Accurate and effective live bacteria microarray patterning on thick polycationic polymer layers co-patterned with HMDS†
Ieong
Wong‡
a,
Xianting
Ding‡
a,
Chunsheng
Wu
b and
Chih-Ming
Ho
*a
aDepartment of Mechanical and Aerospace Engineering, School of Engineering and Applied Science, University of California, Los Angeles 420 Westwood PlazaLos Angeles, CA 90095-1597, USA. E-mail: chihming@ucla.edu; Fax: +1 (310) 206 2302; Tel: +1 (310) 825 9993
bBiosensor National Special Laboratory, MOE Key Laboratory of Biomedical Engineering, Department of Biomedical Engineering, Zhejiang University, Hangzhou, 310027, P. R. China
First published on 3rd July 2012
Abstract
A new bacteria microarray patterning technique is developed by patterning thick polycationic polymers on a glass surface, which generates high-coverage and high-precision E. coli cell patterns. Cell immobilization efficiency is greatly improved, compared to that of the conventional monolayer surface patterning approach. Cell viability tests show very low cytotoxicity of polyethyleneimine (PEI). This advancement should further accelerate biomedical and bacteriological research on the micro scale.
Intersection between micro/nano technology and microbiology in the recent decade has made a significant impact on the research and applications of microorganisms with high spatial resolution.1 Within this context, site-specific immobilization of bacteria is still a subject of substantial interest.1,2 Its key importance in many biomedical and bacteriological research and applications can be found in fundamental studies in cell–cell and cell–surface interactions towards biofilm formation involving quorum sensing,3 fabrication of high-throughput biochemical and drug screening platforms,4 functional biosensors,5 and energy harvesting from motile bacterial biomotors for nano-electro-mechanical devices.6
These technologies require highly accurate spatial placement of live bacterial cells at specific locations, such as transducer electrodes or bacterial microarrays, with microscale or even single-cell resolution. While direct printing of bacteria has been demonstrated using microcontact printing with hydrogel7 or elastomer stamps,8 thermal inkjet methods,9 and bubble-jet10 methods with good viability on agar surfaces, these methods suffer significantly from their low pattern resolution (hundreds of microns). On the other hand, a majority of the high-resolution bacteria patterning platforms are constructed on chemically modified surfaces fabricated using micro/nano surface modification and patterning techniques.4 Using this approach, surface immobilizations of bacteria have been demonstrated on chemically modified surface patterns or microwells through hydrophobic interaction via alkanethiols,11 electrostatic interaction via poly-L-lysine,12 bio-specific interactions via conjugations between antibody and virus,13 antibody and antigen,14 or streptavidin and biotin,15 or chemical attachment via amine- and carboxylic acid-based conjugation.16 In addition, to site-specifically immobilize bacteria at the desired location, the area where cell adhesion is not desired is commonly passivated with a layer of poly(ethylene glycol) (PEG). Such patterned surfaces have been fabricated on glass and silicon surfaces using micro/nano fabrication technologies, such as microcontact printing on PDMS,17 capillary micromolding,13 focused Ga+ ion beam etching,18 photocatalytic oxidation of silane,16 photolithography,19 and low-energy electron beam lithography.20
Recently, the efficiency and reproducibility of a surface modification-based bacteria patterning approach have been questioned.18 Such issues, which were not found in eukaryotic cell patterning, may be attributed to the rigid cell wall of the bacteria allowing only a small contact area with the substrate surface, or the pili and extracellular polymeric substances preventing direct contact to the substrate surface.18 These issues were previously addressed elegantly where bacteria immobilization efficiency could be enhanced through careful selection of an affinity-purified antibody which targets surface antigens at the outer cell wall.18 However, this approach could be complicated by the extra effort, cost, and time required for obtaining the purified antibody, as well as prior knowledge on the genetic and physiological information of the test strains.
Another reason for the low patterning and immobilization efficiency, as has been pointed out recently,21 could be due to the fact that most surface modification methods are based on modifying surfaces with only ultrathin or monolayers of tethering material for bacteria immobilization purposes.21 According to the strains we tested, monolayers of tethering materials, e.g. poly-L-lysine (PLL), do not provide enough attachment force, such as positive electrostatic charge, for bacteria immobilization (data not shown). In fact, in most bacteria surface adhesion force measurement studies using Atomic Force Microscopy (AFM), where secure bacteria attachment on the AFM tips is critical, it is a common practice to coat a thick layer of PLL by completely air-drying PLL drops on the AFM tips without any water rinsing.21
Herein we developed a new surface patterning technique to generate high-coverage bacterial microarrays with high precision, by significantly enhancing bacteria–surface adhesion with a thick polycationic layer patterned with a photolithographic lift-off process (Fig. 1). Besides, to selectively immobilize bacteria at the desired location, a usual practice is to functionalize the area where bacteria immobilization is not desired with a protein resistant layer, mostly PEG. PEG works for bacteria passivation because bacteria generally adhere to a surface through a series of surface interactions which ultimately involve the formation of various protein layers.22 In our work, in contrast to a conventional practice of passivating a surface with a PEG layer for bacteria repulsion purposes, as demonstrated in most previous works,13,17,18,23,24 we exploited a new use of a common photoresist adhesion promoter, hexamethyldisilazane (HMDS), for a surface passivation purpose. This unconventional use of HMDS significantly simplifies the fabrication process.
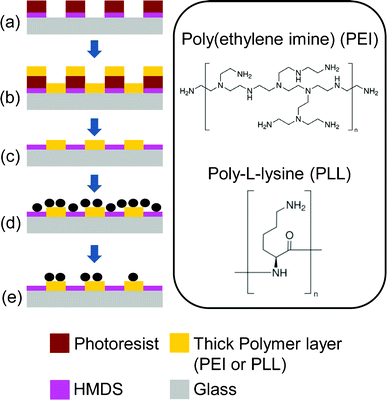 | ||
| Fig. 1 Fabrication process of the cell patterning surface. (a) Glass slides coated with HMDS and patterned with photoresist, (b) polycationic solution incubation, followed by air blow-dry, (c) lift-off the photoresist in acetone and isopropyl alcohol, and blow-dry, (d) cells incubated on the patterned PEI chip, and (e) rinsing off non-specific attached cells with PBS. The inset shows the positively charged amine groups in the molecules. | ||
Fig. 1 shows the fabrication process of the thick polycationic surface patterning (see ESI†). Briefly, oxygen plasma treated glass surfaces were coated with HMDS by evaporation, followed by a conventional photolithography process with positive-tone photoresist (AZ5214) to generate 2 μm holes in the photoresist layer. The samples were then treated with oxygen plasma again to clear off the exposed HMDS layer in the photoresist pattern. This process reveals the bottom hydrophilic silanol groups. An aqueous solution of branched cationic polymers (PEI or PLL) was dispensed evenly onto the sample surface and incubated for 30 min at room temperature, allowing physisorption of the polymer on the negatively charged glass surface. After incubation, excess PEI solution was directly blow dried with a stream of nitrogen gas. A lift-off process in acetone with 10 second ultrasonic pulses was used to remove the photoresist and the polymer layer on it, leaving a condensed layer of polymer co-patterned with HMDS on the glass surface. Water was avoided after the polymer was incubated on the surface, due to the high solubility of the hydrophilic polymer in water. AFM scans of the PEI patterned surface revealed a layer of polymer with a thickness of 100–200 nm co-patterned with HMDS (Fig. 2a). It is worth noting that such a large thickness variation is found on almost every single 2 μm adhesion spot with the center of the spot always thinner due to the lift-off process (see Fig. 2a). Such polymer patterns can be fabricated consistently across samples, which in turn consistently generate high-quality bacteria cell patterns. Therefore, we believe the variation in the cell-adhesion site thickness uniformity might not be a critical factor in affecting the cell attachment. We also believe, theoretically, that the cell-adhesion strength could reach a plateau with increasing layer thickness. In our example, it is possible that the adhesion strength has already reached the plateau at a thickness >100 nm, as evidenced by the high-quality cell patterns across a large area on the substrate (Fig. 3).
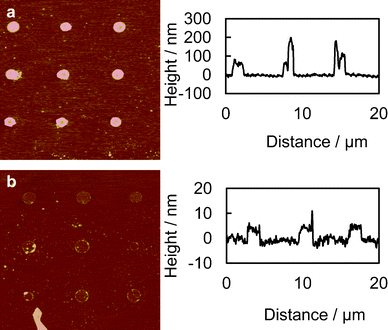 | ||
| Fig. 2 AFM scans of the chemically patterned surfaces. (a) Thick layer of PEI; thickness approximately 100–200 nm and (b) monolayer of PEI patterns, fabricated by rinsing the thick PEI layer patterns with DI water to remove the PEI not directly adsorbed on the glass surface; thickness is approximately 5 nm. Images were processed with the Nanoscope Median filter with an order of 3 × 3 to filter out the high-frequency noise. | ||
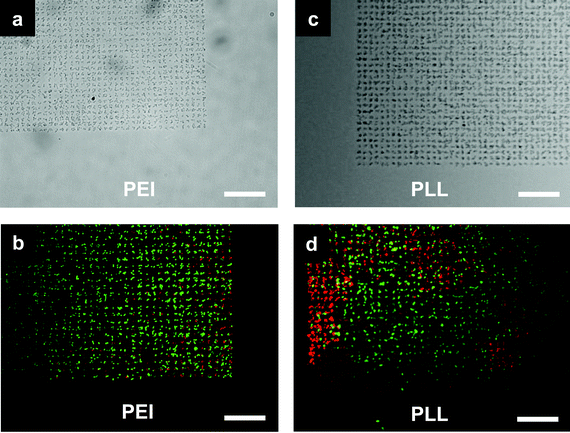 | ||
| Fig. 3 E. coli cells microarray. (a,b) Bacteria adhered to poly(ethyleneimine) (PEI) patterned spots and (c,d) bacteria adhered to poly-L-lysine (PLL) patterned spots. Each spot was 2 μm and was occupied by around 2–3 cells. (b,d) BacLight live/dead assay staining of the cells: live cells fluoresce green and dead cell fluoresce red. PEI patterns generated cell arrays with more than 95% live cells while PLL patterns generated cell arrays of about 75% live cells. Scale bar is 20 μm. | ||
In this study, wild type JCL16 E. coli strain was used as the biological model to test the patterning methodology. Fig. 3 shows the high-specificity patterning of bacteria on the PEI and PLL coated spots while no bacteria could be found on the HMDS passivated surface. The results also revealed a superior passivation ability of HMDS against E. coli binding, in contrast to many protein and mammalian cell patterning studies where HMDS was used to enhance the nonspecific biomolecule physisorption and subsequently used for cell adhesion.25,26 This work also showed an advantage of the pattern fidelity compared to that of conventional monolayer surface patterning approaches for bacteria patterning, especially with its significant improvements in pattern coverage (99% of the patterned regions occupied by cells). As a control, monolayer PEI and PLL patterns fabricated with an additional water rinsing step to dissolve polymeric materials not directly adsorbed on the glass surface generated bacteria arrays with much less coverage (data not shown), as we anticipated, mainly due to the weak surface–cell interaction.21Fig. 2b shows the AFM scans of the samples patterned with monolayers of PEI with a thickness of about 5 nm.
In this work, we unconventionally employed a hydrophobic material, HMDS, as the passivation layer against bacterial cell binding, in contrast to the conventional use of hydrophilic materials, such as PEG. Our study showed that HMDS served as a good candidate material for bacteria repulsion purposes. Indeed, surface hydrophobicity has been found to either enhance or resist surface adhesion, depending on the strains of E. coli, according to other AFM studies.27–29 However, the use of fixated (dead) bacterial cells for force measurements may not represent the same conditions as where live and motile cells are allowed to dynamically interact with the substrate surface. Moreover, short term force measurements with AFM may not be able to address the mechanisms of relatively long term bacteria–surface interactions during the incubation period (30 min) in our experiment.
Using a thick layer of the polycationic polymers to enhance bacteria attachment may raise concerns regarding cytotoxicity towards the bacterial cells.30,31 Since the cytotoxicity may adversely affect the patterned cells for subsequent biological studies, we carried out a live/dead assay (LIVE/DEAD BacLight Viability Kit, Invitrogen) to examine the physiological conditions of the patterned microbes.17 Cell viability condition was indicated by the fluorescent color where live bacterial cells fluoresce green and dead bacterial cells fluoresce red. The incubation time for attaching the cells on the substrate is 30 min, after which the viability test is immediately carried out (see ESI†). The viability test is carried out by following the manufacturer's protocol which comprises a 15 min incubation of the cell pattern in a viability test assay solution, followed with image acquisition under a microscope. Taking into account the image acquisition time, the cells are alive on the pattern for at least 1 h. As presented in Fig. 3b and d, more than 95% of cells were alive on the PEI patterns and 75% on the PLL patterns. Results show that the cell viability is much better on PEI patterns compared to PLL patterns. Studies have shown that PLL has strong cytotoxicity and has been employed as an antimicrobial surface coating.32 However, PEI is not found to be an effective antimicrobial agent until it is alkylated.33,34
Conclusions
In conclusion, we developed a new bacteria patterning technique which can generate high-coverage and high-precision bacteria cell microarrays, by enhancing bacteria–surface adhesion with a thick polycationic layer patterned on a glass surface with a lift-off process. To our knowledge, this is the first work attempting to pattern bacteria on a thick layer of polycationic polymer. The thick PEI and PLL layers were found to be very effective in improving cell attachment. The method generated single-cell microarrays of wild type E. coli with significant improvement in patterning coverage compared to that of a conventional monolayer surface patterning approach. This work also discovered a good passivation ability of HMDS against E. coli binding, in contrast to that of the conventional consensus of hydrophobic material on enhancing nonspecific biomolecule binding. Considering the cytotoxicity possessed by high molecular weight polycationic polymers which may potentially affect the physiological conditions of the patterned bacteria, cell viability tests were carried out and the results show that the cytotoxicity effect was negligible on PEI patterns relative to the PLL patterns. The major advances on bacterial cell immobilization efficiency presented in this work could substantially promote and accelerate biomedical and bacteriological research and applications, which demand high stringency and efficiency for the localization of bacterial cells, such as biosensors, high-throughput drug screening, and biofuel cells.Acknowledgements
The authors thank Prof. James C. Liao at University of California, Los Angeles for providing the E. coli cells. This work was supported by the Center for Cell Control (PN2EY018228) through the NIH Roadmap for Nanomedicine and the Center for Scalable and Integrated Nanomanufacturing (SINAM) under the National Science Foundation (CMMI-0751621).References
- D. B. Weibel, W. R. DiLuzio and G. M. Whitesides, Nat. Rev. Microbiol., 2007, 5, 209 CrossRef CAS.
- C. J. Ingham and J. Vlieg, Lab Chip, 2008, 8, 1604 RSC.
- M. R. Parsek and T. Tolker-Nielsen, Curr. Opin. Microbiol., 2008, 11, 560 CrossRef CAS.
- C. J. Ingham, A. Sprenkels, J. Bomer, D. Molenaar, A. van den Berg, J. Vlieg and W. M. de Vos, Proc. Natl Acad. Sci. U. S. A., 2007, 104, 18217 CrossRef CAS.
- S. Belkin, Curr. Opin. Microbiol., 2003, 6, 206 CrossRef CAS.
- H. C. Berg, Phil. Trans. R. Soc. London B, 2000, 355, 491 CrossRef CAS.
- D. B. Weibel, A. Lee, M. Mayer, S. F. Brady, D. Bruzewicz, J. Yang, W. R. DiLuzio, J. Clardy and G. M. Whitesides, Langmuir, 2005, 21, 6436 CrossRef CAS.
- L. P. Xu, L. Robert, O. Y. Qi, F. Taddei, Y. Chen, A. B. Lindner and D. Baigl, Nano Lett., 2007, 7, 2068 CrossRef CAS.
- P. Calvert, Science, 2007, 318, 208 CrossRef CAS.
- J. Merrin, S. Leibler and J. S. Chuang, PLoS One, 2007, 2, e663 Search PubMed.
- B. Rowan, M. A. Wheeler and R. M. Crooks, Langmuir, 2002, 18, 9914 CrossRef CAS.
- S. Rozhok, Z. F. Fan, D. Nyamjav, C. Liu, C. A. Mirkin and R. C. Holz, Langmuir, 2006, 22, 11251 CrossRef CAS.
- K. Y. Suh, A. Khademhosseini, P. J. Yoo and R. Langer, Biomed. Microdevices, 2004, 6, 223 CrossRef CAS.
- S. W. Howell, H. D. Inerowicz, F. E. Regnier and R. Reifenberger, Langmuir, 2003, 19, 436 CrossRef CAS.
- K. B. Lee, Y. H. Jung, Z. W. Lee, S. Kim and I. S. Choi, Biomaterials, 2007, 28, 5594 CrossRef CAS.
- J. P. Bearinger, L. C. Dugan, L. G. Wu, H. Hill, A. T. Christian and J. A. Hubbell, Biotechniques, 2009, 46, 209 CrossRef CAS.
- S. Rozhok, C. K. F. Shen, P. L. H. Littler, Z. F. Fan, C. Liu, C. A. Mirkin and R. C. Holz, Small, 2005, 1, 445 CrossRef CAS.
- Z. Y. Suo, R. Avci, X. H. Yang and D. W. Pascual, Langmuir, 2008, 24, 4161 CrossRef CAS.
- W. G. Koh, A. Revzin, A. Simonian, T. Reeves and M. Pishko, Biomed. Microdevices, 2003, 5, 11 CrossRef CAS.
- P. Krsko, J. B. Kaplan and M. Libera, Acta Biomater., 2009, 5, 589 CrossRef CAS.
- K. Colville, N. Tompkins, A. D. Rutenberg and M. H. Jericho, Langmuir, 2009, 26, 2639 CrossRef.
- Y. H. An and R. J. Friedman, J. Biomed. Mater. Res., 1998, 43, 338 CrossRef CAS.
- X. Qian, S. J. Metallo, I. S. Choi, H. Wu, M. N. Liang and G. M. Whitesides, Anal. Chem., 2002, 74, 1805 CrossRef CAS.
- A. Razatos, Y. L. Ong, F. Boulay, D. L. Elbert, J. A. Hubbell, M. M. Sharma and G. Georgiou, Langmuir, 2000, 16, 9155 CrossRef CAS.
- N. Li and C.-M. Ho, J. Assoc. Lab. Autom., 2008, 13, 237 CrossRef CAS.
- N. Li and C. M. Ho, Lab Chip, 2008, 8, 2105 RSC.
- J. C. Cau, A. Cerf, C. Thibault, M. Genevieve, C. Severac, J. P. Peyrade and C. Vieu, Microelectron. Eng., 2008, 85, 1143 CrossRef CAS.
- A. Cerf, J.-C. Cau and C. Vieu, Colloids Surf., B, 2008, 65, 285 CrossRef CAS.
- Y. L. Ong, A. Razatos, G. Georgiou and M. M. Sharma, Langmuir, 1999, 15, 2719 CrossRef CAS.
- S. Shima, H. Matsuoka, T. Iwamoto and H. Sakai, J. Antibiot., 1984, 37, 1449 CrossRef CAS.
- M. Vaara and T. Vaara, Antimicrob. Agents Chemother., 1983, 24, 114 CrossRef CAS.
- M. Kar, P. S. Vijayakumar, B. L. V. Prasad and S. S. Gupta, Langmuir, 2010, 26, 5772 CrossRef CAS.
- J. Lin, S. Qiu, K. Lewis and A. M. Klibanov, Biotechnol. Prog., 2002, 18, 1082 CrossRef CAS.
- K. Lewis and A. M. Klibanov, Trends Biotechnol., 2005, 23, 343 CrossRef CAS.
Footnotes |
| † Electronic supplementary information (ESI) available: Full experimental details. See DOI: 10.1039/c2ra20938a/ |
| ‡ These authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2012 |
