Metal-mediated DNA assembly using the ethynyl linked terpyridine ligand†
Thomas
Ehrenschwender
a,
Anna
Barth
b,
Holger
Puchta
b and
Hans-Achim
Wagenknecht
*a
aKarlsruhe Institute of Technology, Institute for Organic Chemistry, Fritz-Haber-Weg 6, 76131 Karlsruhe. E-mail: wagenknecht@kit.edu; Fax: +49-(0)721-608-44825
bKarlsruhe Institute of Technology, Botanical Institute II, Fritz-Haber-Weg 4, 76131 Karlsruhe. E-mail: puchta@kit.edu; Fax: +49-(0)721-608-44874
First published on 16th November 2011
Abstract
The terpyridine ligand directly attached to the 5-position of a uridine allows metal-mediated DNA assembly towards potentially electronically coupled DNA conjugates.
DNA represents an increasingly important tool for the construction of nanoarchitectures or nanoscaled devices due to the predictable Watson–Crick base pairing.1–6 In order to enhance complexity of DNA-based nanostructuring it would be highly desirable to develop additional binding motifs that behave chemically orthogonal to conventional hydrogen bonding of Watson–Crick base pairing that is applied typically as so-called sticky ends. The first alternative, hydrophobic π–π interactions have been used mainly between perylene bisimides as DNA caps to aggregate DNA7 and Y-shaped DNA constructs8 in a reversible fashion. Metal ion–ligand interactions represent the second alternative motif. The latter idea is not new; conjugates of nucleic acids and metal-chelating moieties have been investigated intensively.9–15 Especially the 2,2′:6′,2′′-terpyridine (terpy) ligand is known to efficiently form stable complexes with a broad variety of metal ions16–21 and has been used to assemble oligonucleotides.22–24 However, it is important to point out that all of these studies have been carried out with oligonucleotides that were terminally modified with terpy using a flexible alkyl linker. In supramolecular electronics a strong electronic coupling is provided mainly by acetylene bridges.25 In order to go one step further towards DNA-based nanoelectronics, we present the DNA building block terpy-dU in which the terpy ligand is linked to the 5-position of 2′-deoxyuridinevia the ethynyl bridge. This building block allows internal and terminal terpy-dU modification and thereby provides the basis for metal-mediated DNA assembly.
The synthesis of 2′-deoxyuridine carrying the terpy-acetylene moiety in the 5-position was recently reported.26 Accordingly, the preparation of the corresponding building block was carried out viaSonogashira coupling between DMT-protected 5-iodo-2′-deoxyuridine and 4′-ethynyl-2,2′:6′,2′′-terpyridine followed by standard phosphoramidite formation (see Supporting Information†). The single strands DNA1–DNA4 (Scheme 1) were prepared with one terpy-dU modification in the middle of the sequence and surrounded by T, A, G or C; strands DNA5 and DNA6 carry a terminal terpy-dU label. DNA1 is complementary to DNA2, DNA3 to DNA4, and DNA5 to DNA6 (Fig. 1).
 | ||
| Scheme 1 Structure of terpy-dU in oligonucleotides and sequences of single strands DNA1–DNA6. Duplexes between two modified oligonucleotides are called DNA1-2, DNA3-4 and DNA5-6. Duplexes of only one of the modified oligonucleotides with corresponding unmodified counterstrands are called DNA1Yetc. (with Y = base opposite to terpy-dU, e.g. A in DNA1A). | ||
 | ||
| Fig. 1 Absorption (left) and fluorescence spectra (right) of terpy-dU-modified single strands DNA1, DNA3 and DNA5 and double strands DNA1-2, DNA3-4, DNA5-6; 2.5 μM in Na–Pibuffer at pH 7, 250 mM NaCl, 100 μM EDTA, 20 °C, excitation at 325 nm. | ||
First we studied the influence of a single terpy-dU modification on the melting temperatures (Tm) of double strands (Table 1). If DNA1 and DNA3 are hybridized with completely unmodified counterstrands including A opposite to terpy-dU (yielding double strands DNA1A and DNA3A) the Tm values reveal a strong destabilization (−5.7 °C and −4.1 °C) compared to completely unmodified duplexes. The destabilization is slightly stronger with other bases opposite to terpy-dU (double strands DNA3T, DNA3G and DNA3C) (see Supporting Information†). Obviously, the terpy-dU unit exhibits a small preference for adenine as the counterbase although the metal ligand has been attached via the short ethynyl bridge to the 5-position. In contrast, the duplexes DNA1-2 and DNA3-4 bearing two terpy-dU moieties opposite to each other are stabilized quite significantly (+2.4 °C and +3.6 °C, respectively) compared to completely unmodified duplexes. This is a remarkable result and shows that the hydrophobic interaction of the two terpy unit regains more hybridization energy than the destabilization introduced by the terpy units. Similar results have been obtained with bipyridine pairs27 and binaphthyl pairs28 inside DNA.
| DNA | λ abs [nm] | λ em [nm] | T m [°C] | ΔTm [°C]a |
|---|---|---|---|---|
| a Compared to the unmodified references: Tm = 62.5 °C for DNA1A and Tm = 68.0 °C for DNA5A, each with T instead of terpy-dU. | ||||
| DNA1A | 316 | 378 | 56.8 | −5.7 |
| DNA1-2 | 321 | 390 | 65.9 | +2.4 |
| DNA3A | 316 | — | 63.9 | −4.1 |
| DNA3-4 | 316 | — | 71.6 | +3.6 |
| DNA5A | 316 | 400 | 61.4 | — |
| DNA5-6 | 321 | 400 | 62.7 | — |
The terpy chromophore can be excited selectively at 325 nm yielding a characteristic fluorescence (Fig. 1). Compared to duplex DNA1A the fluorescence of double strand DNA1-2 is quenched. This is the typical result of chromophore aggregation and thereby supports the idea of a hydrophobically interacting terpy “base pair” inside DNA1-2. The fluorescence of duplexes DNA3A and DNA3-4 does not allow this interpretation since it is almost completely quenched, probably due to photoinduced charge transfer processes to adjacent guanines. The fluorescence intensity of DNA5 and DNA5-6 is approximately equal since hydrophobic terpy pairing enforced by the surrounding DNA architecture (as in DNA1-2) is unlikely with terminal modifications.
More importantly, the terpy fluorescence and its quenching can be used to follow and quantify metal ion coordination. We chose Cu2+, Ni2+, Zn2+and Fe2+ as typical representatives, known to form stable complexes with the terpy ligands. First, we examined double strands DNA1A and DNA5A bearing only one terpy-dU in the middle or at the terminus. It is expected that addition of metal ions induces dimerization. From the titration experiments (see Supporting Information†) we calculated the quenched fraction of fluorescence intensity (Fq) at characteristic emission maxima. The results show that fluorescence quenching is complete after addition of 0.5–0.75 equiv. of metal ions (Fig. 2, top). This observation together with the absorption changes (see Supporting Information†) indicate approximately the expected stoichiometry. To further evidence the dimer formation we performed non-denaturing polyacrylamide gel electrophoresis (Fig. 2, bottom). The gels show dimerization of DNA5A and DNA1A in the presence of Ni2+ and Fe2+ by a band of slower mobility. This is a remarkable result by keeping in mind how short the acetylene linkers are between the metal chelators and the nucleic acids on both sides of the complex. On the other hand, dimers of DNA5A and DNA1A in the presence Cu2+ and Zn2+ which are indicated by the fluorescence measurements seem to be not stable enough for non-denaturing gel analysis.
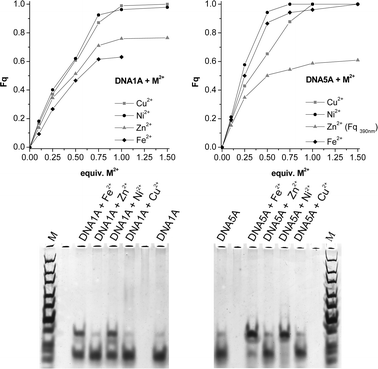 | ||
| Fig. 2 Top: Fluorescence quenching (Fq) for DNA1A (left) and DNA5A (right) upon addition of metal ions; bottom: non-denaturing gel electrophoresis (8% TBM-PAGE) of DNA1A and DNA5A in absence and presence of metal ions after silver staining. | ||
In the second part of this study we performed similar experiments with double strands bearing two terpy-dU units either opposite to each other in the middle (DNA1-2) or at the termini (DNA5-6). It is expected that these DNA probes potentially are forming larger DNA assemblies. The Fq analysis of DNA1-2 (Fig. 3, top) reveals a complete fluorescence quenching after addition of 1.5 equiv. metal ions which is 0.5 equiv. more than expected. The gel analysis (Fig. 3, bottom) shows dimers of DNA1-2 only in the presence of Ni2+ but no larger aggregates. Due to the fact that optical changes clearly indicate metal coordination, it looks reasonable to assume that the two terpy-dU moieties of DNA1-2 are forming a metal-mediated base pair inside the duplex instead of networking between duplexes.
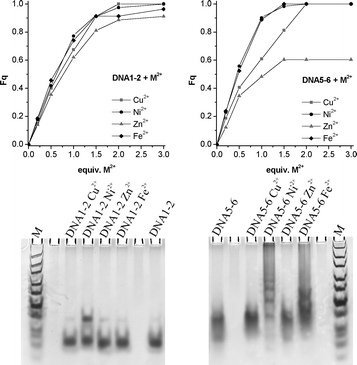 | ||
| Fig. 3 Top: Fluorescence quenching (Fq) for DNA1-2 (left) and DNA5-6 (right) upon addition of metal ions; bottom: non-denaturing gel electrophoresis (8% TBM-PAGE) of DNA1-2 and DNA5-6 in absence and in presence of metal ions after silver staining. | ||
It is important to note that it is problematic to compare Tm values of the metal-ion coordinated samples of DNA1-2 directly with the metal free DNA1-2 since the Tm of the latter duplex revealed an astonishingly stabilized, hydrophobically interacting terpy-dU pair (as discussed above). Compared to a completely unmodified reference double strand, however, DNA1-2 shows significantly higher melting temperatures in the presence of 1 equiv. of Ni2+, Fe2+ and Cu2+ (Table 2). With Ni2+ or Fe2+ two different Tm values are obtained, of which one is even higher than the metal free DNA1-2 (ΔTm positive). The latter observation strongly supports the idea of a metal ion-mediated, internal terpy-dU base pair that interferes with the formation of higher DNA assemblies. The second Tm value at lower temperatures (ΔTm negative) corresponds to duplex assemblies that are conjugated by the terpy-dU complexes, and therefore both hydrophobic and metal-mediated stabilization inside the DNA duplex are lost.
The situation looks different and more straightforward in case of the terminally labeled DNA5-6. The gels show clearly several bands of slower mobility in the presence of Ni2+ and Fe2+ thereby supporting the existence of DNA assemblies larger than dimers. With these metal ions the band of isolated duplexes DNA5-6 has nearly completely vanished and Fq analysis shows complete fluorescence quenching upon addition of slightly more than 1.0 equiv. metal ions, as expected.
In conclusion it became evident from both fluorescence measurements and gel analysis that metal-mediated DNA assemblies do not require long and flexible alkyl chain linkers between the metal chelator and the nucleic acids. Even a short linker, as the acetylene linker, allows dimerization and formation of stable and larger assemblies of terpy-dU-modified DNA in the presence of Ni2+ and Fe2+. Internal metal-mediated base pairing between two terpy-dU modifications interfere with the formation of higher DNA assemblies. This problem can be solved by placing two terpy-dU modifications not exactly opposite to each other in two complementary strands. Higher structures can be formed with doubly terminally labeled DNA. In principal, the short acetylene linkers should provide strong electronic coupling between the metal–ligand complex and the DNA. Hence it is expected that these kind of DNA materials29 have a significant potential for DNA-based nanoelectronics.
Acknowledgements
Financial support by the Deutsche Forschungsgemeinschaft (Wa 1386/12-1), the Center for Functional Nanostructures (CFN) and KIT is gratefully acknowledged.Notes and references
- P. W. K. Rothemund, Nature, 2006, 440, 297–302 CrossRef CAS
.
- F. C. Simmel, Angew. Chem., Int. Ed., 2008, 47, 5884–5887 CrossRef CAS
.
- K. V. Gothelf and T. H. LaBean, Org. Biomol. Chem., 2005, 3, 4023–4037 CAS
.
- E. S. Andersen, M. Dong, M. M. Nielsen, K. Jahn, R. Subramani, W. Mamdouh, M. M. Golas, B. Sander, H. Stark, C. L. P. Oliveira, J. S. Pedersen, V. Birkedal, F. Besenbacher, K. V. Gothelf and J. Kjems, Nature, 2009, 459, 73–77 CrossRef CAS
.
- C. Mao, W. Sun, Z. Shen and N. C. Seeman, Nature, 1999, 397, 144–146 CrossRef CAS
.
- S. P. Liao and N. C. Seeman, Science, 2004, 306, 2072–2074 CrossRef CAS
.
- P. P. Neelakandan, Z. Z. Pan, M. Hariharan, Y. Zheng, H. Weissman, B. Rybtchinski and F. D. Lewis, J. Am. Chem. Soc., 2010, 132, 15808–15813 CrossRef CAS
.
- F. Menacher, V. Stepanenko, F. Würthner and H.-A. Wagenknecht, Chem.–Eur. J., 2011, 17, 6683–6688 CrossRef CAS
.
- H. Yang, C. K. McLaughlin, F. A. Aldaye, G. D. Hamblin, A. Z. Rys, I. Rouiller and H. F. Sleiman, Nat. Chem., 2009, 1, 390–396 CrossRef CAS
.
- K. M. Stewart, J. Rojo and L. W. McLaughlin, Angew. Chem., Int. Ed., 2004, 43, 5808–5811 CrossRef CAS
.
- K. M. Stewart and L. W. McLaughlin, J. Am. Chem. Soc., 2004, 126, 2050–2057 CrossRef CAS
.
- D. Mitra, N. Di Cesare and H. F. Sleiman, Angew. Chem., Int. Ed., 2004, 43, 5804–5808 CrossRef CAS
.
- K. V. Gothelf, A. Thomsen, M. Nielsen, E. Clo and R. S. Brown, J. Am. Chem. Soc., 2004, 126, 1044–1046 CrossRef CAS
.
- M. Göritz and R. Krämer, J. Am. Chem. Soc., 2005, 127, 18016–18017 CrossRef
.
- M. Kalek, A. S. Madsen and J. Wengel, J. Am. Chem. Soc., 2007, 129, 9392–9400 CrossRef CAS
.
- S. Leininger, B. Olenyuk and P. J. Stang, Chem. Rev., 2000, 100, 853–907 CrossRef CAS
.
- L. Zapata, K. Bathany, J. M. Schmitter and S. Moreau, Eur. J. Org. Chem., 2003, 1022–1028 CrossRef CAS
.
- R. B. Martin and J. A. Lissfelt, J. Am. Chem. Soc., 1956, 78, 938–940 CrossRef CAS
.
- R. H. Holyer, C. D. Hubbard, S. F. A. Kettle and R. G. Wilkins, Inorg. Chem., 1966, 5, 622–625 CrossRef CAS
.
- R. Cali, E. Rizzarelli, S. Sammartano and G. Siracusa, Transition Met. Chem., 1979, 4, 328–332 CrossRef CAS
.
- G. U. Priimov, P. Moore, L. Helm and A. E. Merbach, Inorg. React. Mech., 2001, 3, 1–23 CAS
.
- K. M. Stewart and L. W. McLaughlin, Chem. Commun., 2003, 2934–2935 RSC
.
- S. Ghosh, I. Pignot-Paintrand, P. Dumy and E. Defrancq, Org. Biomol. Chem., 2009, 7, 2729–2737 CAS
.
- J. S. Choi, C. W. Kang, K. Jung, J. W. Yang, Y. G. Kim and H. Y. Han, J. Am. Chem. Soc., 2004, 126, 8606–8607 CrossRef CAS
.
- A. P. H. J. Schenning and E. W. Meijer, Chem. Commun., 2005, 3245–3258 RSC
.
- L. Kalachova, R. Pohl and M. Hocek, Synthesis, 2009, 105–112 RSC
; L. Kalachova, R. Pohl and M. Hocek, Org. Biomol. Chem, 2011 10.1039/c1ob06359f
.
- C. Brotschi and C. J. Leumann, Angew. Chem., Int. Ed., 2003, 42, 1655–1658 CrossRef CAS
.
- S. Hainke and O. Seitz, Angew. Chem., Int. Ed., 2009, 48, 8250–8253 CrossRef CAS
.
- J. R. Burns, J. Zekonyte, G. Siligardi, R. Hussain and E. Stulz, Molecules, 2011, 16, 4912–4922 CrossRef CAS
.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c1ob06421e |
| This journal is © The Royal Society of Chemistry 2012 |
