such as magnetic resonance imaging (MRI), and they offer superior sensitivity and high spatial resolution (compared to MRI, see Fig. 3).29 Whilst mass spectrometry (MS) has mass appeal for metabolomics for example30 and can be used for chemical imaging of tissues and cells31,32 – including in 3D33 – as well as being combined with vibrational spectroscopy,34 imaging MS is a destructive analysis and the practical spatial resolution for MALDI-MS is 50–200 μm and for SIMS is 1 μm, thus Raman spectroscopy has considerable opportunities to shed light on disease. | ||
| Fig. 3 A figurative (as opposed to fibre-optic) bundle of some of the key terms and demonstrated capabilities of in vivo Raman imaging mentioned throughout this article. | ||
In any review of biomedical imaging, and particularly one focussed on Raman spectroscopy, it would be a heedless omission not to include any one of a number of studies by the Sunney Xie group. A major contributor to biomedical imaging, based at Harvard University (http://bernstein.harvard.edu), this group's body of work includes both coherent Raman scattering techniques,35 comprising of coherent anti-Stokes Raman scattering (CARS) microscopy36–38 and stimulated Raman scattering (SRS) microscopy.39 Descriptions of these techniques can be found in Table 1 and it should also be noted that the signal from CARS is non-linearly proportional to species concentration whilst the signal from SRS is linearly proportional.
| Technique | Acronym | Short definition | References |
|---|---|---|---|
| Surface enhanced Raman spectroscopy | SERS | Requires close proximity/adsorption onto a roughened metal surface, a colloidal solution or a roughened electrode (usually Ag or Au). Enhancement explained by two processes; an electromagnetic enhancement effect (thought to dominate) and a charge transfer mechanism, known as chemical enhancement. Has a fluorescence quenching effect. Can tune to a specific chromophore for additional resonance enhancement (known as SERRS). Enhancements over normal Raman scattering of typically 103 to 106 | 19, 122, 136 and 137 |
| Coherent anti-Stokes Raman spectroscopy | CARS | A multiphoton form of Raman spectroscopy based on a non-linear conversion of two lasers into a coherent high intensity beam in the anti-Stokes region. The emission is usually many orders of magnitude greater than spontaneous Raman scattering. Useful for obtaining spectra of fluorescing samples. Nonresonant background can complicate spectral assignment, limit sensitivity, and affect quantitative interpretation | 27, 38, 41, 45 and 138 |
| Stimulated Raman scattering | SRS | A multiphoton technique analogous to stimulated emission, where two lasers coincide on a sample. The sample is excited by colinear and tightly focused pump and Stokes beams. When the differences in known frequencies match a molecular vibration in the sample, the Stokes beam intensity increases and pump beam intensity decreases as a result of the coherent excitation of molecular vibration. Unlike CARS, it does not exhibit a nonresonant background and is a good technique for sensitive, high spatial resolution 3D imaging | 25, 26, 39, 61, 63 and 139 |
| Spatially offset Raman spectroscopy | SORS | Raman spectra are collected from locations spatially separated from the point of laser illumination on the sample surface. SORS allows for the isolation of chemically rich spectral information from distinct substructures or layers and through other barriers, not accessible via spontaneous Raman. Typical wavelengths used for biological tissue are 785 or 830 nm. Ideal for detecting disease in cells and tissue underlying other tissue types, such as bone through skin, cancer cells through muscle and lipid tissue for example | 25, 26, 39, 61, 63 and 139 |
As briefly mentioned above, the signal from spontaneous Raman scattering is known to be weak, with potentially long integration times, which impacts by significantly reducing imaging speed (an important factor as in vivo samples are in constant motion at the micro scale). Although these nonlinear methods were first invented in the 1960s, coherent Raman scattering techniques can be said to have very recently surmounted any technical difficulties and imaging speed hurdles by enhancing the Raman signal level by up to five orders of magnitude,39 and image collection speeds by three orders of magnitude. Indeed, on the subject of speed, one of the more memorable articles propelling CARS microscopy as a rapid method over a decade ago (for the identification of bacterial spores), had the unforgettable acronym FAST-CARS (femtosecond adaptive spectroscopic techniques for coherent anti-Stokes Raman spectroscopy).40
More recent reports have highlighted some of the potential limitations of CARS,29 which has been said to make quantitative interpretation and applications other than lipid imaging41 unnecessarily challenging. These limitations can be particularly noticeable in the fingerprint region where spectral signals are said to be congested39 and separating out weak from strong signals in this region is thus more problematic for CARS microscopic imaging. That being said, CARS microscopy has been successfully demonstrated as a biomedical imaging method for a range of ex vivo biological samples such as brain structures,38 colon tissue,42 and arterial tissue27 for example, as well as in vivo studies including sciatic nerve tissue43 and atherosclerotic plaque deposits44 in animals, with a recent report also demonstrating the use of CARS microscopy in skin imaging of humans in vivo.45
Recent articles within the last half decade have explored SRS as an alternative imaging technology, which, unlike CARS, does not exhibit a nonresonant background and so is inherently more quantitative, though like other nonlinear multiphoton techniques (including CARS), does allow for sensitive, high spatial resolution 3D imaging.46 Whilst initial reports demonstrated stimulated Raman scattering's advantages over CARS (e.g. a lack of nonresonant background complications), it was said to not be suitable for bioimaging due to sample photodamage from excessive laser power46 and had a limited spatial and spectral resolution as well as a slower image acquisition rate than CARS.47 These challenges were rapidly overcome by the Xie group the following year using a multifaceted approach which lowered peak rates by three orders of magnitude! (resulting in no photodamage), optimised spectral resolution, increased sensitivity by four orders of magnitude than the previous year's report and surpassed the detection limit previously stated for CARS microscopy.46
Not content with these and other leaps forward, three bioimaging applications were then elegantly presented by Xie and co-workers. The first application monitored and imaged the uptake of polyunsaturated omega-3 fatty acids by living human cancer cells, specifically eicosapentaenoic acid (EPA). Using the Raman band at 3015 cm−1 (attributable to unsaturated fatty acids), and in contrast, the Raman band at 2920 cm−1 whose peak intensity is similar for saturated and unsaturated fatty acids, they were able to use SRS to follow the uptake of EPA by living cells, concluding that EPA is taken up by the cells and more strongly enriched in lipid droplets compared to other cellular organelles.46 The second application presented the potential for tissue imaging without the requirement for staining, highlighting the 3D sectioning capability and subcellular resolution of SRS, using the CH2 stretching vibration at 2845 cm−1 (see Table 2 for Raman band frequencies of biological interest).
| Band frequency (cm−1) | Vibration mode | Assignment |
|---|---|---|
| Protein secondary structure | ||
| 930–950 | N–Cα–C stretch | Skeletal stretch/α-helix |
| 1235–1259 | N–H and C–H band | Amide III/β-sheet |
| 1260 | N–H and C–H band | Amide III/disordered |
| 1300–1340 | N–H and C–H band | Amide III/α-helix |
| 1650–1655 | H-bonded C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretch O stretch | Amide I/α-helix |
| 1670–1680 | H-bonded C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretch O stretch | Amide I/β-sheet and β-barrel |
| Amino acid residues | ||
| 508–545 | S–S stretch | Trans and gauche conformers |
| 655 | C–S stretch | Gauche conformer |
| 704 | C–S stretch | Trans conformers |
| 835/857 | H-bonding of indole ring | Tyrosine Fermi doublet |
| 875–880 | H-bonding of indole ring | Tryptophan orientation |
| 1008/1034 | Phenyl ring | Phenylalanine |
| 1551–1556 | Indole ring | Tryptophan |
| 1605 | Phenyl ring | Phenylalanine |
| 1615 | Indole ring | Tyrosine |
| RNA and DNA | ||
| 813–816 | O–P–O stretching | A-form helix |
| 914–925 | C–O and C–C stretching | Ribose–phosphate |
| 1095 | PO2 symmetric stretch | B-DNA and Z-DNA marker |
| 1135, 1235, 1395 | Ring stretch | Uracil |
| 1174, 1325, 1370 | Ring stretch | Guanine |
| 1245, 1275 | Ring stretch | Cytosine |
| 1256, 1514 | Ring stretch | Adenine |
| 1671 | C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretch O stretch | Thymine |
| Sugars | ||
| 1000–1200 | C–O and C–C stretch | β-D-Glucose, D-(+)dextrose |
| 1025, 1047, 1155 | C–O stretch from CH2OH of carbohydrates | Glycogen |
| 1267 | NH2 rocking | GlcNAc and GalNAc |
| 1300–1500 | CH2 and CH2OH deformations | β-D-Glucose, D-(+)dextrose |
| Lipids | ||
| 891, 908 | CH2 rocking | Fatty acid chain length |
| 1080 | PO2-symmetric stretch | Phospholipids |
| 1259 | PO2-asymmetric stretch | Phospholipids |
| 1296 | C–C stretch | Unbranched saturated fatty acids |
| 1660 | C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C stretch C stretch | Unsaturated lipid bonds |
| 2873, 2931, 2961 | C–H stretch | Acyl chains of lipids |
| 2888, 2926 | CH2 asymmetric stretch | Saturated lipid bonds |
| 3009 | H–C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C stretch C stretch | Unsaturated lipid bonds |
This was demonstrated in a variety of mouse tissues; from neuron bundles in corpus callosum (highlighting myelin sheaths), thick brain tissue, and from depth profiles of ear tissue, in addition to comparative SRS and CARS images of stratum corneum (visually illustrating how the nonresonant background from CARS can complicate image interpretation). Finally, this seminal series of experiments showed the use of SRS to monitor drug delivery, namely deuterated dimethyl sulfoxide (DMSO), a skin penetration enhancer and retinoic acid (RA) used to treat a range of skin conditions and acute promyelocytic leukemia. Drug delivery into fresh mouse skin was monitored by tuning into the vibrations for DMSO at 670 cm−1 and RA at 1570 cm−1 (as well as lipids of subcutaneous fats at 2845 cm−1).46 This series of experiments elegantly demonstrated the potential for SRS as a new approach for studying pharmacokinetics in situ, sensitive label-free imaging, and molecular sensing in 3D in living cells and tissue.
Whilst the potential of SRS had been so ably demonstrated, the series of experiments above had been undertaken ex vivo. Well aware of the importance of in vivo optical imaging in biomedicine, and perhaps acutely mindful of the challenges required to achieve this goal, as well as the fact that SRS imaging had not yet been accomplished in vivo (in animals or humans), an article was published in 2010 demonstrating that in vivo SRS was not only possible (both in mice and humans), but that it could also be performed at video-rate speeds.29 This high-speed imaging was achieved by modulating the intensity of the Stokes beam at 20 MHz and the group building their own all-analogue lock-in amplifier with a response time of ∼100 ns (whereas this was previously limited by commercially available 100 μs lock-in amplifiers).29 This new custom-built system raster scanned across a sample with a line rate of 8 kHz (100 ns per pixel at 512 × 512 pixels with up to 25 frames per s), increasing imaging speed by three orders of magnitude. In addition, to significantly increase the collection of backscattered light from in vivo samples further in-house modifications were undertaken, such as changing the geometry of the photodetector and microscope objective. This involved exciting light from the objective through an aperture in the detector placed between the microscope objective and the sample, and specially designing a filter to block the modulated Stokes beam whilst transmitting the pump beam.29
The effectiveness of this new video-rate SRS imaging technique was then demonstrated through a series of experiments such as skin imaging in living mice. The CH3 stretching vibration at 2950 cm−1 was shown to mainly highlight proteins, as well as residual lipid structures and individual red blood cells in the viable epidermis, whilst the water signal at 3250 cm−1 was coincident with sebaceous glands in both positive and negative contrast. In vivo imaging of drug penetration of trans-retinol was demonstrated in living mouse skin, visualising with SRS imaging and 3D depth projection that penetration of the topically applied drug occurs along the hair shaft. This pathway was not observed in previous experiments with excised fresh tissue,46 which was said to further highlight the importance of in vivo imaging, as it can lead to insights into the transport mechanisms of small molecules in living organisms.
Finally, a series of in vivo SRS imaging experiments were performed on human skin of a volunteer's forearm showing cell layers of the viable epidermis to a depth of 50 μm, and the boundary (via observation of varying nuclear sizes) between the viable epidermis and stratum corneum when tuned into the CH3 stretching vibration at 2950 cm−1. The penetration-enhancing small molecule DMSO (which was deuterated) was applied to the skin, and its accumulation in the hair shaft and lack of complete penetration into the hair itself was imaged using the C–D stretching vibration at 2125 cm−1. With image acquisition times of 150 ms and 37 ms respectively, these blur-free high-speed in vivo vibrational imaging experiments demonstrated the diagnostic potential of this technology in humans.29
Stem cells
Stem cells,48 stem cell therapy,49,50 and stem cell engineering51 are extremely important and highly topical areas of scientific research with huge potential benefits for the treatment of a wide range of diseases and biomedical applications. The ability to identify the phenotypic purity of live cells is absolutely crucial (and a noninvasive optical method would be highly beneficial), as excessive proliferation of unwanted phenotypes (i.e., uncontrolled differentiation) following transplantation could result in undesirable consequences, such as tissue overgrowth and tumour formation for example.52,53 One of the recent studies by the Notingher group applied Raman spectroscopy to determine if it could be used as a label-free, noninvasive method to detect and image intrinsic chemical differences that could be used as molecular markers among highly heterogeneous stem cells populations; specifically, cardiomyocytes (CMs) derived from human embryonic stem cells (hESCs).52Using a custom-built instrument, Raman detection and imaging of molecular markers specific to hESC-derived CMs was carried out, along with retrospective phenotypic identification of all cells via immunofluorescence imaging integrated with the Raman microscope. Multivariate statistical analysis of Raman spectra and cross-validation methods were used to develop a model (from 50 CMs and 40 non-CMs within the same heterogeneous populations) to determine the true accuracy of phenotypic identification of CMs, the sensitivity and specificity parameters, and select discriminatory Raman bands. Raman spectral images corresponding to the Raman bands identified by both the multivariate model and immunostaining of the same cells allowed for the accurate assignment of Raman molecular markers. The conclusions drawn from the results were that spectral differences were mainly attributable to glycogen and myofibrils, with glycogen being responsible for discrimination of CMs (with a band assignment at 860 cm−1), and myofibril proteins providing a lesser contribution (with a band assignment at 938 cm−1).52 This study demonstrated the potential of Raman spectroscopy for noninvasive phenotypic identification of stem cells, though the authors themselves stated that it was not yet practical for medical applications due to their long spectral acquisition times.
However, a later study by the same group on the same hESC-derived cardiomyocytes reduced spectral acquisition times by a hundredfold, from minutes per cell to 5 s per cell, without the need for raster scanning. When incorporating high-powered commercial lasers, this could be further reduced to cell sorting speeds of approximately 10 cells per s.54 In addition, a recent study by the same group has successfully applied Raman to identify, image and quantify the differentiation status of live neural stem cells in vitro, where this time the spectral differences were said to be related to cytoplasmic RNA.55 A number of other interesting studies involving Raman analysis of stem cell populations have also appeared in the literature very recently56–59 and this area of interest, not surprisingly, continues to grow and flourish.
As well as cellular differentiation, the location of specific drugs inside cells is also of interest as this may allow the elucidation of the pharmaceutical's site of action. A recent study addressing this enabled mapping the site of action of the HIV protease inhibitors indinavir and lopinavir in cervical carcinoma cells expressing the E6 oncogene from human papilloma virus (HPV).60 This study demonstrated that indinavir undergoes enhanced nuclear accumulation in E6 expressing cells, indicating this as the site of action for this compound against the HPV. Further interesting studies showing the plethora of Raman imaging applications include a report on stimulated Raman photoacoustic imaging,61 surface enhanced Raman scattering (SERS) imaging using nanotags in live mice, as a potential multiplexed imaging detection method for multiple biomarkers in living subjects associated with a specific disease,62 quantitative multiplex SRS imaging,63 noninvasive time-course imaging of apoptotic cells,64 and multivariate image reconstruction methods for Raman hyperspectral datasets.65 For those specifically interested in coherent nonlinear optical imaging, which includes stimulated Raman scattering, the reader is directed to a recent review by Min et al.,66 others include a review of CARS microscopy,37 as well as gold nanoparticles and imaging in medicine.67
Endoscopy
The clinical potential for in vivo Raman endoscopy has been the subject of research for over a decade, since the first published report of in vivo Raman spectra of human gastrointestinal tissue measured during routine clinical endoscopy in 2000.68 In the same year work by the Stone group incorporated Raman spectroscopy and clinical endoscopy to discriminate between normal, dysplastic and cancerous laryngeal tissue.69 Nick Stone's group, based in the UK (http://www.exeter.ac.uk/) have played a major role in pioneering the field of clinical optical diagnostics with (in addition to other vibrational spectroscopies) well received studies using Raman spectroscopy/endoscopy to analyse a range of diseases/disorders such as Barrett's oesophagus70 and bladder and prostate cancer.71 One of the recent studies by the Stone group involved the evaluation of the suitability of a custom-built fibre-optic Raman probe for the potential in vivo diagnosis of early onset oesophageal neoplasia. Whilst this involved ex vivo sampling, the results clearly demonstrated the potential for the rapid and accurate differentiation between benign tissue, Barrett's oesophagus and both dysplastic and malignant tissue.The culmination of this study was a custom-built confocal Raman probe constructed by the group. This novel probe had been reported by the group previously,72 although by the time of the latest study, the probe had undergone some modifications which had improved its performance for spectral acquisition during oesophageal endoscopy. The 90 cm long, 2.7 mm diameter fibre-optic probe was designed to fit into the instrument channel of a standard clinical endoscope and to have direct contact with oesophageal epithelial tissue. The optics had been modified by incorporating a graded index lens at the tip (and the output power regulated to 60 mW), so that a sampling depth of 100 to 200 μm could be ensured. Collecting spectra from this depth meant that signals from deeper tissue structures would not obscure those collected from mucosal abnormalities such as early neoplastic changes,73 and that these abnormalities could be quickly classified in timescales suitable to a clinical setting. The performance of the probe ex vivo had been evaluated for translational use for in vivo sample collection, with low laser power at the probe tip and short spectral acquisition times said to enable its routine use for oesophageal endoscopy and paving the way for in vivo clinical trials.73 This group have recently published a review on in vitro and in vivo Raman spectroscopy as a potential routine tool for the rapid, noninvasive, early diagnosis of lesions and preventing development of cancer in the oesophagus.74
Another group who have made a significant contribution to the field of Raman endoscopy in recent years is headed by Zhiwei Huang and based at the Optical Imaging Laboratory in Singapore (http://www.bioeng.nus.edu.sg/optbioimaging/huang/). Several of these studies utilised image-guided Raman endoscopy, which the group reported for the first time in 2009.75 Whilst image-guided endoscopy is by no means novel,76,77 the integration of image-guided techniques with Raman endoscopy is relatively recent. The first report on this technique involved integrating Raman spectroscopy with trimodal imaging techniques (white-light reflectance, autofluorescence and narrow-band) and the development of a novel 1.8 mm Raman probe which filtered out interference from fluorescence as well as interference from silica from within optical fibres. This was demonstrated via the rapid collection of Raman spectra (<1 s) and the corresponding endoscopic images of different locations of the upper gastrointestinal tract of a healthy volunteer in real-time and in vivo.75
Since then this group have published several articles using image-guided Raman endoscopy, demonstrating its potential as an in vivo real-time detection method for a variety of diagnostic applications. These studies have included, perhaps not surprisingly, the in vivo diagnosis of oesophageal cancer using this technique in conjunction with biomolecular modelling.78 This involved collecting spectra from 75 oesophageal tissue sites from 27 patients of normal tissue (squamous mucosa) and malignant tumours. The cancerous tissue was said to show distinct Raman signals mainly associated with cell proliferation, lipid reduction, abnormal nuclear activity and neovasculation. To estimate the biochemical composition of oesophageal tissue, biomolecular modelling was employed using six basis reference spectra from actin, collagen type I, DNA, histones, triolein and glycogen. This allowed for the construction of a linear discriminant analysis (LDA) model with a sensitivity of 97% and specificity of 95.2% for the in vivo diagnosis of oesophageal cancer.78 These results have since been said to be extremely promising, but that image-guided Raman endoscopy is yet to be used to detect dysplasia or the early onset of cancer.73
Nevertheless, the Huang group have previously demonstrated the potential of Raman spectroscopy for the detection of dysplasia, with a ball-lens fibre-optic probe, in the high wavenumber region (HW) (2800–3700 cm−1) for the in vivo detection of cervical dysplasia (a HW Raman probe was first presented by Gerwin Puppels and co-workers for in vitro measurements of brain tissue in 200579). The perceived main advantages of HW Raman were said to be a significant reduction in fluorescence and background signal from optical fibres, more intense Raman signals (compared to the fingerprint region), and the possibility of an unfiltered single fibre Raman probe design for in vivo clinical use.80 In addition to this study, several more recent reports have continued to investigate this area.81–83
The in vivo detection of epithelial neoplasia in the stomach using image-guided Raman endoscopy has also been demonstrated, and significant differences between normal and cancerous gastric tissue were reported.84 More recently, a study has been published showing the development of an online automated spectral diagnostics system integrated with image-guided Raman endoscopy for real-time in vivo diagnosis during endoscopic examination.85 This system was built on a database of 2465 normal and 283 cancerous gastric tissue spectra acquired from 305 patients, with the system employing a variety of diagnostic algorithms. Other so-called in vivo real-time applications recently reported have involved transnasal image-guided Raman endoscopy of the larynx and nasopharynx, which the authors hoped would pave the way for realizing early diagnosis and detection of cancers and precancers of the head and neck.86 All of these studies show promise and many have similarities in terms of rapid spectral acquisition times (absolutely crucial in in vivo studies), and novel developments in optical engineering, but there remains more to accomplish, such as the ability not only to discriminate cancer, but to classify and grade both cancerous and precancerous cells and tissue.87,88
Recent developments
In terms of noninvasive biomedical Raman other very exciting developments are spatially offset Raman spectroscopy (SORS)89 as well as transmission Raman, both techniques directly resulting from research involving depth profiling using Raman Kerr-gating methods.90 SORS was invented and developed in the Central Laser Facility of the Rutherford Appleton Laboratory (http://www.clf.rl.ac.uk) in 2005 by Pavel Matousek and readers are directed to an excellent overview of this fast moving field by Matousek and Stone.91The central difference in SORS is that Raman spectra are collected from different locations, spatially separated (offset) from the point of laser excitation on the sample (Fig. 4). As a consequence of photon diffusion processes within tissue the resultant Raman spectra contain different relative contributions from different depths within the sample and thus allows for the highly accurate chemical analysis of subsurface objects of interest.89 These spectra acquired from different spatial offsets are processed to reveal pure Raman spectra of subcomponents from separate depth locations within tissue.91 SORS can be applied in a number of Raman collection and beam delivery geometries including single point collection, ring, and other pattern illumination.91 In respect to biomedical imaging this approach is said to be particularly useful when configured as inverse SORS, where the sample is illuminated by an adjustable ring-shaped laser beam (generated by a conical (axicon) lens) and the Raman light collected via fibres in the centre of the ring.92 The radius of this ring is said to define the spatial offset and as it is adjustable, this can be optimised to suit both the scattering properties and dimensions of each sample and covers a wider illumination zone on a sample surface than conventional SORS.91 SORS is said to be effective at tissue depths in excess of 500 μm, which is well beyond the accessible range of conventional confocal Raman spectroscopy. Applications can be as diverse and wide-ranging as noninvasive detection of pharmaceuticals through packaging,93 detection of hidden explosives and drug precursors behind opaque plastics and garments,21,94–96 as well as agricultural and food product analysis.97
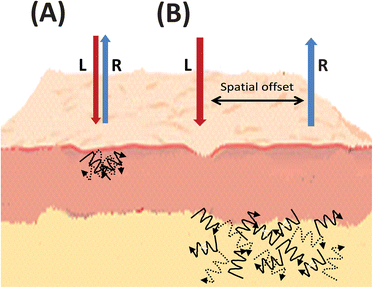 | ||
| Fig. 4 Simplified graphical representation of (A) spontaneous Raman compared to (B) spatially offset Raman scattering, illustrating the spatial offset and deeper subsurface probing in skin tissue. L = laser light, R = Raman light. | ||
From the outset, Matousek and collaborators immediately realised the diagnostic potential of SORS with its ability for noninvasive subsurface probing of for example bone through skin89 and dermatology studies.92 This foresight was quickly realised with the first SORS spectra of ex vivo bone collected in 200698,99 from animal and human cadavers, and in vivo SORS demonstrated by Matousek during the same year.92 There have been several studies since including accurate in vivo assessment of bone composition using the carbonate (1070 cm−1) -to-phosphate (958 cm−1) ratio through the skin of live mice,100in vivo evaluation of bone grafts,101,102 transcutaneous in vivo monitoring of glucocorticoid induced osteoarthritis103 and in vivo measurement and evaluation of subtle changes in bone composition.104
A significant body of pioneering work forwarding Raman spectroscopy as a tool for breast cancer diagnosis was undertaken in the George R. Harrison Spectroscopy Lab at MIT (http://web.mit.edu/spectroscopy/) under the directorship of Michael S. Feld. This included identifying chemical differences in microcalcifications from benign and malignant breast lesions,105,106 demonstrating the real time capabilities of an in vivo Raman system during femoral bypass and breast lumpectomy surgeries,107 and also involved testing of spectral diagnostic algorithms for breast cancer diagnosis.108 The results from the Raman research on the chemical composition and identification of the different types of microcalcifications has led directly to others investigating the potential of SORS as a possible clinical adjunct to mammography, for the noninvasive diagnosis of breast cancer. As the changing concentration of carbonate substitution for phosphate ions in the calcium hydroxide lattice in microcalcifications may relate to the process of tumour cell metastasis, and the ability to measure the magnitude of this (as well as soft tissue signals) by Raman, could indicate the potential progression of this cancer.109
Research by Stone, Matousek, and collaborators, demonstrated the proof-of-principle of SORS for potential in vivo breast cancer diagnosis in a model system using three calcification standards overlaid with various preparations of animal tissue (i.e. chicken breast tissue, with and without skin). For this work they utilised a continuous wave 827 nm laser with a spatial offset for collection of 3 mm, which enabled the probing and collection of Raman spectra from calcifications through up to 10 mm of tissue.109 Previous results by this group using Kerr-gated Raman techniques had achieved penetration depths of 1 mm in comparison.90 This SORS study demonstrated the collection of high quality Raman spectra and biochemical information measured through 8.7 mm of tissue, identifying the difference between three calcification standards.
Subsequent experiments by the same group applied transmission Raman spectroscopy in combination with chemometrics in similar model systems collecting Raman calcification signals from depths of 20 mm, this time through porcine tissue, as a breast tissue ‘phantom’.110 This was said to reach the lower range for clinically relevant breast tissue thicknesses from mammographic screening (1.9 cm), with further work said to be required to reach the top of this range (5 cm).91 Whilst these studies utilised the carbonate-to-phosphate band ratio (similar to the SORS bone studies above100), a more recent study showed that the intense Raman phosphate band at 960 cm−1 broadens and shifts as carbonate concentration (from calcium hydroxyapatites) increases in the calcifications.111 This study was said to pave the way toward a new generation of noninvasive breast cancer screening methods based around SORS and transmission Raman spectroscopy.111 Some very recent studies by the Mahadevan-Jansen group have investigated SORS for the real-time, intraoperative assessment of breast cancer tumour margins.112 This same group then developed and tested a SORS probe with multiple source detector offset limits specific to this type of analysis, and published results acquired from 35 freeze–thaw breast cancer samples in vitro.113
Variants of SORS have also recently been used to augment analysis and include in vivo transcutaneous glucose sensing in rats114 and further demonstrated the accuracy and functionality of this in a later study.115 These studies employed what they term surface enhanced SORS (SESORS), first published in 2010,116 and presented at the Federation of Analytical Chemistry and Spectroscopy Societies (FACSS) meeting in the USA during the same year. SESORS marry SERS techniques using nanoparticles and nanosurfaces with the subsurface probing of SORS, with a further study exploring this technique's potential for Raman imaging.117
On the subject of SERS, which has not been mentioned in any great detail here, many studies have appeared in the literature during the last few years related to disease detection and we would like to highlight just a modest selection of these. They of course include the multiplexed imaging of SERS nanotags in vivo already mentioned above,62 as well as other multiplexing studies, such as multiplex single nucleotide polymorphism (SNPs) genotyping coupled to SERS,118 multiplexed in vivo cancer detection using SERS NIR nanotags, demonstrating the excellent sensitivity, stability and tumour specificity of three bioconjugated nanotags,119 recent reviews on the area of SERS multiplexed detection for disease diagnostics,120 as well as SERS cancer detection and imaging and the potential of SERS agents for targeted drug delivery and photothermal therapy.121
Several SERS studies involving immunoassays, including cancer detection,122 detection of a potential pancreatic cancer marker in serum,123 and on-chip immunoassays using hollow gold nanospheres.124 Fluorescent SERS gold co-functionalized nanorod probes for in vivo imaging of lymph node mapping and tumor targeting in mice,125 high sensitivity in vivo detection of inflammation using gold nanoclusters conjugated to monoclonal antibodies126 and the use of functionalised nanoparticles and SERS for the detection of DNA relating to disease.127 With the recent interest concerning the potential resurgence in microbial disease, as well as bioterrorism, an article demonstrating a portable quantitative SERS system for detection of Bacillus spores at levels significantly lower than those previously reported for SERS has been published.128Fig. 5 shows SERS spectra of a biomarker from the spores of Bacillus, a species of which Bacillus anthracis is the cause of the acute, and mostly lethal, disease anthrax. The detection limit of this dipicolinic acid biomarker is estimated at 5 ppb (30 nM).128
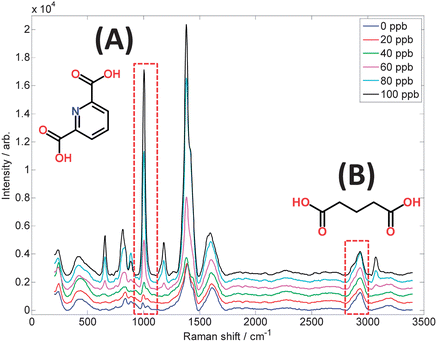 | ||
| Fig. 5 SERS spectra of dipicolinic acid, a biomarker for Bacillus spores, measured using citrate-reduced silver colloid and a portable 633 nm Raman spectrometer. The concentration range 0–100 ppb in the sample volumes used is of equivalent magnitude to the infective dose of inhalation anthrax. Highlighted are: (A) the pyridine ring breathing vibration and structure of dipicolinate at 1006 cm−1, and (B) the C–H stretching vibration and structure of glutaric acid at 2934 cm−1, which is included as an internal standard to allow accurate quantification. | ||
In addition to those primary articles and reviews already mentioned above, other recently published reviews and books on an array of Raman-based topics which may be of interest include those concerning in vivo and in vitro analysis,129 Raman scattering in pathology,130 optical tumour margin identification in the larynx,131 data-classification algorithms for spectral analysis,132 recent advances in gold nanoparticle based assays for detecting and identifying microbes,133 and emerging Raman applications and techniques in biomedical and pharmaceutical fields.134
Concluding remarks
We hope that whilst only an introductory overview to some of the more recent work in this field, the range and scope of the studies shown here elegantly demonstrate the development and exploitation of Raman spectroscopy as a medical diagnostic tool. We believe this truly is an exciting field, where those at the forefront are propelling it forwards and are simply not content to remain within the confines of current knowledge or commercially available optics. These studies highlight the necessary interdisciplinary nature of this field, with inspirational contributions from a range of analytical scientists and technologists including clinicians, biologists, chemists, optical engineers, as well as statisticians and chemometricians. These multidisciplinary inputs and fresh approaches are invigorating this field, unlocking the doors to new insights,135 adding to knowledge, and opening up new dimensions and avenues of study, as well as potential clinical applications, for biomedical Raman spectroscopy.Acknowledgements
DIE would like to thank Analyst for their kind invitation to write this article and the patience of all in the editorial team during its construction, as well as Prof. Paul O'Brien FRS, Dr Steve O'Hagan and Howbeer Muhamad Ali for assistance with graphics and software. RG, LA and DPC are very grateful to the UK BBSRC, EPSRC, Avacta group plc., and industrial members of the Bioprocessing Research Industry Club (BRIC) for funding this work.References
- M. T. Masoud and R. Masoud, Br. Med. J., 2006, 332, 120 CrossRef.
- I. Newton, Phil. Trans., 1671, 6, 3075–3087 CrossRef.
- A. R. Hall, All was light: An introduction to Newton's Opticks, Clarendon Press, Oxford, 1993 Search PubMed.
- R. RichardsKortum and E. SevickMuraca, Annu. Rev. Phys. Chem., 1996, 47, 555–606 CrossRef CAS.
- R. Singh and F. Riess, Curr. Sci., 1998, 75, 965–971 Search PubMed.
- J. Strutt, Philos. Mag., 1899, 47, 375–394 Search PubMed.
- Anon, American Chemical Society International Historic Chemical Landmarks, The Raman Effect, http://portal.acs.org/portal/acs/corg/content?_nfpb=true&_pageLabel=PP_SUPERARTICLE&node_id=515&use_sec=false&sec_url_var=region1&__uuid=59c1b22b-50f3-4af9-b4b5-d5ad83faf161, accessed 27 March 2013.
- C. V. Raman, Nature, 1921, 108, 367 CrossRef.
- C. V. Raman, Nature, 1921, 108, 402–403 CrossRef CAS.
- C. V. Raman, Nature, 1922, 109, 444–445 CrossRef CAS.
- C. V. Raman and K. S. Krishnan, Nature, 1928, 121, 501–502 CrossRef CAS.
- D. I. Ellis, Bioanalysis, 2011, 3, 1189–1194 CrossRef CAS.
- D. I. Ellis and R. Goodacre, Analyst, 2006, 131, 875–885 RSC.
- N. B. Colthup, L. H. Daly and S. E. Wiberly, Introduction to Infrared and Raman Spectroscopy, Academic Press, New York, 1990 Search PubMed.
- P. Hendra, C. Jones and G. Warnes, Fourier Transform Raman Spectroscopy, Ellis Horwood, Chichester, 1991 Search PubMed.
- B. Schrader, Infrared and Raman spectroscopy: methods and applications, Verlag Chemie, Weinheim, 1995 Search PubMed.
- D. I. Ellis, W. B. Dunn, J. L. Griffin, J. W. Allwood and R. Goodacre, Pharmacogenomics, 2007, 8, 1243–1266 CrossRef CAS.
- D. I. Ellis, G. G. Harrigan and R. Goodacre, in Metabolic Profiling: Its Role in Biomarker Discovery and Gene Function Analysis, ed. R. Goodacre and G. G. Harrigan, Kluwer Academic, Boston, 2003, pp. 111–124 Search PubMed.
- R. M. Jarvis, A. Brooker and R. Goodacre, Anal. Chem., 2004, 76, 5198–5202 CrossRef CAS.
- R. M. Jarvis and R. Goodacre, Anal. Chem., 2004, 76, 40–47 CrossRef CAS.
- B. Cletus, W. Olds, E. L. Izake, S. Sundarajoo, P. M. Fredericks and E. Jaatinen, Anal. Bioanal. Chem., 2012, 403, 255–263 CrossRef CAS.
- M. E. Farrell, E. L. Holthoff and P. M. Pellegrino, in Chemical, Biological, Radiological, Nuclear, and Explosives, ed. A. W. Fountain, SPIE-Int. Soc. Optical Engineering, Bellingham, 2012, vol. 8358 Search PubMed.
- D. I. Ellis, V. L. Brewster, W. B. Dunn, J. W. Allwood, A. P. Golovanov and R. Goodacre, Chem. Soc. Rev., 2012, 41, 5706–5727 RSC.
- D. I. Ellis, D. Broadhurst, S. J. Clarke and R. Goodacre, Analyst, 2005, 130, 1648–1654 RSC.
- C. W. Freudiger, W. Min, G. R. Holtom, B. W. Xu, M. Dantus and X. S. Xie, Nat. Photonics, 2011, 5, 103–109 CrossRef CAS.
- B. G. Saar, L. R. Contreras-Rojas, X. S. Xie and R. H. Guy, Mol. Pharmaceutics, 2011, 8, 969–975 CrossRef CAS.
- H. W. Wang, T. T. Le and J. X. Cheng, Opt. Commun., 2008, 281, 1813–1822 CrossRef CAS.
- R. Manoharan, Y. Wang and M. S. Feld, Spectrochim. Acta, Part A, 1996, 52, 215–249 CrossRef.
- B. G. Saar, C. W. Freudiger, J. Reichman, C. M. Stanley, G. R. Holtom and X. S. Xie, Science, 2010, 330, 1368–1370 CrossRef CAS.
- W. B. Dunn, A. Erban, R. J. M. Weber, D. J. Creek, M. Brown, R. Brietling, T. Hankemeier, R. Goodacre, S. Neumann, J. Kopka and M. R. Viant, Metabolomics, 2013, 9, S44–S66 CrossRef.
- E. G. Armitage, H. L. Kotze and N. P. Lockyer, Metabolomics, 2013, 9, S102–S109 CrossRef.
- P. M. Angel and R. M. Caprioli, Biochemistry, 2012 DOI:10.1021/bi301519p.
- J. S. Fletcher and J. C. Vickerman, Anal. Chem., 2013, 85, 610–639 CrossRef CAS.
- R. Masyuko, E. J. Lanni, J. V. Sweedler and P. W. Bohn, Analyst, 2013, 138, 1924–1939 RSC.
- Coherent Raman Scattering Microscopy, ed. J. X. Cheng and X. S. Xie, CRC Press, Abingdon, Oxon, UK, 2012 Search PubMed.
- W. M. Tolles, J. W. Nibler, J. R. McDonald and A. B. Harvey, Appl. Spectrosc., 1977, 31, 253–271 CrossRef CAS.
- J. X. Cheng and X. S. Xie, J. Phys. Chem. B, 2004, 108, 827–840 CrossRef CAS.
- C. L. Evans, X. Y. Xu, S. Kesari, X. S. Xie, S. T. C. Wong and G. S. Young, Opt. Express, 2007, 15, 12076–12087 CrossRef CAS.
- X. Zhang, M. B. J. Roeffaers, S. Basu, J. R. Daniele, D. Fu, C. W. Freudiger, G. R. Holtom and X. S. Xie, ChemPhysChem, 2012, 13, 1054–1059 CrossRef CAS.
- M. O. Scully, G. W. Kattawar, R. P. Lucht, T. Opatrny, H. Pilloff, A. Rebane, A. V. Sokolov and M. S. Zubairy, Proc. Natl. Acad. Sci. U. S. A., 2002, 99, 10994–11001 CrossRef CAS.
- T. Hellerer, C. Axang, C. Brackmann, P. Hillertz, M. Pilon and A. Enejder, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 14658–14663 CrossRef CAS.
- C. Krafft, A. A. Ramoji, C. Bielecki, N. Vogler, T. Meyer, D. Akimov, P. Rosch, M. Schmitt, B. Dietzek, I. Petersen, A. Stallmach and J. Popp, J. Biophotonics, 2009, 2, 303–312 CrossRef.
- T. B. Huff and J. X. Cheng, J. Microsc., 2007, 225, 175–182 CrossRef CAS.
- C. Matthaus, S. Dochow, G. Bergner, A. Lattermann, B. F. M. Romeike, E. T. Marple, C. Krafft, B. Dietzek, B. R. Brehm and J. Popp, Anal. Chem., 2012, 84, 7845–7851 CrossRef.
- H. G. Breunig, R. Buckle, M. Kellner-Hofer, M. Weinigel, J. Lademann, W. Sterry and K. Konig, Microsc. Res. Tech., 2012, 75, 492–498 CrossRef.
- C. W. Freudiger, W. Min, B. G. Saar, S. Lu, G. R. Holtom, C. W. He, J. C. Tsai, J. X. Kang and X. S. Xie, Science, 2008, 322, 1857–1861 CrossRef CAS.
- E. Ploetz, S. Laimgruber, S. Berner, W. Zinth and P. Gilch, Appl. Phys. B: Lasers Opt., 2007, 87, 389–393 CrossRef CAS.
- F. M. Watt and B. L. M. Hogan, Science, 2000, 287, 1427–1430 CrossRef CAS.
- I. Klimanskaya, N. Rosenthal and R. Lanza, Nat. Rev. Drug Discovery, 2008, 7, 131–142 CrossRef CAS.
- D. A. Robinton and G. Q. Daley, Nature, 2012, 481, 295–305 CrossRef CAS.
- S. F. Badylak, D. J. Weiss, A. Caplan and P. Macchiarini, Lancet, 2012, 379, 943–952 CrossRef.
- F. C. Pascut, H. T. Goh, N. Welch, L. D. Buttery, C. Denning and I. Notingher, Biophys. J., 2011, 100, 251–259 CrossRef CAS.
- B. E. Reubinoff, M. F. Pera, C. Y. Fong, A. Trounson and A. Bongso, Nat. Biotechnol., 2000, 18, 399–404 CrossRef CAS.
- F. C. Pascut, H. T. Goh, V. George, C. Denning and I. Notingher, J. Biomed. Opt., 2011, 16, 045002 CrossRef.
- A. Ghita, F. C. Pascut, M. Mather, V. Sottile and I. Notingher, Anal. Chem., 2012, 84, 3155–3162 CrossRef CAS.
- A. Downes, R. Mouras, P. Bagnaninchi and A. Elfick, J. Raman Spectrosc., 2011, 42, 1864–1870 CrossRef CAS.
- S. O. Konorov, H. G. Schulze, C. G. Atkins, J. M. Piret, S. A. Aparicio, R. F. B. Turner and M. W. Blades, Anal. Chem., 2011, 83, 6254–6258 CrossRef CAS.
- S. O. Konorov, H. G. Schulze, J. M. Piret, R. F. B. Turner and M. W. Blades, J. Raman Spectrosc., 2011, 42, 1135–1141 CrossRef CAS.
- H. G. Schulze, S. O. Konorov, N. J. Caron, J. M. Piret, M. W. Blades and R. F. B. Turner, Anal. Chem., 2010, 82, 5020–5027 CrossRef CAS.
- D. H. Kim, R. M. Jarvis, J. W. Allwood, G. Batman, R. E. Moore, E. Marsden-Edwards, L. Hampson, I. N. Hampson and R. Goodacre, Anal. Bioanal. Chem., 2010, 398, 3051–3061 CrossRef CAS.
- V. V. Yakovlev, H. F. Zhang, G. D. Noojin, M. L. Denton, R. J. Thomas and M. O. Scully, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 20335–20339 CrossRef CAS.
- C. L. Zavaleta, B. R. Smith, I. Walton, W. Doering, G. Davis, B. Shojaei, M. J. Natan and S. S. Gambhir, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 13511–13516 CrossRef CAS.
- D. Fu, F. K. Lu, X. Zhang, C. Freudiger, D. R. Pernik, G. Holtom and X. S. Xie, J. Am. Chem. Soc., 2012, 134, 3623–3626 CrossRef CAS.
- A. Zoladek, F. C. Pascut, P. Patel and I. Notingher, J. Raman Spectrosc., 2011, 42, 251–258 CrossRef CAS.
- M. Miljkovic, T. Chernenko, M. J. Romeo, B. Bird, C. Matthaus and M. Diem, Analyst, 2010, 135, 2002–2013 RSC.
- W. Min, C. W. Freudiger, S. J. Lu and X. S. Xie, in Annual Review of Physical Chemistry, ed. S. R. Leone, P. S. Cremer, J. T. Groves and M. A. Johnson, Annual Reviews, Palo Alto, 2011, vol. 62, pp. 507–530 Search PubMed.
- E. Boisselier and D. Astruc, Chem. Soc. Rev., 2009, 38, 1759–1782 RSC.
- M. G. Shim, L. Song, N. E. Marcon and B. C. Wilson, Photochem. Photobiol., 2000, 72, 146–150 CAS.
- N. Stone, P. Stavroulaki, C. A. Fulljames, M. Birchall and H. Barr, in Biomedical Spectroscopy: Vibrational Spectroscopy and Other Novel Techniques, ed. A. MahadevanJansen and G. J. Puppels, SPIE-Int. Soc. Optical Engineering, Bellingham, 2000, vol. 1, pp. 120–128 Search PubMed.
- C. Kendall, N. Stone, N. Shepherd, K. Geboes, B. Warren, R. Bennett and H. Barr, J. Pathol., 2003, 200, 602–609 CrossRef.
- P. Crow, A. Molckovsky, N. Stone, J. Uff, B. Wilson and L. M. Wongkeesong, Urology, 2005, 65, 1126–1130 CrossRef CAS.
- J. C. C. Day, R. Bennett, B. Smith, C. Kendall, J. Hutchings, G. M. Meaden, C. Born, S. Yu and N. Stone, Phys. Med. Biol., 2009, 54, 7077–7087 CrossRef CAS.
- L. M. Almond, J. Hutchings, C. Kendall, J. C. C. Day, O. A. C. Stevens, G. R. Lloyd, N. A. Shepherd, H. Barr and N. Stone, J. Biomed. Opt., 2012, 17, 081421 CrossRef.
- L. M. Almond, J. Hutchings, N. Shepherd, H. Barr, N. Stone and C. Kendall, J. Biophotonics, 2011, 4, 685–695 CrossRef.
- Z. W. Huang, S. K. Teh, W. Zhen, J. H. Mo, K. Lin, X. Z. Shao, K. Y. Ho, M. Teh and K. G. Yeoh, Opt. Lett., 2009, 34, 758–760 CrossRef.
- X. D. Li, S. A. Boppart, J. Van Dam, H. Mashimo, M. Mutinga, W. Drexler, M. Klein, C. Pitris, M. L. Krinsky, M. E. Brezinski and J. G. Fujimoto, Endoscopy, 2000, 32, 921–930 CrossRef CAS.
- R. L. P. Rhoten, M. G. Luciano and G. H. Barnett, Neurosurgery, 1997, 40, 632–637 CAS.
- M. S. Bergholt, W. Zheng, K. Lin, K. Y. Ho, M. Teh, K. G. Yeoh, J. B. Y. So and Z. Huang, Technol. Cancer Res. Treat., 2011, 10, 103–112 CAS.
- L. F. Santos, R. Wolthuis, S. Koljenovic, R. M. Almeida and G. J. Puppels, Anal. Chem., 2005, 77, 6747–6752 CrossRef CAS.
- J. H. Mo, W. Zheng, J. J. H. Low, J. Ng, A. Ilancheran and Z. W. Huang, Anal. Chem., 2009, 81, 8908–8915 CrossRef CAS.
- S. Duraipandian, W. Zheng, J. Ng, J. J. H. Low, A. Ilancheran and Z. W. Huang, Anal. Chem., 2012, 84, 5913–5919 CrossRef CAS.
- A. F. Garcia-Flores, L. Raniero, R. A. Canevari, K. J. Jalkanen, R. A. Bitar, H. S. Martinho and A. A. Martin, Theor. Chem. Acc., 2011, 130, 1231–1238 CrossRef CAS.
- K. Lin, D. L. P. Cheng and Z. W. Huang, Biosens. Bioelectron., 2012, 35, 213–217 CrossRef CAS.
- Z. W. Huang, S. K. Teh, W. Zheng, K. Lin, K. Y. Ho, M. Teh and K. G. Yeoh, Biosens. Bioelectron., 2010, 26, 383–389 CrossRef CAS.
- S. Duraipandian, M. S. Bergholt, W. Zheng, K. Y. Ho, M. Teh, K. G. Yeoh, J. B. Y. So, A. Shabbir and Z. W. Huang, J. Biomed. Opt., 2012, 17, 081418 CrossRef.
- M. S. Bergholt, K. Lin, W. Zheng, D. P. C. Lau and Z. W. Huang, J. Biomed. Opt., 2012, 17, 077002 CrossRef.
- M. J. Baker, E. Gazi, M. D. Brown, J. H. Shanks, N. W. Clarke and P. Gardner, J. Biophotonics, 2009, 2, 104–113 CrossRef CAS.
- M. J. Baker, E. Gazi, M. D. Brown, J. H. Shanks, P. Gardner and N. W. Clarke, Br. J. Cancer, 2008, 99, 1859–1866 CrossRef CAS.
- P. Matousek, I. P. Clark, E. R. C. Draper, M. D. Morris, A. E. Goodship, N. Everall, M. Towrie, W. F. Finney and A. W. Parker, Appl. Spectrosc., 2005, 59, 393–400 CrossRef CAS.
- R. Baker, P. Matousek, K. L. Ronayne, A. W. Parker, K. Rogers and N. Stone, Analyst, 2007, 132, 48–53 RSC.
- P. Matousek and N. Stone, J. Biophotonics, 2013, 6, 7–19 CrossRef CAS.
- P. Matousek, E. R. C. Draper, A. E. Goodship, I. P. Clark, K. L. Ronayne and A. W. Parker, Appl. Spectrosc., 2006, 60, 758–763 CrossRef CAS.
- C. Eliasson and P. Matousek, Anal. Chem., 2007, 79, 1696–1701 CrossRef CAS.
- C. Eliasson, N. A. Macleod and P. Matousek, Anal. Chem., 2007, 79, 8185–8189 CrossRef CAS.
- E. L. Izake, B. Cletus, W. Olds, S. Sundarajoo, P. M. Fredericks and E. Jaatinen, Talanta, 2012, 94, 342–347 CrossRef CAS.
- L. Lee, A. Frisby, R. Mansson and R. J. Hopkins, in Optics and Photonics for Counterterrorism and Crime Fighting VII Optical Materials in Defence Systems Technology VIII and Quantum-Physics-Based Information Security, ed. C. Lewis, D. Burgess, R. Zamboni, F. Kajzar, A. A. Szep, M. T. Gruneisen, M. Dusek and J. G. Rarity, SPIE-Int. Soc. Optical Engineering, Bellingham, 2011, vol. 8189 Search PubMed.
- D. T. Yang and Y. B. Ying, Appl. Spectrosc. Rev., 2011, 46, 539–560 CrossRef.
- M. V. Schulmerich, K. A. Dooley, M. D. Morris, T. M. Vanasse and S. A. Goldstein, J. Biomed. Opt., 2006, 11, 060502 CrossRef.
- M. V. Schulmerich, W. F. Finney, V. Popescu, M. D. Morris, T. M. Vanasse and S. A. Goldstein, in Biomedical Vibrational Spectroscopy III: Advances in Research and Industry, ed. A. MahadevanJansen and W. H. Petrich, SPIE-Int. Soc. Optical Engineering, Bellingham, 2006, vol. 6093, p. O930 Search PubMed.
- M. V. Schulmerich, J. H. Cole, J. M. Kreider, F. Esmonde-White, K. A. Dooley, S. A. Goldstein and M. D. Morris, Appl. Spectrosc., 2009, 63, 286–295 CrossRef CAS.
- P. I. Okagbare, F. W. L. Esmonde-White, S. A. Goldstein and M. D. Morris, Analyst, 2010, 135, 3142–3146 RSC.
- P. I. Okagbare, F. W. L. Esmonde-White, S. A. Goldstein and M. D. Morris, in Photonic Therapeutics and Diagnostics VII, ed. N. Kollias, B. Choi, H. Zeng, H. W. Kang, B. E. Knudsen, B. J. F. Wong, J. F. R. Ilgner, K. W. Gregory, G. J. Tearney, L. Marcu, H. Hirschberg, S. J. Madsen, A. Mandelis, A. MahadevanJansen and E. D. Jansen, SPIE-Int. Soc. Optical Engineering, Bellingham, 2011, vol. 7883 Search PubMed.
- J. R. Maher, J. Inzana, M. Takahata, H. A. Awad and A. J. Berger, in Biomedical Vibrational Spectroscopy V: Advances in Research and Industry, ed. A. MahadevanJansen and W. Petrich, SPIE-Int. Soc. Optical Engineering, Bellingham, 2012, vol. 8219 Search PubMed.
- P. I. Okagbare, D. Begun, M. Tecklenburg, A. Awonusi, S. A. Goldstein and M. D. Morris, J. Biomed. Opt., 2012, 17, 090502 CrossRef.
- A. S. Haka, K. E. Shafer-Peltier, M. Fitzmaurice, J. Crowe, R. R. Dasari and M. S. Feld, Cancer Res., 2002, 62, 5375–5380 CAS.
- A. S. Haka, K. E. Shafer-Peltier, M. Fitzmaurice, J. Crowe, R. R. Dasari and M. S. Feld, Proc. Natl. Acad. Sci. U. S. A., 2005, 102, 12371–12376 CrossRef CAS.
- J. T. Motz, S. J. Gandhi, O. R. Scepanovic, A. S. Haka, J. R. Kramer, R. R. Dasari and M. S. Feld, J. Biomed. Opt., 2005, 10, 031113 CrossRef.
- A. S. Haka, Z. Volynskaya, J. A. Gardecki, J. Nazemi, R. Shenk, N. Wang, R. R. Dasari, M. Fitzmaurice and M. S. Feld, J. Biomed. Opt., 2009, 14, 054023 CrossRef.
- N. Stone, R. Baker, K. Rogers, A. W. Parker and P. Matousek, Analyst, 2007, 132, 899–905 RSC.
- N. Stone and P. Matousek, Cancer Res., 2008, 68, 4424–4430 CrossRef CAS.
- M. M. Kerssens, P. Matousek, K. Rogers and N. Stone, Analyst, 2010, 135, 3156–3161 RSC.
- A. Mahadevan-Jansen, M. D. Keller, E. Vargis, B. Caldwell, T. Q. Nguyen, N. D. Granja, M. Sanders and M. C. Kelley, Spectroscopy, 2011, 48–58 CAS.
- M. D. Keller, E. Vargis, N. D. Granja, R. H. Wilson, M. A. Mycek, M. C. Kelley and A. Mahadevan-Jansen, J. Biomed. Opt., 2011, 16, 077006 CrossRef.
- J. M. Yuen, N. C. Shah, J. T. Walsh, M. R. Glucksberg and R. P. Van Duyne, Anal. Chem., 2010, 82, 8382–8385 CrossRef CAS.
- K. Ma, J. M. Yuen, N. C. Shah, J. T. Walsh, M. R. Glucksberg and R. P. Van Duyne, Anal. Chem., 2011, 83, 9146–9152 CrossRef CAS.
- N. Stone, K. Faulds, D. Graham and P. Matousek, Anal. Chem., 2010, 82, 3969–3973 CrossRef CAS.
- N. Stone, M. Kerssens, G. R. Lloyd, K. Faulds, D. Graham and P. Matousek, Chem. Sci., 2011, 2, 776–780 RSC.
- A. J. Lowe, Y. S. Huh, A. D. Strickland, D. Erickson and C. A. Batt, Anal. Chem., 2010, 82, 5810–5814 CrossRef CAS.
- K. K. Maiti, U. S. Dinish, A. Samanta, M. Vendrell, K. S. Soh, S. J. Park, M. Olivo and Y. T. Chang, Nano Today, 2012, 7, 85–93 CrossRef CAS.
- J. A. Dougan and K. Faulds, Analyst, 2012, 137, 545–554 RSC.
- M. Vendrell, K. K. Maiti, K. Dhaliwal and Y.-T. Chang, Trends Biotechnol., 2013, 31, 249–257 CrossRef CAS.
- J. H. Granger, M. C. Granger, M. A. Firpo, S. J. Mulvihill and M. D. Porter, Analyst, 2013, 138, 410–416 RSC.
- G. F. Wang, R. J. Lipert, M. Jain, S. Kaur, S. Chakraboty, M. P. Torres, S. K. Batra, R. E. Brand and M. D. Porter, Anal. Chem., 2011, 83, 2554–2561 CrossRef CAS.
- H. Chon, C. Lim, S. M. Ha, Y. Ahn, E. K. Lee, S. I. Chang, G. H. Seong and J. Choo, Anal. Chem., 2010, 82, 5290–5295 CrossRef CAS.
- J. Qian, L. Jiang, F. H. Cai, D. Wang and S. L. He, Biomaterials, 2011, 32, 1601–1610 CrossRef CAS.
- R. McQueenie, R. Stevenson, R. Benson, N. MacRitchie, I. McInnes, P. Maffia, K. Faulds, D. Graham, J. Brewer and P. Garside, Anal. Chem., 2012, 84, 5968–5975 CrossRef CAS.
- D. Graham, R. Stevenson, D. G. Thompson, L. Barrett, C. Dalton and K. Faulds, Faraday Discuss., 2011, 149, 291–299 RSC.
- D. P. Cowcher, X. Yun and R. Goodacre, Anal. Chem., 2013, 85, 3297–3302 CrossRef CAS.
- S. Wachsmann-Hogiu, T. Weeks and T. Huser, Curr. Opin. Biotechnol., 2009, 20, 63–73 CrossRef CAS.
- Z. J. Smith, T. R. Huser and S. Wachsmann-Hogiu, Anal. Cell. Pathol., 2012, 35, 145–163 CAS.
- O. R. Hughes, N. Stone, M. Kraft, C. Arens and M. A. Birchall, Head Neck-J. Sci. Spec. Head Neck, 2010, 32, 1544–1553 Search PubMed.
- C. Krafft, G. Steiner, C. Beleites and R. Salzer, J. Biophotonics, 2009, 2, 13–28 CrossRef CAS.
- M. A. Syed and S. H. A. Bokhari, J. Biomed. Nanotechnol., 2011, 7, 229–237 CrossRef CAS.
- Emerging Raman Applications and Techniques in Biomedical and Pharmaceutical Fields (Biological and Medical Physics, Biomedical Engineering), ed. P. Matousek and M. D. Morris, Springer, Heidelberg, 2010 Search PubMed.
- D. I. Ellis and R. Goodacre, Curr. Opin. Biotechnol., 2012, 23, 22–28 CrossRef CAS.
- R. A. Alvarez-Puebla and L. M. Liz-Marzan, Small, 2010, 6, 604–610 CrossRef CAS.
- X. M. Qian and S. M. Nie, Chem. Soc. Rev., 2008, 37, 912–920 RSC.
- H. F. Wang, Y. Fu, P. Zickmund, R. Y. Shi and J. X. Cheng, Biophys. J., 2005, 89, 581–591 CrossRef CAS.
- K. J. Blow and D. Wood, IEEE J. Quantum Electron., 1989, 25, 2665–2673 CrossRef CAS.
- H. Abramczyk, B. Brozek-Pluska, J. Surmacki, J. Jablonska and R. Kordek, J. Mol. Liq., 2011, 164, 123–131 CrossRef CAS.
- J. M. Benevides, S. A. Overman and G. J. Thomas, J. Raman Spectrosc., 2005, 36, 279–299 CrossRef CAS.
- J. De Gelder, K. De Gussem, P. Vandenabeele and L. Moens, J. Raman Spectrosc., 2007, 38, 1133–1147 CrossRef CAS.
- B. Singh, R. Gautam, S. Kumar, B. N. V. Kumar, U. Nongthomba, D. Nandi, G. Mukherjee, V. Santosh, K. Somasundaram and S. Umapathy, Curr. Sci., 2012, 102, 232–244 CAS.
- Z.-Q. Wen, J. Pharm. Sci., 2007, 96, 2861–2878 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2013 |
