Membrane disrupting antimicrobial peptide dendrimers with multiple amino termini†
Michaela
Stach
a,
Noélie
Maillard
a,
Rameshwar U.
Kadam
a,
David
Kalbermatter
a,
Marcel
Meury
c,
Malcolm G. P.
Page
b,
Dimitrios
Fotiadis
c,
Tamis
Darbre
a and
Jean-Louis
Reymond
*a
aDepartment of Chemistry and Biochemistry, University of Berne, Freiestrasse 3, CH-3012, Berne, Switzerland. E-mail: jean-louis.reymond@ioc.unibe.ch; Fax: +41 31 631 80 57; Tel: +41 31 631 43 25
bBasilea Pharmaceutica Ltd., Grenzacherstrasse 487, PO Box, 4005 Basel, Switzerland
cInstitute of Biochemistry and Molecular Medicine, University of Berne, Bühlstrasse 28, 3012, Berne, Switzerland
First published on 23rd November 2011
Abstract
Antimicrobial peptide dendrimer H1 Leu8(Lys-Leu)4(Lys-Phe)2Lys-LysNH2 (Lys = branching lysine) was identified by screening a 6750-membered combinatorial library by the bead-diffusion assay. Sequence variations also revealed dendrimer bH1 Leu8(Dap-Leu)4(Dap-Phe)2Dap-LysNH2 (Dap = branching 2,3-diaminopropanoic acid) as a more potent analog. H1 and bH1 showed good antimicrobial activities mediated by membrane disruption (MIC = 2–4 μg mL−1 on Bacillus subtilis and Escherichia coli) but low hemolytic activity (MHC = 310 μg mL−1 respectively >2000 μg mL−1).
Many antimicrobial peptides (AMPs) act by disrupting microbial membranes.1 These include linear α-helical amphipathic peptides,2cyclic peptides and peptoids,3 foldamers,4 various amide oligomers,5 and multivalent lysine dendrimers appended with linear AMPs of various lengths.6,7 In all of these cases the multiple positive charges necessary for membrane disruption are brought about by the side chains of basic amino acids.8 Herein we report the discovery of antimicrobial peptide dendrimers such as H1 and bH1 in which positive charges are provided by the multiple amino termini at the dendrimer periphery (Fig. 1). This new type of AMP acts as membrane disrupting agent and shows remarkably low hemolytic activity.
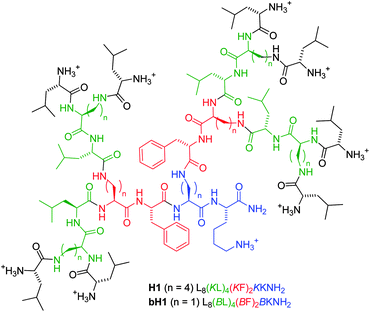 | ||
| Fig. 1 Structure of antimicrobial peptide dendrimers H1 and bH1. | ||
We recently developed the chemistry of peptide dendrimers incorporating variable amino acids within the dendrimer branches.9 In contrast to the polylysine dendrimers which are limited to the multivalent display of linear peptides,10 our peptide dendrimers vary in the nature and length of the dendrimer core, intermediate and terminal branches. This diversity can be exploited to obtain peptide dendrimers acting as enzyme11 and metalloprotein models,12 and in glycosylated version as biofilm inhibitors13 and drug delivery agents.14
We wondered if the multiple amino termini created by the ramified structure of our peptide dendrimers might serve as the source of positive charges in a new type of dendritic AMPs in which the nature of the amino acids within the branches would enable a membrane disrupting activity by bringing about hydrophobic groups. To test this hypothesis, a diverse 6750-membered combinatorial library L of G3 peptide dendrimers X48(KX3)4(KX2)2KX1 (X = variable amino acid, K = branching L-lysine, Fig. 2A), a sequence type which is particularly resistant to proteolysis,15 was prepared by split-and-mix solid phase peptide synthesis (SPPS)16 on a photolabile resin suitable for off-bead assays.17
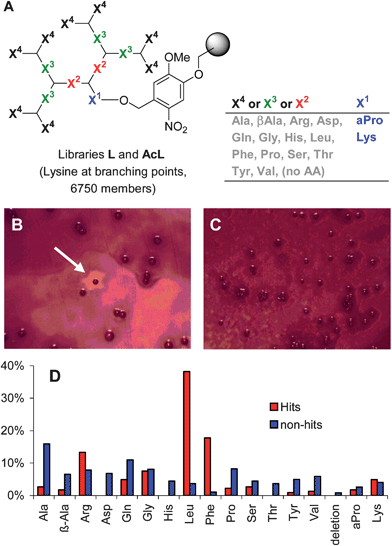 | ||
| Fig. 2 Screening for dendrimer AMPs. (A) Structure of peptide dendrimer libraries L and AcL. (B) Antimicrobial screening showing active beads from library L after staining the B. subtilis bacteria with MTT (thiazolyl blue tetrazolium bromide). (C) Inactive beads in library AcL. (D) Overall composition of hits and non-hits at variable positions X1–X4, calculated from the total occurrence of each amino acidTAA = 8X4 + 4X3 + 2X2 + X1. The sequences of 15 hits and 51 non-hits were determined. Sequences are shown in Tables S1 and S2†. | ||
Library L with free amino termini and the corresponding N-acetylated library AcL were subjected to antimicrobial activity screening18 using a bead-diffusion assay19 with an adapted protocol20 compatible with bead decoding by amino acid analysis (AAA).21,22 In this assay the beads are photolyzed in the absence of solvent and spread onto an agar plate inoculated with bacterial lawn. Peptides diffuse into the agar resulting in a clearing zone around beads carrying AMPs, whose sequence can be determined by AAA of the peptide left on the bead.
Upon screening of dendrimer library L, three to five beads per agar plate (0.1% of approximately 5000 beads) were surrounded by a clearing zone (Fig. 2B). The assay with acetylated library AcL did not give any hits (Fig. 2C). The active beads from library L contained predominantly the hydrophobic residues Leu and Phe (>10-fold enrichment over control), with a consensus for sequence Leu8(Lys-Leu/Phe)4(Lys-Phe/Pro)2Lys-LysNH2, while control inactive beads gave random sequences (Fig. 2D and Tables S1 and S2†). Taken together, these results showed that the combination of free amino termini with hydrophobic side chains was essential for obtaining an antimicrobial activity in these dendrimers.
The positive hits H1, H2 and H3, all carrying eight N-terminal leucines at position X4, were resynthesized by SPPS, together with their N-acetylated analogs as controls Ac1, Ac2 and Ac3. All products were obtained in good yields after purification by preparative HPLC. The following analogs of the strongest hits H1 and H2 were also prepared: (a) diastereoisomers dH1 and dH2 with D-amino acids at all non-branching positions; (b) isoleucine analogs iH1 and iH2; (c) an alanine scan series AX1–AX4 corresponding to hit H1; (d) analogs with the shorter 2,3-diaminopropanoic (Dap) branching unit instead of lysine bH1 and bH2 (Table 1).
| No | Sequencea | Yieldb | MS calc./obsdc |
|---|---|---|---|
| a B indicates a branching 2,3-diaminopropanoic acid and K a branching lysine. Standard one-letter codes are used with L-amino acids in upper case and D-amino acids in lower case. b Yield given taking the amino acid content determined by amino acid analysis into account. c Sequence: ILPWKWPWWPWRRNH2. | |||
| H1 | (L)8(KL)4(KF)2KKNH2 | 45.6 mg/8% | 2693.9/2694.0 |
| Ac1 | (AcL)8(KL)4(KF)2KKNH2 | 9.5 mg/6% | 3030.0/3031.0 |
| H2 | (L)8(KF)4(KP)2KKNH2 | 62.5 mg/14% | 2729.8/2730.0 |
| Ac2 | (AcL)8(KF)4(KP)2KKNH2 | 18.3 mg/12% | 3065.9/3066.7 |
| H3 | (L)8(KQ)4(KF)2KKNH2 | 73.1 mg/12% | 2753.8/2754.0 |
| Ac3 | (AcL)8(KQ)4(KF)2KKNH2 | 76.8 mg/44% | 3089.9/3091.0 |
| dH1 | (l)8(Kl)4(Kf)2KkNH2 | 76.2 mg/12% | 2693.9/2694.0 |
| iH1 | (I)8(KI)4(KF)2KKNH2 | 33.6 mg/1% | 2693.9/2694.0 |
| dH2 | (l)8(Kf)4(Kp)2KkNH2 | 42.0 mg/10% | 2729.8/2730.0 |
| iH2 | (I)8(KF)4(KP)2KKNH2 | 38.2 mg/5% | 2729.8/2730.0 |
| AX1 | (L)8(KL)4(KF)2KANH2 | 27.2 mg/5% | 2638.6/2637.7 |
| AX2 | (L)8(KL)4(KA)2KKNH2 | 33.4 mg/10% | 2543.5/2542.5 |
| AX3 | (L)8(KA)4(KF)2KKNH2 | 14.8 mg/4% | 2527.3/2526.6 |
| AX4 | (A)8(KL)4(KF)2KKNH2 | 23.2 mg/8% | 2359.0/2358.2 |
| bH1 | (L)8(BL)4(BF)2BKNH2 | 21.8 mg/7% | 2401.1/2400.0 |
| bH2 | (L)8(BF)4(BP)2BKNH2 | 33.5 mg/12% | 2437.1/2436.0 |
| indo | Indolicidin c | 47.5 mg/9% | 1906.1/1906.0 |
The purified hits H1 and H2 showed the expected antimicrobial activity against B. subtilis. Dendrimer H3 with glutamine instead of Leu/Phe in position X3 was not active showing the importance of a hydrophobic internal shell for activity. The N-acetylated controls Ac1–Ac3 were also inactive, consistent with the absence of active beads in the N-acetylated library, once more highlighting the importance of the multiple free N-termini preferentially from N-terminal leucine residues for antimicrobial activity. Although they were slightly less active, the diastereoisomers dH1/dH2 and isomers iH1/iH2 showed comparable activities to H1/H2. In the alanine scan series, mutation of the outer eight or four leucines in the G3 (AX4) or G2 (AX3) branches reduced activity, confirming that hydrophobic residues were needed for activity in combination with the multiple positive charges provided by the free N-termini (Table 2).
| Cpd. | MICaB. subtilisBR151 | MICaE. coliDH5α | MICaP. aeruginosaPA01 | MHC b erythrocytes |
|---|---|---|---|---|
| a Minimal inhibitory concentration (MIC) values are given in μg mL−1 and were determined following ref. 23. b Minimal hemolytic concentration (MHC) values are given in μg mL−1 and were determined following ref. 24. All measurements were done in triplicate for MIC and duplicate for MHC. Shown are average values. Ac1, Ac2 and Ac3 showed MIC > 100 μg mL−1 against B. subtilisBR151 and were not evaluated further. | ||||
| H1 | 2.0 | 21 | >72 | 310 |
| H2 | 2.0 | 19 | 66 | >10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
| H3 | >100 | >100 | >100 | 250 |
| dH1 | 2.5 | 15 | 60 | >10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
| iH1 | 4.4 | 79 | >100 | 20 |
| dH2 | 4.4 | 11 | 60 | >10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
| iH2 | 2.5 | 18 | 61 | >10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
| AX1 | 2.0 | 15 | ≥100 | 31 |
| AX2 | 4.5 | >100 | 67 | >2000 |
| AX3 | 67 | >100 | >100 | >2000 |
| AX4 | >100 | >100 | 67 | 1000 |
| bH1 | 2.9 | 3.9 | 18 | >2000 |
| bH2 | 3.1 | 15 | 20 | >2000 |
| indo | 3.0 | 22 | >100 | 16 |
Several of the dendrimers also showed good activities against E. coli and P. aeruginosa, in particular the Dap-branched analogs bH1 and bH2, which were also more compact (rH ≈ 1.15 nm from diffusion NMR) than the lysine branched dendrimers (rH = 1.30–1.40 nm, see ESI†). Profiling against an additional 17 strains showed further interesting activities for H1, iH1 and bH1 (Table 3). Hemolysis by the dendrimers was generally weak compared to the linear control peptide indolicidin, except in AX1 lacking the core lysine residue and iH1 containing isoleucines. The Dap branched analog bH1 combined a broader activity than H1 with a weaker hemolytic activity, suggesting a useful therapeutic window. Similarly low hemolysis has been observed previously in antimicrobial lysine dendrimers.7
| Bacterium | PB | AP | H1 | iH1 | bH1 |
|---|---|---|---|---|---|
| a Minimal inhibitory concentration (MIC) values are given in μg mL−1 of the weighed lyophilized solid as TFA salt and were determined following the standard broth microdilution method recommended by the Clinical and Laboratory Standards Institute (M7-A7). The concentration range was 0.06 to 32 μg mL−1. Polymyxin B (PB) and ampicillin (AP) were used as positive controls. The peptides did not show measurable inhibition (MIC > 32 μg mL−1) on the following strains: S. aureus (42080, ATCC29213), En. faecalis (ATCC29212, Van B E80-8), E. coli (ATCC25922), P. aeruginosa (ATCC27853, PA01(HS), 8S/H, K799/WT, K799/61). Peptides Ac1, H3, Ac2, H2, iH2 and bH2 were not active in any of the strains tested (MIC > 32 μg mL−1). AX1–AX4 were not measured in this series. | |||||
| S. aureus ATCC25923 | ≥32 | 0.13 | 24 | >32 | ≥32 |
| S. aureus 887 | 16 | >32 | 12 | 16 | 32 |
| S. epidermidis ATCC14990 | >32 | 1 | 32 | 32 | 24 |
| S. epidermidis J147 | >32 | 1.5 | 24 | >32 | 24 |
| En . faecium ATCC19434 | >32 | 4 | ≥32 | 24 | >32 |
| En . faecium Van B E38-10 | >32 | >32 | 32 | 8 | >32 |
| E. coli HB101 (PAT266) | 2 | >32 | 32 | >32 | 32 |
| E. coli DC2 | 4 | 1 | >32 | >32 | 32 |
The peptide dendrimers were generally less active on Gram negative bacteria, which together with the activity of the dendrimer stereoisomers and the requirement for a combination of cationic charges and hydrophobic residues pointed towards membrane disruption as a possible mechanism of action. The effect was evidenced for dendrimer H1 using synthetic unilamellar vesicles assembled from E. colilipids. Dendrimer H1 at 25 μg mL−1 effected a complete disassembly of the vesicles as observed by electron microscopy, while the vesicles were stable in the presence of the N-acetylated analog Ac1 at the same concentration (Fig. 3).
 | ||
| Fig. 3 Negative-stain transmission electron microscopy of untreated and dendrimer-treated E. colilipid vesicles: untreated (A), 25 μg mL−1H1-treated (B) and Ac1-treated (C) liposomes. The scale bars represent 100 nm. | ||
The interaction of the peptide dendrimers with the membrane probably involves electrostatic binding of the cationic N-terminal protonated amines with the anionic phospholipid head group and insertion of hydrophobic amino acid side chains into the lipid bilayer. A molecular dynamics (MD) simulation of dendrimer H1 in the presence of a phosphorylcholine (POPC) lipid bilayer over 100 ns indeed supports this hypothesis (Fig. 4). A more extended simulation will be required to see if the interaction leads to the formation of pores as observed in MD studies of amphiphilic membrane disrupting AMPs, cell penetrating peptides, and G3–G7 PAMAM dendrimers.25
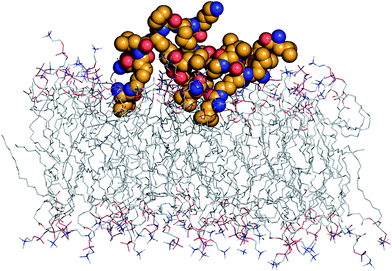 | ||
| Fig. 4 MD simulation of peptide dendrimer H1:POPC membrane interaction. The model represents the last frame obtained from 100 ns MD simulation using NPγT ensemble at 300 K using the OPLS-AA force field. Six of the eight N-terminal leucine protonated amines form H-bonds with anionic phospholipid head groups, while their side chains have inserted into the lipid bilayer. | ||
In summary, G3 peptide dendrimers with hydrophobic amino acids in their branches and multiple free amino termini as the sole source of positive charges possess significant antimicrobial activities against Gram positive and Gram negative bacteria. The activities of H1 and bH1 compare well with those of linear peptides such as indolicidin, but also of previously reported dendritic peptides (best MIC ≈ 1–20 μg mL−1, various bacterial strains, see ref. 6 and 7). These peptide dendrimers represent a new class of AMPs since previous peptide based linear or dendritic AMPs all relied on amino acid side chains as the source of positive charges. The evidence for significant activity variations upon amino acid exchanges suggests that it should be possible to optimize activities against specific strains by further exploration of the sequence space, which might open a new opportunity to develop non-toxic antimicrobial agents.
Acknowledgements
This work was supported financially by the University of Berne, the Swiss National Science Foundation, and the European Union FP7-ITN-238434.Notes and references
- (a) E. B. Hadley and R. E. Hancock, Curr. Top. Med. Chem., 2010, 10, 1872–1881 CAS; (b) J. Davies and D. Davies, Microbiol. Mol. Biol. Rev., 2010, 74, 417–433 CrossRef CAS; (c) D. M. Bowdish, D. J. Davidson and R. E. Hancock, Curr. Protein Pept. Sci., 2005, 6, 35–51 CrossRef CAS.
- (a) Z. Oren and Y. Shai, Biopolymers, 1998, 47, 451–463 CrossRef CAS; (b) S. Zhu, A. Aumelas and B. Gao, J. Med. Chem., 2011, 54, 1091–1095 CrossRef CAS.
- (a) S. Fernandez-Lopez, H. S. Kim, E. C. Choi, M. Delgado, J. R. Granja, A. Khasanov, K. Kraehenbuehl, G. Long, D. A. Weinberger, K. M. Wilcoxen and M. R. Ghadiri, Nature, 2001, 412, 452–455 CrossRef CAS; (b) D. Comegna, M. Benincasa, R. Gennaro, I. Izzo and F. De Riccardis, Bioorg. Med. Chem. Lett., 2010, 18, 2010–2018 CrossRef CAS.
- P. Claudon, A. Violette, K. Lamour, M. Decossas, S. Fournel, B. Heurtault, J. Godet, Y. Mely, B. Jamart-Gregoire, M. C. Averlant-Petit, J. P. Briand, G. Duportail, H. Monteil and G. Guichard, Angew. Chem., Int. Ed., 2010, 49, 333–336 CAS.
- (a) I. S. Radzishevsky, T. Kovachi, Y. Porat, L. Ziserman, F. Zaknoon, D. Danino and A. Mor, Chem. Biol., 2008, 15, 354–362 CrossRef CAS; (b) B. P. Mowery, A. H. Lindner, B. Weisblum, S. S. Stahl and S. H. Gellman, J. Am. Chem. Soc., 2009, 131, 9735–9745 CrossRef CAS.
- (a) S. P. Liu, L. Zhou, R. Lakshminarayanan and R. W. Beuerman, Int. J. Pept. Res. Ther., 2011, 16, 199–213 CrossRef; (b) X. Chen, M. Zhang, C. Zhou, N. R. Kallenbach and D. Ren, Appl. Environ. Microbiol., 2011, 77, 4878–4885 CrossRef CAS.
- (a) J. P. Tam, Y. A. Lu and J. L. Yang, Eur. J. Biochem., 2002, 269, 923–932 CrossRef CAS; (b) J. Janiszewska, J. Swieton, A. W. Lipkowski and Z. Urbanczyk-Lipkowska, Bioorg. Med. Chem. Lett., 2003, 13, 3711–3713 CrossRef CAS; (c) Z. Liu, A. W. Young, P. Hu, A. J. Rice, C. Zhou, Y. Zhang and N. R. Kallenbach, ChemBioChem, 2007, 8, 2063–2065 CrossRef CAS; (d) S. Hou, C. Zhou, Z. Liu, A. W. Young, Z. Shi, D. Ren and N. R. Kallenbach, Bioorg. Med. Chem. Lett., 2009, 19, 5478–5481 CrossRef CAS.
- (a) M. N. Melo, R. Ferre and M. A. Castanho, Nat. Rev. Microbiol., 2009, 7, 245–250 CrossRef CAS; (b) K. A. Brogden, Nat. Rev. Microbiol., 2005, 3, 238–250 CrossRef CAS; (c) M. R. Yeaman and N. Y. Yount, Pharmacol. Rev., 2003, 55, 27–55 CrossRef CAS.
- (a) J. Kofoed and J.-L. Reymond, Curr. Opin. Chem. Biol., 2005, 9, 656–664 CrossRef CAS; (b) T. Darbre and J.-L. Reymond, Acc. Chem. Res., 2006, 39, 925–934 CrossRef CAS; (c) T. Darbre and J.-L. Reymond, Curr. Top. Med. Chem., 2008, 8, 1286–1293 CrossRef CAS.
- (a) L. Crespo, G. Sanclimens, M. Pons, E. Giralt, M. Royo and F. Albericio, Chem. Rev., 2005, 105, 1663–1681 CrossRef CAS; (b) K. Sadler and J. P. Tam, Rev. Mol. Biotechnol., 2002, 90, 195–229 CrossRef CAS.
- (a) A. Esposito, E. Delort, D. Lagnoux, F. Djojo and J. L. Reymond, Angew. Chem., Int. Ed., 2003, 42, 1381–1383 CrossRef CAS; (b) S. Javor and J. L. Reymond, J. Org. Chem., 2009, 74, 3665–3674 CrossRef CAS; (c) R. Biswas, N. Maillard, J. Kofoed and J. L. Reymond, Chem. Commun., 2010, 46, 8746–8748 RSC; (d) N. A. Uhlich, T. Darbre and J. L. Reymond, Org. Biomol. Chem., 2011, 9, 7071–7084 RSC.
- (a) P. Sommer, N. A. Uhlich, J. L. Reymond and T. Darbre, ChemBioChem, 2008, 9, 689–693 CrossRef CAS; (b) N. A. Uhlich, P. Sommer, C. Buhr, S. Schurch, J. L. Reymond and T. Darbre, Chem. Commun., 2009, 6237–6239 RSC; (c) N. A. Uhlich, A. Natalello, R. U. Kadam, S. M. Doglia, J. L. Reymond and T. Darbre, ChemBioChem, 2010, 11, 358–365 CrossRef CAS.
- (a) E. M. Johansson, S. A. Crusz, E. Kolomiets, L. Buts, R. U. Kadam, M. Cacciarini, K. M. Bartels, S. P. Diggle, M. Camara, P. Williams, R. Loris, C. Nativi, F. Rosenau, K. E. Jaeger, T. Darbre and J. L. Reymond, Chem. Biol., 2008, 15, 1249–1257 CrossRef CAS; (b) E. M. V. Johansson, R. U. Kadam, G. Rispoli, S. A. Crusz, K.-M. Bartels, S. P. Diggle, M. Camara, P. Williams, K.-E. Jaeger, T. Darbre and J.-L. Reymond, Med. Chem. Commun., 2011, 2, 418–420 RSC; (c) R. U. Kadam, M. Bergmann, M. Hurley, D. Garg, M. Cacciarini, M. A. Swiderska, C. Nativi, M. Sattler, A. R. Smyth, P. Williams, M. Cámara, A. Stocker, T. Darbre and J. L. Reymond, Angew. Chem., Int. Ed., 2011, 50, 10631–10635 CrossRef CAS.
- (a) D. Lagnoux, T. Darbre, M. L. Schmitz and J. L. Reymond, Chem.–Eur. J., 2005, 11, 3941–3950 CrossRef CAS; (b) E. M. Johansson, J. Dubois, T. Darbre and J. L. Reymond, Bioorg. Med. Chem., 2010, 18, 6589–6597 CrossRef CAS.
- P. Sommer, V. S. Fluxa, T. Darbre and J. L. Reymond, ChemBioChem, 2009, 10, 1527–1536 CrossRef CAS.
- (a) A. Furka, F. Sebestyen, M. Asgedom and G. Dibo, Int. J. Pept. Protein Res., 1991, 37, 487–493 CrossRef CAS; (b) K. S. Lam, S. E. Salmon, E. M. Hersh, V. J. Hruby, W. M. Kazmierski and R. J. Knapp, Nature, 1991, 354, 82–84 CrossRef CAS; (c) R. A. Houghten, C. Pinilla, S. E. Blondelle, J. R. Appel, C. T. Dooley and J. H. Cuervo, Nature, 1991, 354, 84–86 CrossRef CAS; (d) K. S. Lam, R. Liu, S. Miyamoto, A. L. Lehman and J. M. Tuscano, Acc. Chem. Res., 2003, 36, 370–377 CrossRef CAS.
- (a) N. Maillard, R. Biswas, T. Darbre and J.-L. Reymond, ACS Comb. Sci., 2011, 13, 310–320 CrossRef CAS; (b) N. Maillard, T. Darbre and J. L. Reymond, J. Comb. Chem., 2009, 11, 667–675 CrossRef CAS.
- (a) S. E. Blondelle and K. Lohner, Curr. Pharm. Des., 2010, 16, 3204–3211 CrossRef CAS; (b) R. Rathinakumar and W. C. Wimley, FASEB J., 2010, 24, 3232–3238 CrossRef CAS.
- (a) J. M. Quillan, C. K. Jayawickreme and M. R. Lerner, Proc. Natl. Acad. Sci. U. S. A., 1995, 92, 2894–2898 CrossRef CAS; (b) K. R. Oldenburg, K. T. Vo, B. Ruhland, P. J. Schatz and Z. Yuan, J. Biomol. Screening, 1996, 1, 123–130 CrossRef CAS; (c) J. L. Silen, A. T. Lu, D. W. Solas, M. A. Gore, D. MacLean, N. H. Shah, J. M. Coffin, N. S. Bhinderwala, Y. Wang, K. T. Tsutsui, G. C. Look, D. A. Campbell, R. L. Hale, M. Navre and C. R. DeLuca-Flaherty, Antimicrob. Agents Chemother., 1998, 42, 1447–1453 CAS; (d) C. K. Jayawickreme, H. Sauls, N. Bolio, J. Ruan, M. Moyer, W. Burkhart, B. Marron, T. Rimele and J. Shaffer, J. Pharmacol. Toxicol. Methods, 1999, 42, 189–197 CrossRef CAS; (e) D. J. Burns, J. L. Kofron, U. Warrior and B. A. Beutel, Drug Discovery Today, 2001, 6, S40–S47 CrossRef CAS.
- V. S. Fluxa, N. Maillard, M. G. P. Page and J.-L. Reymond, Chem. Commun., 2011, 47, 1434–1436 RSC.
- (a) A. Clouet, T. Darbre and J.-L. Reymond, Angew. Chem., Int. Ed., 2004, 43, 4612–4615 CrossRef CAS; (b) N. Maillard, A. Clouet, T. Darbre and J.-L. Reymond, Nat. Protoc., 2009, 4, 132–142 CrossRef CAS.
- (a) J. Kofoed and J. L. Reymond, J. Comb. Chem., 2007, 9, 1046–1052 CrossRef CAS; (b) J. Kofoed and J. L. Reymond, Chem. Commun., 2007, 4453–4455 CAS.
- I. Wiegand, K. Hilpert and R. E. Hancock, Nat. Protoc., 2008, 3, 163–175 CrossRef CAS.
- N. P. Chongsiriwatana, J. A. Patch, A. M. Czyzewski, M. T. Dohm, A. Ivankin, D. Gidalevitz, R. N. Zuckermann and A. E. Barron, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 2794–2799 CrossRef CAS.
- A. Gurtovenko, J. Anwar and I. Vattulainen, Chem. Rev., 2010, 110, 6077–6103 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available: Details of library design, synthesis and assays. See DOI: 10.1039/c1md00272d |
| This journal is © The Royal Society of Chemistry 2012 |
