DOI:
10.1039/C2RA20085F
(Paper)
RSC Adv., 2012,
2, 7146-7155
TMAO and sorbitol attenuate the deleterious action of atmospheric pressure non-thermal jet plasma on α-chymotrypsin
Received
18th November 2011
, Accepted 25th May 2012
First published on 28th May 2012
Abstract
Enzymes exhibit a substantial degree of structural variability in the folded state and they are very sensitive to environmental changes. In order to investigate the effect of environmental changes on enzymes, we have studied the effect of cold atmospheric pressure plasma jets (APPJ) on α-chymotrypsin (CT). The APPJ consists of microdischarges, which are ejected from porous alumina through a 1 mm hole, reaching the CT samples and resulting in changes in the CT conformation. Furthermore, to gain insight into the contribution of the co-solvents, such as osmolytes (1 M trimethylamine N-oxide (TMAO), 1 M proline, 1 M betaine, 1 M sorbitol and 1 M glycerol) and the denaturant (1 M urea), on CT against the APPJ action, we monitored the associated structural changes in α-chymotrypsin (CT) using circular dichroism (CD), fluorescence and NMR measurements. Contrasting results are obtained from experimental data in the case of TMAO and urea, which allow us to infer that TMAO is able to attenuate strongly the deleterious action of APPJ on CT. Furthermore, the deleterious action of urea is enhanced in the presence of APPJ. The alterations in the secondary structure of this β/β protein, as quantified by the CD spectra, show reasonable enhancement for the β-strands in the presence of osmolytes as compared to buffer, even after the treatment with APPJ.
1 Introduction
The possibility to exploit the versatility of atmospheric pressure non-thermal plasmas has steadily drawn increasing attention.1–3 Atmospheric pressure plasma jets (APPJ) have emerged as one of the most promising tools in surface treatment and various biological applications.4–6 Many plasma jet devices producing atmospheric pressure plasma plumes have been explored with thermally sensitive materials for varied medical applications.7–12 This is because the atmospheric pressure plasma is operated in open air, which overcomes the limitation imposed by the vacuum-based plasma. Moreover, the utilization of atmospheric air not only reduces the complexity of the device but also enhances the production of reactive species, such as hydroxyl radicals, atomic oxygen and nitric oxide.7,10,11 From an economics point of view, it may also be desirable to utilize gases that are less expensive, such as air, for the applications of non-thermal atmospheric plasma jest. However, the interaction studies of APPJ with DNA have shown that APPJ damages DNA.13,14 Similar effects of plasma on enzyme structure have also been observed by Dudak et al., which supports the above studies.15 Consequently, treating enzymes with plasma technology modifies the surface structure of ovalbumin and improves its solubility and functionality.16 These experimental results from various research groups13–16 emphasise that the structural modification of biomolecules following treatment with plasma may vary from biomolecule to biomolecule. However, the biological applications need to be explored due to a lack of data on plasma interactions with biomolecules.
To gain more insight into the APPJ action on biomolecules, we have studied the effect of APPJ on enzymes with and without co-solvents. Due to the APPJ action there is a rapid physiological stress on the enzyme; however, the enzyme needs to maintain its natively folded structure for proper functioning. However, these physiological stresses are overcome in the presence of co-solvents. Detailed structural information and the function of enzymes in co-solvents is critical for understanding their metabolic role and their use as industrial biocatalysts. Under the physiological stress of APPJ, many proteins/enzymes can be degraded with or without co-solvents and this varies from biomolecule to biomolecule. This phenomenon is being studied with proteolytic enzyme α-chymotrypsin (CT), which has promising industrial applicability.17,18,19 CT, a valuable biological substance, can be used for interpreting the mechanism of protein folding or unfolding upon the addition of co-solvents.19–22 The nature of the co-solvent governs the stability and properties of proteins and also the structural effects via bimolecular interactions between the functional groups present in the proteins and co-solvent particles.23–25
Naturally occurring osmolytes help the cells to respond to osmotic stress.19,20,23–25 Urea is a non-compatible osmolyte, which is also a well known denaturant. In our previous studies, we have systematically showed the effects of osmolytes or denaturants on CT.19,20 Nonetheless, there have been no documentation of the influence of APPJ on CT with or without co-solvents. In light of these considerations, the mechanistic basis of changes in the conformation of CT in the presence of specially designed air–plasma jet devices (APPJ) (operating at a low frequency in our laboratory) are explored. In addition, five osmolytes, namely 1 M trimethylamine N-oxide (TMAO), 1 M proline, 1 M betaine, 1 M sorbitol and 1 M glycerol, and a denaturant (1 M urea) have been chosen to study the APPJ action on CT in the presence of co-solvents. We also investigated the thermodynamic folding properties as a function of the co-solvent and utilised spectroscopic techniques, such as CD, fluorescence spectroscopy and NMR to gain a deeper insight. We are further motivated to estimate the changes in the Gibbs free energy of unfolding (ΔGU) at 25 °C (which is a better indicator of the global protein stability than the melting temperature (Tm), changes in the enthalpy (ΔH) and changes in the heat capacity (ΔCp)).
2 Experimental
2.1 Materials
α-Chymotrypsin (CT) from bovine pancreas type II, which is essentially salt free (molecular weight: 25 kDa), was obtained from Sigma–Aldrich (USA). The osmolytes and denaturant (TMAO, proline, betaine, sorbitol, glycerol and urea, respectively) were purchased from Sigma–Aldrich (USA). All materials, with a high purity, were used without further purification. The phosphate buffer solution of pH 7.2 was prepared using distilled deionized water at 18.3 MΩ.
2.2 Circular dichroism spectroscopy
CD spectroscopic studies were performed using a J-715 spectrophotometer (Jasco, UK) equipped with a Peltier system for temperature control. CD calibration was performed using (1S)-(+)-10-camphorsulfonic acid (Aldrich, Milwaukee, WI), which exhibits a 34.5 M cm−1 molar extinction coefficient at 285 nm and 2.36 M cm−1 molar ellipticity (θ) at 295 nm. The sample was pre-equilibrated at the desired temperature for 15 min and the scan speed was fixed for adaptative sampling (error of 0.01) with a response time of 1 s and a 1 nm bandwidth. The secondary and tertiary structures of CT were monitored using a 1.0 cm path length cuvette. The concentration for the secondary and tertiary structures of CT were 0.1 mg ml−1 and 1 mg ml−1, respectively, and each spectrum is an average of six spectra. Each sample spectrum was obtained by subtracting an appropriate blank media without CT from the experimental enzyme spectrum. Thermal denaturation studies were carried out at a heating rate of 1 °C min−1. This scan rate is found to provide adequate time for equilibration. The sample was heated from 20 to 80 °C. The change in absorbance at 288 nm was observed with increasing temperature. After denaturation, the sample was immediately cooled down to measure the conformational changes of the protein. The error in the melting temperature (Tm) did not exceed 0.1 °C. The estimated relative uncertainties in (ΔH), (ΔCp) and (ΔGU) are around 2–5% of the reported values.21,22,26
The values of map N and map D for the enzyme are obtained through the linear fitting of the pre- and post-transition data. The folded and unfolded conformations are present at significant concentrations and fF + fU = 1, where fF and fU represent the fraction of protein present in the folded and unfolded conformations, respectively, under the conditions where y is being measured. Combining the equations gives:
| |  | (1) |
The equilibrium constant (K) and the free energy change (ΔG) can be calculated using:
| | 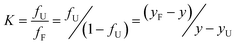 | (2) |
and
| | | ΔG = −RT lnK = − RT ln[((yF−y)/(y−yU))] | (3) |
| |  | (4) |
where R is the gas constant and T is the absolute temperature. The values of yF and yU in the transition region are obtained by extrapolating from pre- and post-transition regions. The plot of ΔH vs. Tm at each concentration of the osmolytes provides the value of ΔCp, the change in the constant pressure heat capacity,26 where ΔCp = (ΔH/Tm). A thermodynamic analysis of the thermal denaturation of S Con A in ILs additionally permits us to calculate the Gibbs free energy of unfolding at 25 °C (ΔGu), which is a better indication of global protein folding studies using the Gibbs–Helmholtz equation 19,20 as follows:
| | | ΔGU (T) = ΔH[−(T/Tm)] − ΔCp[(Tm−T) + Tln(T/Tm)] | (5) |
2.3 Fluorescence spectroscopy
Steady-state fluorescence measurements were conducted with a Cary Eclipse spectrofluorometer (Varian optical spectroscopy instruments, Mulgrave, Victoria, Australia) equipped with thermostat cell holders and the temperature was kept constant by a circulating water bath using a Peltier device attached to the sample holder of the fluorometer.21 The excitation wavelength was set at 290 nm to evaluate the contribution of the tryptophan residues to the overall fluorescence emission.27,28 The experiments were performed at 25 °C using a 1 cm sealed cell and excitation and emission slit widths of 5 nm. The spectra were corrected for the background signal. Both the change in the fluorescence intensity and the shift in the fluorescence maximum wavelength were recorded to monitor the unfolding transition.
2.4 NMR studies
A Bruker–Biospin 500 instrument was used for recording the spectra. The spectra are the result of 256 scans, with a 1 s delay time at 303.4 K and were processed with Bruker Topspin version 2.1 software. A 40 mg ml−1 concentration of the CT solution was used for the NMR spectroscopic analysis with and without 1 M TMAO. About 10% of 2H2O was added to provide an internal-field-frequency lock signal.
2.5 Atmospheric pressure plasma device (APPJ)
The conventional representation of the cold atmospheric pressure plasma jet is shown in Scheme 1. The key components of the plasma jet system are the electrodes, the dielectrics and a high-voltage power supply. The ac power supply was a commercially available transformer for neon light operated at 60 Hz and was connected to the two electrodes. The voltage controller regulates the primary voltage of the high voltage transformer. The inner electrode was a typical injection needle made of stainless steel with an inner diameter of 1.2 mm and a thickness of 0.2 mm; it is tightly covered with a quartz tube with an outer diameter of 3.2 mm. Porous alumina, 10 mm in diameter and 20 mm in length, was machined for the inner electrode, through which the quartz tube was inserted so that the tip of the inner electrode and the inner surface of the porous alumina were in contact. The outer electrode was fabricated from stainless steel and had a somewhat conical shape; it was centrally perforated with a hole of 1 mm through which the plasma jet was ejected to the surrounding ambient air. The porous alumina with the inner electrode were installed within the outer electrode. The discharge gap (dg) is the distance (in this work, dg = 2 mm) between the tips of the porous alumina and the inner electrode. It can be adjusted by controlling the depth at which the inner electrode is inserted into the porous alumina. The inner surface of the outer electrode and the tip of the porous alumina are also in contact. Air is injected into the injection needle and is then ejected through the 1 mm hole in the outer electrode via the porous alumina. The alumina used in this work had approximately 30 vol% porosity and an average pore diameter of 100 μm. Once air was introduced through the inner electrode and the high-voltage ac power was applied, a discharge was fired in the porous alumina between the electrodes and a long plasma jet, reaching lengths up to several centimeters, was ejected to the open air.11
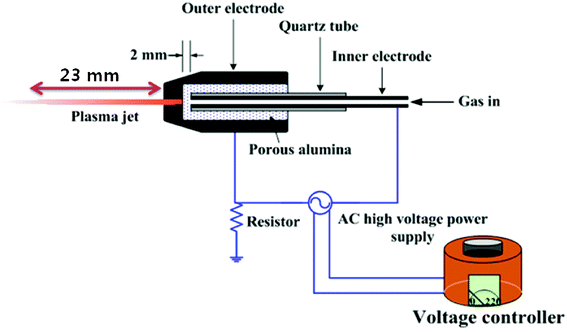 |
| | Scheme 1 A schematic depiction of the cold atmospheric pressure plasma jet (APPJ) device | |
We have used compressed atmospheric dry air with a purity of 99.9999%. This dry air had a flow rate of 3 L min−1, which was adjusted and measured by a mass flow controller. The plasma parameters (electron temperature and plasma density) were measured. In the measurement of the electron temperature, a high speed image intensified camera was used to obtain the temporal propagation distance versus the elapsed time in our experiment. Once we had an average value of the temporal distance (Δx) versus the time interval (Δt), the average plasma propagation speed for ionization front was obtained by  = 0.7 cm μs−1. This plasma speed can be expressed by
= 0.7 cm μs−1. This plasma speed can be expressed by  . Here, the electron temperature kTe can be experimentally measured to be ∼ 1.7 eV under dry air. In addition, the plasma density can be obtained from
. Here, the electron temperature kTe can be experimentally measured to be ∼ 1.7 eV under dry air. In addition, the plasma density can be obtained from  , where I is measured electric current, e is the electron charge,
, where I is measured electric current, e is the electron charge,  = ∼ 0.7 cm μs−1 is the plasma propagation velocity determined by the high speed image camera and σ is the radial profile of the plasma jet determined by the experiment. From these parameters, an electron temperature of 1.7 eV and a plasma density of 3.3 × 1012 cm−3 can be estimated. The ionization rate was about 10−7, meaning that this plasma belongs to cold plasma, which is almost room temperature since most of the air molecules are not ionized but are in a neutral state.
= ∼ 0.7 cm μs−1 is the plasma propagation velocity determined by the high speed image camera and σ is the radial profile of the plasma jet determined by the experiment. From these parameters, an electron temperature of 1.7 eV and a plasma density of 3.3 × 1012 cm−3 can be estimated. The ionization rate was about 10−7, meaning that this plasma belongs to cold plasma, which is almost room temperature since most of the air molecules are not ionized but are in a neutral state.
The emission spectra were recorded as illustrated in Fig. 1. We observed emission lines from a molecular NO β, γ system between 200 and 250 nm and a superoxide anion O2* at 245 nm, as well as an emission line at 307 nm from OH molecules (Fig. 1a). It is noted in this experiment that the VUV lines in the range of 120 nm–200 nm are easily absorbed by the atmospheric oxygen molecules; however, the emission lines beyond 200 nm easily pass through the oxygen molecules in atmospheric air, which enables us to investigate the emission spectrum between 200 nm and 1000 nm (Fig. 1b). We observed that the emission spectra emitted from the molecular N2 (C–B) second positive system can be seen at wavelengths between 300 nm and 400 nm. In addition, the lines from molecular N2+ (B–X) are also shown between 450 nm and 600 nm and those for N2 (B–A) are shown between 600 nm and 700 nm. It is also noted that the emission lines from the O2 first negative system are observed at wavelengths between 470 nm and 620 nm, typically 500.6 nm in this experiment. The spectra of the N2 molecular first positive system appear from 700 nm to 900 nm. In addition, it is especially noted that the oxygen atom O lines at 777. 6 nm and 844.8 nm can be observed in this experiment.
 |
| | Fig. 1 The emission spectra of cold atmospheric pressure plasma jets (APPJ): (a) 200 to 1000 nm and (b) 200 to 500 nm. | |
Sample preparation
Enzyme stability was analyzed by incubating 2 ml screw-capped vials in 0.05 M sodium phosphate buffer pH 7.2 solutions in the presence and absence of co-solvents (1 M TMAO, 1 M proline, 1 M betaine, 1 M sorbitol, 1 M glycerol and 1 M urea) at 25 °C for 4 h to attain complete equilibrium. All samples were prepared at an enzyme concentration of 1 mg mL−1 in 1 M concentrations of the co-solvent for the CD experiments. After completely dissolving the enzyme in the solution, the mixture was filtered with a 0.45 μm disposal filter (Millipore, Millex-GS) through a syringe before performing the measurements. After incubation of CT with or without the co-solvent for 4 h, the samples were treated at distances of 23 mm from the APPJ tip for 5 min. Three samples were treated for each condition to minimize the error.
Results and discussion
3.1 Thermodynamic analysis using CD
CD spectroscopy is a spectroscopic technique used to elucidate the structure and conformational changes of proteins. A thermal unfolding CD curve was undertaken according to the two state equilibrium model N ↔ U of CT. Our previous work demonstrates the existence of the U state in equilibrium with the N state.21,22 Taking the two-state model into consideration, the thermodynamic profile at a wavelength of 288 nm is caused by alterations in the local environment of the aromatic residues.21,22 Thermodynamic parameters can be related more directly to the structural arrangements of enzymes and to their interactional solvent effects, which help in anticipating the contributions of protein stability. The elementary thermodynamic forces that underlie the structure and function of biomolecules are quintessential for chemical technology. Explicitly, the Gibbs free energy change (ΔGU) is useful to describe global protein folding, while the enthalpy change (ΔH), the heat capacity change (ΔCp) and the melting temperature (Tm) are useful in measuring the protein stability in terms of the noncovalent forces of the different structural states. It is evident from Fig. 2a that the exposure of CT to APPJ, results in changes in the Tm as compared to the control. In addition to this, there is a decrease in thermodynamic profile, including Tm and ΔGU of CT in buffer after the APPJ treatment. These results prove that APPJ action leads to changes in the native conformation of CT.
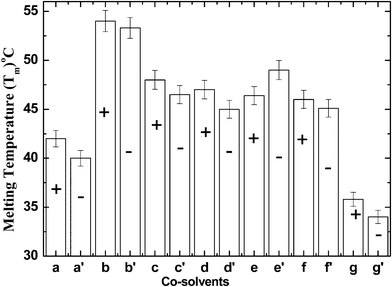 |
| | Fig. 2 The variation of the Tm values of CT in buffer, osmolytes or denaturant with APPJ treatment (−) and without APPJ treatment (+): buffer (a), buffer + APPJ (a′), 1 M TMAO (b), 1 M TMAO + APPJ (b′), 1 M proline (c), 1 M proline + APPJ (c′), 1 M betaine (d), 1 M betaine + APPJ (d′), 1 M sorbitol (e), 1 M sorbitol + APPJ (e′), 1 M glycerol (f), 1 M glycerol + APPJ (f′), 1 M urea (g) and 1 M urea + APPJ (g′). | |
It is well known that naturally occurring osmolytes protect enzymes from external stress. Our thermodynamic experimental data shows that all osmolytes stabilize the native structure of CT and the denaturant (urea) denatures CT without APPJ action; hence, these results corroborate previous results.19,20 In the presence of co-solvents, we used osmolytes/denaturant to observe the effect of APPJ on CT. We used 1 M TMAO, 1 M proline, 1 M betaine, 1 M sorbitol, 1 M glycerol as the osmolytes, along with 1 M urea as the denaturant. After the treatment of CT with APPJ for 5 min, there is a variation in the thermodynamic profile of CT in the presence of osmolytes and denaturant, as illustrated in Fig. 2 and Table 1. The ΔGU value gives the stability of the enzyme in the folded state; hence, after APPJ exposure, an increase in ΔGU for sorbitol signifies the increased stability of the CT structure. The ΔGU results show that the stabilizing action of co-solvents against APPJ exposure varies from osmolyte to osmolyte, where the efficiency of stabilization follows the trend TMAO > proline > sorbitol > betaine > glycerol (Table 2). Interestingly, the Tm of the folding induced by the osmolytes reveals that TMAO is the strongest stabilizer, while proline, sorbitol and betaine are moderate stabilizers; however, glycerol is established as a weak stabilizer. All osmolytes are able to counter the effects of APPJ on the enzyme. Surprisingly, the action of APPJ on sorbitol is reversed; it stabilizes CT to a greater extent after treatment (Fig. 2e′ ). Before APPJ exposure, the Tm value of CT in sorbitol was 46.4 °C, whereas after treatment with APPJ for 5 min the Tm value increased to 49 °C. Here, we focused on denaturants, such as urea, in presence of AAPJ. Urea is considered to be a non-ionic chaotrope and has a tendency to break the hydrogen bonds within an enzyme and preferentially interact with the enzyme surface, thus appearing to be bound. The enzyme is noted to be preferentially bound, which eventually leads to the denaturation of CT. In the presence of APPJ, the denaturation action increases, which results in a decrease in Tm to 34.0 °C and ΔGU to 7.51 kJ. mol−1 (Table 1). Our thermodynamic studies reveal that osmolytes have a counteracting effect and also reverse the effect on the CT structure promoted by APPJ, which in turn can be due to the highly unfavourable interaction of the osmolytes with the functional groups of CT, whereas the denaturation action increases in the presence of urea.24
Table 1 The melting temperature (Tm), the change in enthalpy (ΔH) and the change in the heat capacity (ΔCp) are determined by CD, and the calculated change in the Gibbs free energy (ΔGU) in the unfolded state at 25 °C for CT in different solvent mediaa
| Sample |
T
m (°C) |
ΔH (kJ mol−1) |
ΔGU (kJ mol−1) |
ΔCp (kJ mol−1 C−1) |
|
Each value is the average of three measurements. The error in Tm does not exceeds ± 0.1 °C. The estimated relative uncertainties in ΔH, ΔCp and ΔGU are around 2% of the reported values.
|
| Buffer |
42.0 |
424 |
18.15 |
10.09 |
| Buffer + APPJ |
40.0 |
299 |
11.72 |
7.10 |
| 1 M TMAO |
54.0 |
705 |
45.20 |
13.05 |
| 1 M TMAO + APPJ |
53.3 |
700 |
43.90 |
13.11 |
| 1 M proline |
48.0 |
684 |
36.90 |
14.25 |
| 1 M proline + APPJ |
46.5 |
670 |
34.40 |
14.40 |
| 1 M betaine |
47.0 |
620 |
32.00 |
13.19 |
| 1 M betaine + APPJ |
45.0 |
600 |
29.16 |
13.31 |
| 1 M sorbitol |
46.4 |
570 |
29.10 |
10.20 |
| 1 M sorbitol + APPJ |
49.0 |
575 |
32.08 |
11.73 |
| 1 M glycerol |
46.0 |
480 |
24.30 |
10.30 |
| 1 M glycerol + APPJ |
45.1 |
460 |
22.36 |
10.20 |
| 1 M urea |
35.8 |
305 |
9.03 |
8.51 |
| 1 M urea + APPJ |
34.0 |
296 |
7.51 |
8.70 |
Table 2 The secondary structure composition of α-chymotrypsin determined from far-UV CD spectra in different solvent media at 25 °C
| Sample |
α-sheet (%) |
β-sheet (%) |
Random (%) |
Total (%) |
| Buffer |
8% |
45% |
47% |
100% |
| Buffer + APPJ |
9% |
36% |
56% |
100% |
| 1 M glycerol |
7% |
47% |
46% |
100% |
| 1 M glycerol + APPJ |
8% |
45% |
47% |
100% |
| 1 M sorbitol |
2% |
47% |
48% |
100% |
| 1 M sorbitol + APPJ |
5% |
48% |
47% |
100% |
| 1 M TMAO |
2% |
51% |
47% |
100% |
| 1 M TMAO + APPJ |
2% |
50% |
48% |
100% |
| 1 M betaine |
2% |
51% |
47% |
100% |
| 1 M betaine + APPJ |
4% |
49% |
47% |
100% |
| 1 M urea |
10% |
43% |
46% |
100% |
| 1 M urea + APPJ |
12% |
35% |
53% |
100% |
3.2 CD analysis for the secondary and tertiary structure of CT
To quantify the conformational modifications, changes in the 3D structure of an enzyme can be related to the changes observed in the CD spectrum. To study the mechanism of events regarding the osmolytes' role in the enhancement of the stability of the CT structure before and after AAPJ treatment, we used CD spectroscopy for further analysis. The far-UV CD spectra of the enzyme indicate that the protein has a particular secondary structure in each co-solvent (Fig. 3a to f ). The CD spectrum of CT in buffer has a minimum at ≈202 nm and no positive band.27 CT is a type of all-β protein characterized by a CD spectrum that resembles one of a random coil conformation. The crystalline structure data shows that this kind of protein consists of antiparallel β-pleated sheets, which are either highly distorted or form very short irregular strands.27,28 The CD spectra of CT in buffer, TMAO, sorbitol and glycerol have no positive CD signal, whereas in betaine it shows positive bands at 201 and 225 nm, as illustrated in Fig. 3c, while proline has a very high positive absorption band 20 (data not shown here). The far-UV CD spectra data helps us to estimate the β-structure using K2d software29 and these results are shown in Table 2. The observed results are in agreement with those reported in previous research.20,27,28 It can be clearly observed from Table 2 that the drop in the β-value is increased in buffer as compared to osmolytes for treated CT; whereas, in urea, the β-value decreases more as compared to treated CT in buffer. The β-structure remains the same before and after treatment with APPJ in the presence of TMAO, while in urea it shows a drastic decrease in the β-structure after APPJ treatment (Table 2). Comparison of the ellipticity of CT in osmolytes with or without treatment (as illustrated in Fig. 3b) reveals that, except for TMAO, no other osmolytes follow the same ellipticity pattern after treatment. So, this suggests that denaturation increases with the plasma treatment. Moreover, even the presence of osmolytes (except TMAO) are not able to control the denaturation action due to the APPJ effect, which results in the loss of the secondary structure of CT (Fig. 3c to f). With sorbitol, the secondary structure of CT is more stable after treatment (Table 2).
 |
| | Fig. 3 Far-UV CD spectra of CT in (a) buffer, (b) 1 M TMAO, (c) 1 M betaine, (d) 1 M sorbitol, (e) 1 M glycerol and (f) 1 M urea at 25 °C, where the co-solvents are used with or without treatment with APPJ for 5 min (red and black, respectively). | |
To ascertain the effect of the osmolytes and denaturant on CT in the APPJ treated samples, we performed a CD spectral analysis in near-UV region (240–300 nm). The results are illustrated in Fig. 4a to g . This spectral region is dominated by the contribution of aromatic residues (e.g., Trp and Tyr) and disulfide chromophores, which give rise to broad, but weak signals.20,27,28 The intensity of the 240–300 nm bands is affected by the local conformation changes around the chromatophores. The near-UV CD spectrum of CT in buffer with or without osmolytes represents the contribution of the Tyr and Trp residues, which are responsible for the peaks and shoulders between 270 and 300 nm, and Phe residues, which strongly contribute to the bands in the 258–270 nm region. Fig. 4a shows the CD spectrum of CT in buffer solutions and the maxima are identified at 254, 289 and 296 nm, which may be assigned to the Trp residues of CT.25,26 After the treatment of CT in urea with the plasma jet, there is decrease in the ellipticity of the peaks and the changes are more prominent in the 286 nm peak due to the denaturation of the native CT structure. Fig. 4b shows that, except for TMAO, no other osmolytes are able to counteract the APPJ action on CT. Hence, we can conclude that the structural information of CT in TMAO is unaffected by APPJ exposure. These results can again be attributed to a compact conformation, where the internal aromatic residues remain in the hydrophobic core of the protein at a reduced distance.
 |
| | Fig. 4 Near-UV CD spectra of CT in (a) buffer, (b) 1 M TMAO, (c) 1 M proline, (d) 1 M betaine, (e) 1 M sorbitol, (f) 1 M glycerol and (g) 1 M urea at 25 °C, where the co-solvents are used with or without treatment with APPJ for 5 min (red and black, respectively). | |
3.3 Fluorescence analysis
Further insight into ability of the osmolytes to counteract the denaturing effect of APPJ on CT was also required. So, we have performed fluorescence spectroscopy to investigate the environment surrounding the fluorophore residues (e.g., Trp, Tyr or Phe) of the protein as illustrated in Table 3. The denaturation process can be studied by observing the changes in the maximum intensity of fluorescence (Imax) and the maximum emission wavelength (Emax) as well as the increased polarity of the Trp residues of the protein.20,25,26 The Trp residue has a strong Stokes shift, which is dependent on the solvent environment. As the co-solvent is changed, there is a change in the maximum emission wavelength of Trp, which indicates that the Trp Emax depends on the co-solvent environment. Table 3 shows the initial fluorescence spectra of native CT in all the assayed media (buffer, 1 M TMAO, 1 M proline, 1 M betaine, 1 M sorbitol, 1 M glycerol and 1 M urea) at 25 °C and it also summarizes the spectral data for the enzyme after 5 min APPJ treatment in all the co-solvents. The maximum intensities for all the spectra were normalized with respect to the spectrum obtained for native CT in buffer at 25 °C. As can be seen, the enzyme exhibits the same initial Emax ≈ 344.5 nm in buffer (Fig. 5a), whereas in the presence of other co-solvents, such as 1 M proline, 1 M glycerol, 1 M TMAO and 1 M betaine, it is found to be ≈340.0 nm and the samples in 1 M sorbitol and 1 M urea were red shifted to ≈347.5 nm (Fig. 5b). After the treatment with APPJ for 5 min, it was noticed that the fluorescence spectra of CT showed changes for both the Emax and Imax parameters as compared to untreated CT in all co-solvents. After the APPJ treatment of CT, red shifts were observed in all media except TMAO and sorbitol (Fig. 5b, Table 3). In TMAO, we observed no changes in the Emax and Imax parameters, while in sorbitol there were changes in Emax (blue shifted) and Imax (a decrease). This suggests that, after the APPJ treatment of CT in the presence of sorbitol, the structure of CT was more stable. In this way, the evolution of both Imax and Emax in aqueous media corresponds to an usual unfolding process of CT, which enhances the exposure of the Trp residues to the bulk solvent (the Emax of the free Trp residue in aqueous solution is 350 nm).28 In a hydrophobic environment (i.e. buried within the core of the protein), Trp has a high quantum yield and therefore a high fluorescence intensity is observed. In contrast, in a hydrophilic environment (i.e. exposed to solvent) the quantum yield decrease and a low fluorescence intensity is observed. Hence, fluorescence analysis also supports the CD spectral data, which indicated a change in the native CT conformation of the treated samples in the presence of osmolytes. Before the treatment of CT with osmolytes, all osmolytes showed a blue shift (except sorbitol) and in the denaturant (urea) a red shift was observed as compared to the buffer. After APPJ exposure of CT in all osmolytes and denaturant, the samples showed red shifts as compared to their native spectra, except for TMAO and sorbitol.
 |
| | Fig. 5 Fluorescence spectra of CT in co-solvents with and without treatment with APPJ for 5 min: (a) buffer (black), buffer + APPJ (red), 1 M proline (blue), 1 M proline + APPJ (cyan), 1 M glycerol (magenta), 1 M glycerol + APPJ (dark green); (b) 1 M TMAO (green), 1 M TMAO + APPJ (orange), 1 M betaine (magenta), 1 M betaine + APPJ (pink), 1 M sorbitol (blue), 1 M sorbitol + APPJ (dark cyan), 1 M urea (black) and 1 M urea + APPJ (red). | |
Table 3 The fluorescence intensity of CT in co-solvents with and without APPJ exposure for 5 min
A systematic correlation, drawn from the results of the CD and the fluorescence (Fig. 3–5), suggest that the osmolytes stabilize the enzyme via the formation of a flexible and more compact 3D structure, which can be better related to the preservation behaviour of the essential water shell.16,20,26 However, the osmolytes are not able to stabilize the compact structure of CT after treatment with APPJ. This may be due to the plasma irradiations, which excite the molecule of the osmolytes and, consequently, the excited state of the osmolytes lose their protective shield around the CT structure. As a result, there is an alteration in the thermodynamic and spectroscopic profile of CT in co-solvents, which subsequently lead to a decrease in the thermodynamic and spectroscopic profile. It is surprising that TMAO attenuates the APPJ action (Scheme 2), while it stabilizes CT after APPJ exposure in the presence of sorbitol as compared to the control.
 |
| | Scheme 2 A schematic depiction of the preferential hydration of CT in the presence of TMAO and the denaturation of CT without TMAO, during the treatment of CT with a cold atmospheric pressure plasma jet. | |
3.4 NMR studies to show the TMAO action against APPJ
1H NMR studies were also performed to assess the APPJ action on CT in the presence or absence of TMAO. From the results, it was found that TMAO is able to attenuate the deleterious action of APPJ on CT (Fig. 6). Strong hydrogen bonds, akin to those found in water, are observed with TMAO, which results in a shift of the proton resonance peaks downfield by about 2 ppm. However, weaker hydrogen bonds, akin to those found in carbonyls and amino protons, shift the resonance downfield by approximately 1 ppm.21,22,30
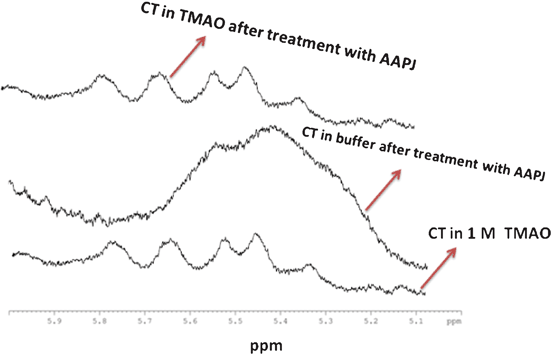 |
| | Fig. 6
1H NMR data for the counteractivity of CT against APPJ in the presence of 1 M TMAO. | |
Furthermore, the NMR peaks do not show any significant change in the aromatic region following APPJ treatment (data not shown). The NMR peaks between 5 and 6 ppm are absent in the treated sample of CT (Fig. 6); however, the peaks match for CT in TMAO before and after the APPJ action. Hence, we have concluded that TMAO acts as a protective co-solvent for CT against the action of APPJ.
4 Conclusions
CD, fluorescence and NMR investigations of CT dynamics after treatment with APPJ for 5 min reveal that there is a deformation of the CT structure. Furthermore, osmolytes can control the Tm and ΔGU values but they are not able to maintain the conformation of CT adequately (with the exception of TMAO). Sorbitol stabilizes the structure of CT to a greater extent in the presence of APPJ, as compared to the untreated samples.
Therefore, our results form an important basis for understanding the functional role of osmolytes and denaturants to counter the APPJ action. Moreover, the results obtained from the above study may prove beneficial in the understanding of osmolyte and denaturant effects in various biological applications. In addition, the study supports the potential application of APPJ in various fields, including biotechnology, biophysical chemistry and plasma medicine.
Acknowledgements
We are very thankful to Dr. Key-Sun Kim, center for neuroscience, KIST, Korea, for providing the CD instrument. We gratefully acknowledge SRC program of National Research Foundation of Korea (NRF) Grant funded by the Korean Government (MEST) (No. 20100029418) and in part by Kwangwoon University 2011 and 2012. PA gratefully thank PBRC for providing laboratory and financial assistance for research and PV thanks to CSIR, New Delhi, through the grant No. 01(2343)/09/EMR-II for financial assistance.
References
- M. Laroussi, I. Alexeff, J. P. Richardson and F. F. Dyer, IEEE Trans. Plasma Sci., 2002, 30, 158–159 CrossRef CAS.
- G. Cho, H. Lim, J.H. Kim, D. J. Jin, G. C. Kwon, E. H. Choi and H. S. Uhm, IEEE Trans. Plasma Sci., 2011, 39, 1234–1238 CrossRef.
- E. H. Choi, J. C. Ahn, M. W. Moon, J. G. Kim, M. C. Choi, C. G. Ryu, S. H. Choi, T. S. Cho, Y. Jung, G. S. Cho and H. S. Uhm, IEEE Trans. Plasma Sci., 2011, 39, 3288–3299 CrossRef.
- M. Moisan, J. Barbeau and S. Moreau, Int. J. Pharm., 2001, 226, 1–21 CrossRef CAS.
- H. X. Liu, J. R. Chen, L. Q. Yang and Y. Zhou, Appl. Surf. Sci., 2008, 254, 1815–1821 CrossRef CAS.
- X. Zhang, J. Huang, X. Liu, L. Peng, L. Guo, G. Lv, W. Chen, K. Feng and Si-ze Yang, J. Appl. Phys., 2009, 105, 063302–063307 CrossRef.
- J. F. Kolb, A.-A. H. Mohamed, R. O. Price, R. J. Swanson, A. Bowman, R. L. Chiavarini, M. Stacey and K. H. Schonenbach, Appl. Phys. Lett., 2008, 92, 241501–241503 CrossRef.
- Y. C. Hong and H. S. Uhm, Appl. Phys. Lett., 2006, 89, 221504–221506 CrossRef.
- X. Zhang, M. Li, R. Zhou, K. Feng and S. Yang, Appl. Phys. Lett., 2008, 93, 021502–021504 CrossRef.
- X. T. Deng, J. J. Shi and M. G. Kong, J. Appl. Phys., 2007, 101, 074701–074709 CrossRef.
- Y. C. Hong, W. S. Kang, Y. B. Hong, W. J. Yi and H. S. Uhm, Phys. Plasmas, 2009, 16, 123502–123506 CrossRef.
- Y. C. Hong, S. C. Cho and H. S. Uhm, Appl. Phys. Lett., 2007, 90, 141501–141503 CrossRef.
- S. Ptasinska, B. Bahnev, A. Stypczynska, M. Bowden, N. St. J. Braithwaite and N. J. Mason, J. Phys.: Conf. Ser., 2009, 194, 152029 CrossRef.
- D. O'Connell, L. J. Cox, W. B. Hyland, S. J. McMahon, S. Reuter, W. G. Graham, T. Gans and F. J. Currell, Appl. Phys. Lett., 2011, 98, 043701–043703 CrossRef.
-
F. C. Dudak, J. Kousal, U. O. S. Seker, I. H. Boyaci, A. Choukourov, H. Biederman, Proc. 28th ICPIG, Prague, 2007 DOI:10.1021/bp025665e.
- C. H. Gao, T. J. Herald and P. L. Miuno, J. Food Sci., 2001, 66, 89–94 CrossRef CAS.
- D. Constantinescu, H. Weingdärtner and C. Herrmann, Angew. Chem., Int. Ed., 2007, 46, 8887–8889 CrossRef CAS.
- W. Wei and N.D. Danielson, Biomacromolecules, 2011, 12, 290–297 CrossRef CAS.
- P. Venkatesu, M. J. Lee and H. M. Lin, J. Phys. Chem. B, 2009, 113, 5327–5338 CrossRef CAS.
- P. Attri, P. Venkatesu and M. J. Lee, J. Phys. Chem. B, 2010, 114, 1471–1478 CrossRef CAS.
- P. Attri, P. Venkatesu, A. Kumar and N. Byrne, Phys. Chem. Chem. Phys., 2011, 13, 17023–17026 RSC.
- P. Attri, P. Venkatesu and A. Kumar, Phys. Chem. Chem. Phys., 2011, 13, 2788–2796 RSC.
- P. Attri and P. Venkatesu, Thermochim. Acta, 2011, 526, 143–150 CrossRef CAS.
- P. H. Yancey, M. E. Clark, S. C. Hand, R. D. Bowlus and G. N. Somero, Science, 1982, 217, 1214–1222 CAS.
- C. C. Lenky, C. J. McEntyre and M. Lever, Anal. Biochem., 2012, 420, 7–12 CrossRef CAS.
- C. N. Pace and D. V. Laurents, Biochemistry, 1989, 28, 2520–2525 CrossRef CAS.
- E. V. Kudryashova, A. K. Gladilin, A. V. Vakurov, F. Heizt, A. V. Levashov and V. V. Mozhaev, Biotechnol. Bioeng., 1997, 55, 267–277 CrossRef CAS.
- T. de Diego, P. Lozano, S. Gmouh, M. Vaultier and J. L. Iborra, Biotechnol. Bioeng., 2004, 88, 916–924 CrossRef CAS.
- C. Perez-Iratxeta and M. A. Andrade-Navarro, BMC Struct. Biol., 2008, 8, 25–27 CrossRef.
- G. Robillard and R. G. Shulman, J. Mol. Biol., 1972, 71, 507–511 CrossRef CAS.
|
| This journal is © The Royal Society of Chemistry 2012 |
Click here to see how this site uses Cookies. View our privacy policy here. 
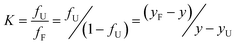


 = 0.7 cm μs−1. This plasma speed can be expressed by
= 0.7 cm μs−1. This plasma speed can be expressed by  . Here, the electron temperature kTe can be experimentally measured to be ∼ 1.7 eV under dry air. In addition, the plasma density can be obtained from
. Here, the electron temperature kTe can be experimentally measured to be ∼ 1.7 eV under dry air. In addition, the plasma density can be obtained from  , where I is measured electric current, e is the electron charge,
, where I is measured electric current, e is the electron charge,  = ∼ 0.7 cm μs−1 is the plasma propagation velocity determined by the high speed image camera and σ is the radial profile of the plasma jet determined by the experiment. From these parameters, an electron temperature of 1.7 eV and a plasma density of 3.3 × 1012 cm−3 can be estimated. The ionization rate was about 10−7, meaning that this plasma belongs to cold plasma, which is almost room temperature since most of the air molecules are not ionized but are in a neutral state.
= ∼ 0.7 cm μs−1 is the plasma propagation velocity determined by the high speed image camera and σ is the radial profile of the plasma jet determined by the experiment. From these parameters, an electron temperature of 1.7 eV and a plasma density of 3.3 × 1012 cm−3 can be estimated. The ionization rate was about 10−7, meaning that this plasma belongs to cold plasma, which is almost room temperature since most of the air molecules are not ionized but are in a neutral state.






