What yeast and cardiomyocytes share: ultradian oscillatory redox mechanisms of cellular coherence and survival†
David
Lloyd
*a,
Sonia
Cortassa
b,
Brian
O'Rourke
b and
Miguel A.
Aon
b
aSchool of Biosciences, Cardiff University, Museum Avenue, Cardiff CF10 3AT Wales, UK. E-mail: lloydd@cardiff.ac.uk; Fax: +44(0) 2920 874305; Tel: +44(0)2920 842649
bJohns Hopkins University, School of Medicine, 1059 Ross Bldg., 702 Rutland Av., Baltimore, MD 21205, USA
First published on 5th December 2011
Abstract
The coherent and robust, yet sensitively adaptable, nature of organisms is an astonishing phenomenon that involves massive parallel processing and concerted network performance at the molecular level. Unravelling the dynamic complexities of the living state underlines the essential operation of ultradian oscillations, rhythms and clocks for the establishment and maintenance of functional order simultaneously on fast and slower timescales. Non-invasive monitoring of respiration, mitochondrial inner membrane potentials, and redox states (especially those of NAD(P)H, flavin, and the monochlorobimane complex of glutathione), even after more than 50 years research, continue to provide both new insights and biomedical applications. Experiments with yeast and in cardiac cells reveal astonishing parallels and similarities in their dynamic biochemical organization.
Insight, innovation, integrationInsight is lacking about the overall integration, coordination and coherence of living systems, especially with respect to temporal organisation. Here we have developed a method whereby yeast or cardiomyocyte single cells are observed by two photon laser scanning fluorescence microscopy that enables mitochondrial membrane potential, NAD(P)H, and glutathione redox states, as well as reactive O2 species to be monitored simultaneously in identical fields. Concerted oscillatory performance (period range sec to min) was observed in both systems, emphasizing the central role of redox balance. Membrane inlet mass spectrometry on self-synchronised yeasts also indicated coordinated and self-similar multiple time scales for respiratory oscillations (period range min to h). These features are central for spatiotemporal synchrony and network coherence. |
Introduction
The enormous complexity of biological organization in time is still one of the least understood as a functional and integrated continuum (Fig. 1). Even though parts of the temporal span have been well-studied, e.g. the millisecond and daily (circadian) domains, the co-ordination of fast and slow reactions, pathways and processes is not yet clarified.1 For much of the last century the emphasis on balanced controls on both physiological functions, protein turnover, and biochemical pathways has led to the prominent concept of homeostasis. We now know that these earlier views on the self-regulation of open systems did not sufficiently emphasize that steady-states are not always precisely attained but rather achievable only within limits. Simplistic interpretation of data from homogenates of heterogeneous cell populations, where details of spatio-temporal structure and function are lost due to the methodology and filtered out by inadequate frequency of sampling often ignores the rich dynamics of the system. Important advances in non-invasive physical techniques and biological imaging have transformed our ability to study live single cells and to reveal the dynamic complexity of the cellular network. Now it becomes ever more evident that steady-states are the exception rather than the rule. The living state is homeodynamic rather than homeostatic2,3 and non-linearity of functional activities lead to a plethora of oscillatory, rhythmic and timed outputs.4,5 A distinction between these three classes of periodic processes is easily made. Thus an oscillator may be highly damped only to fade into a stationary state within a few cycles: such a waveform is typical for a signal (e.g. a Ca2+-oscillation),6 that has to be triggered and later switched off. A rhythm is a self-sustained oscillation, e.g. the heart-beat, that perpetuates itself automatically for the whole lifetime of the animal. A biological clock is a rhythm that has a built-in temperature compensating mechanism.7,8 There may be many different ways of achieving this, but many routes arise from kinetic similarity of feedback in opposing reactions.9Fig. 2 shows some examples of oscillators, rhythms and clocks characterized in biological systems; they span many time domains from <ms to years.![The temporal structure of biological organization as characterized by the relaxation times of biological processes. The different time scales exhibited by living systems are shown by the typical relaxation times (after perturbation) in decades on a logarithmic scale. This way of representation accounts for the broad temporal scale spanned by the occurrence of biological processes, i.e. from picoseconds [10−12 s] to many years. The biological processes indicated represent many of the main molecular and cellular functional properties as well as those related to conformational changes in molecules (e.g. following ligand interactions), metabolism, energy transduction, solute transport, action potentials in neurons and cardiomyocytes, macromolecule polymerization and cell growth and division. (Reproduced from Lloyd and Rossi (2008) in Ultradian Rhythms from Molecules to Mind).](/image/article/2012/IB/c1ib00124h/c1ib00124h-f1.gif) | ||
| Fig. 1 The temporal structure of biological organization as characterized by the relaxation times of biological processes. The different time scales exhibited by living systems are shown by the typical relaxation times (after perturbation) in decades on a logarithmic scale. This way of representation accounts for the broad temporal scale spanned by the occurrence of biological processes, i.e. from picoseconds [10−12 s] to many years. The biological processes indicated represent many of the main molecular and cellular functional properties as well as those related to conformational changes in molecules (e.g. following ligand interactions), metabolism, energy transduction, solute transport, action potentials in neurons and cardiomyocytes, macromolecule polymerization and cell growth and division. (Reproduced from Lloyd and Rossi (2008) in Ultradian Rhythms from Molecules to Mind). | ||
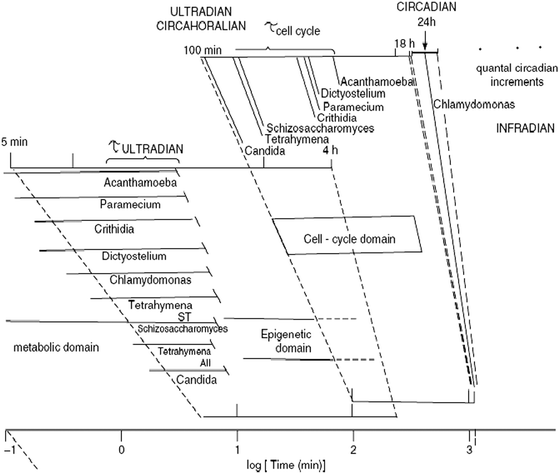 | ||
| Fig. 2 Periodic phenomena across temporal scales. The period of cyclic processes in the ultradian (<24 h), circadian (24 h) and infradian (>24 h) time domains are shown in a logarithmic scale. Several clocks (temperature-compensated biological mechanisms) can be found in the ultradian temporal domain, in addition to the ubiquitous circadian clock. A prominent ultradian clock is the circahoralian (about 1 h period) that is indicated as occurring in many lower eukaryotes and is always of shorter duration that the cell division cycle, as shown in the figure. (Reproduced from Lloyd and Rossi (2008) in Ultradian Rhythms from Molecules to Mind). | ||
The core of rhythmic organization in yeast and heart muscle cells
Evidence gathered over the past 15 years has indicated that a network of redox processes located across and between the mitochondria and cytosol of yeasts (Saccharomyces cerevisiae,1,4,5Candida albicans10,11) and cardiomyocytes obtained from small mammals, mice and guinea pigs,12,13 show remarkable similarities.14Cardiomyocytes are separated from yeasts by 1.2 billion years of evolution and have approximately 5X greater gene numbers. Both cell types are exquisitely (but differently) specialized to survive and perform effectively their very different functions. Thus it is quite astonishing that they still have so much in common in their basic redox balancing machinery.14 However it should not be concluded that this indicates that they constitute a special case, but rather that their core requirements for maintenance of cellular coherence are similar at least at a functional level. It seems likely that these highly conserved features will not prove different in other cells or organisms.
Fig. 3 indicates common features not only between widely-separated taxa, but also across widely-based time domains.14–16 The scheme shows that generation of rhythms entails the cycling of cytoplasmic and mitochondrial proteins between their oxidized and reduced states, mainly driven by ROS and NAD(P)H and reflected in the redox potentials of the thiol pools.15,17,18Mitochondria are the main source of ROS produced by the respiratory chain; oxidative stress results from an imbalance between ROS production and ROS scavenging. The glutathione redox potential, and the absolute concentrations of reduced (GSH) and oxidised (GSSG) glutathione, modulate glutathione reductase (GR) and transhydrogenase (THD). Although in yeast it has not been confirmed that THD plays a part, thioredoxin and glutaredoxin play a crucial role. In turn, the redox status in the mitochondrial matrix represented by NADH and NADPH pools,19 which are interconverted by THD activity, drives GSH regeneration.15 Mitochondrial GSH is also replenished via cytoplasmic import of reducing power through carriers and metabolic shuttle systems.20–22 In cardiomyocytes, mitochondrial oscillations are triggered when a threshold level of ROS is attained12,13 which happens when the glutathione redox potential oxidizes to GSH![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) GSSG ratios between 150
GSSG ratios between 150![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 to 100
1 to 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. A critical ratio of GSH-GSSG of 50
1. A critical ratio of GSH-GSSG of 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 elicits the opening of the permeability transition pore, cell hyper-contraction, and death.15,18 In yeast, numerous processes (magenta) have proven to be oscillatory and we propose that ensembles of oscillators are coupled via this primordial mechanism.5,14 In yeast, these processes may be modulated by illumination, temperature changes or chemical perturbation. Perturbation analysis of the yeast ultradian system utilising NO+ donors19 confirms the central role of this redox system; the numbers on Fig. 3 represent the site of perturbation. The assay, biological significance, physiology and pathophysiology of mitochondrial glutathione have been reviewed.20–22 The pivotal roles of the GSH/GSSG redox couple in the generation of oscillatory respiration in yeast has been confirmed.23
1 elicits the opening of the permeability transition pore, cell hyper-contraction, and death.15,18 In yeast, numerous processes (magenta) have proven to be oscillatory and we propose that ensembles of oscillators are coupled via this primordial mechanism.5,14 In yeast, these processes may be modulated by illumination, temperature changes or chemical perturbation. Perturbation analysis of the yeast ultradian system utilising NO+ donors19 confirms the central role of this redox system; the numbers on Fig. 3 represent the site of perturbation. The assay, biological significance, physiology and pathophysiology of mitochondrial glutathione have been reviewed.20–22 The pivotal roles of the GSH/GSSG redox couple in the generation of oscillatory respiration in yeast has been confirmed.23
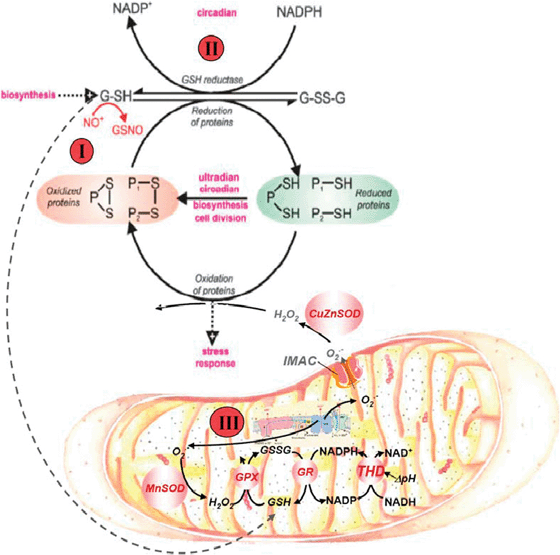 | ||
| Fig. 3 Redox cycling of intracellular thiols at the core of yeast and heart rhythmicity. The scheme shows that generation of rhythms entails the cycling of cytoplasmic and mitochondrial proteins between their oxidized and reduced states mainly driven by ROS and the redox potential of the thiols pool. Mitochondria are the main source of ROS produced by the respiratory chain; oxidative stress results from an imbalance between ROS production and ROS scavenging. The glutathione and thioredoxin redox potentials, and the absolute concentrations of reduced (GSH) and oxidized (GSSG) glutathione, modulate ROS emission in mitochondria. Mitochondrial GSH and thioredoxin (Trx2) regeneration in the matrix are essential for keeping the ROS balance. In yeast, numerous processes (magenta I–III) have proven to be oscillatory and we propose that ensembles of oscillators are coupled via this primordial mechanism. In heart, the redox cycling involves at least glutathione, Trx2, and NAD(P)H couples in the mitochondrial matrix, as modulators of ROS generation. Perturbation analysis of the yeast ultradian system utilising NO+ donors, 5-nitro-2-furaldehyde(I), D,L-butathionine (S,R)-sulphoximine(II) or protonophores(III), confirms the central role of this redox system. The numbers on the figure represent the site of perturbation. (Reproduced from Aon MA, Roussel MR, Cortassa S, B. O'Rourke, Murray DB, et al. (2008) The Scale-Free Dynamics of Eukaryotic Cells. PLoS ONE 3(11): e3624. doi: 10.1371/journal.pone.0003624). | ||
The history of the concept and evidence for the pivotal role of the cycling of low molecular weight thiols and protein thiols during ultradian cell-division cycle, and circadian rhythmicity goes back to the 1930's. Key advances were made with the pioneering research by Rapkine24 employing experiments on sea urchin eggs. Subsequent theoretical treatment and mathematical modelling led to an unified basic understanding of the fundamental significance of oscillations in living systems.
Spatial organization of yeast and heart mitochondrial networks
Reconstruction of three-dimensional images from serial optical sections of yeast indicates a single complex-reticulate ‘mega-mitochondrion’ together with smaller cylindrical organelles (Fig. 4a):25 progress through the reproductive budding cycle is attended by a succession of fusion and fission events. Two-photon laser scanning fluorescence microscopy reveals the autofluorescence of the organism as an extremely sensitive and useful measure of its redox state (Fig. 4b). Two photon microscopy of isolated cardiomyocytes (Fig. 4c and d)12 and of the epicardium of an isolated perfused guinea pig heart18 after pre-loading or perfusion with 100 nM tetramethyrhodamine-methyl ester (TMRM), a membrane potential sensitive fluorophore, shows an even more intricate mitochondrial organization. The mitochondria are densely packed in parallel rows along the length of the cell forming an extensive network of discrete units12,26–28 that extends throughout the pericardiumvia interlinked individual cells through gap junctions. In addition to monitoring inner mitochondrial membrane electrochemical potential (ΔΨm, Fig. 4c), two other fluorophores have proven to be useful for simultaneously interrogating the redox status of intramitochondrial processes. These are a chloromethyl-derivative of dichlorodihydrofluorescein (CM-DCF) measuring reactive O2 species (ROS) mainly as H2O2, and mono-chlorobimane (MCB), a probe for the tripeptide, reduced glutathione (GSH). Glutathione bimane, GSB, the adduct formed by GSH and MCB, has been demonstrated to be a sensitive indicator of the cytoplasmic GSH![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) GSSG ratio (Fig. 5). Interestingly, ΔΨm loss correlated with oxidation of the NAD(P)H pool to ∼20% of the baseline level. This threshold corresponded to rapid depletion of the mitochondrial GSH pool just before ΔΨm collapse in cells loaded with MCB (Fig. 5b and c).
GSSG ratio (Fig. 5). Interestingly, ΔΨm loss correlated with oxidation of the NAD(P)H pool to ∼20% of the baseline level. This threshold corresponded to rapid depletion of the mitochondrial GSH pool just before ΔΨm collapse in cells loaded with MCB (Fig. 5b and c).
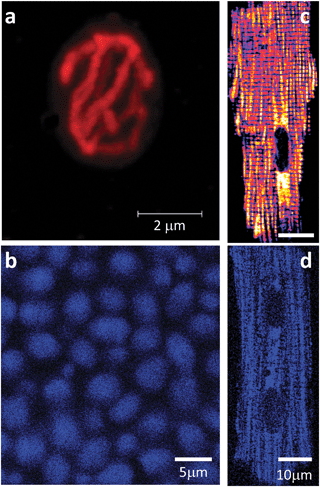 | ||
| Fig. 4 Mitochondria in yeast and cardiomyocytes. (a,b) Confocal microscopy of a Saccharomyces cerevisiaecell from an aerobically-grown culture loaded in (a) with tetramethyl rhodamine ethyl ester (TMRE). (b) autofluorescence (NAD(P)H) during perfusion of untreated tethered organisms with 5mM glucose in aerated Na phosphate-buffered saline. (c,d) Two photon laser scanning fluorescence microscopy of a freshly isolated cardiomyocyte loaded with (c) TMRE or after (d) autofluorescence. Notice the reticulate or lattice organization of the mitochondrial network in yeast and heart, respectively. | ||
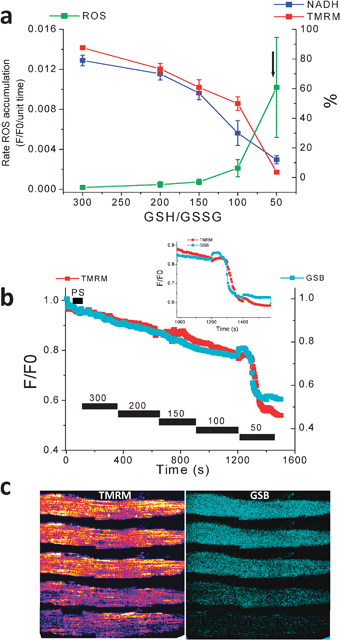 | ||
Fig. 5 ROS production, and ΔΨm, NADH, and GSH status as a function of GSH![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) GSSG ratio in saponin-permeabilized cardiomyocytes. Myocytes were resuspended and loaded with TMRM (100 nM) and CM-H2DCFDA (2 μM) for at least 20 min. After loading, the excess dye was washed out, and the cells were briefly superfused with a permeabilizing solution and then continuously perfused with an intracellular solution containing different GSH/GSSG ratios as indicated. ΔΨm, oxidation of the ROS probe, and NADH redox state were simultaneously monitored using two-photon fluorescence excitation. (a) rate of oxidation of the ROS probe (F/F0/unit time), NADH (in % of initial fluorescence before permeabilization), and tetramethyl rhodamine methyl ester, TMRM (ΔΨm in % of initial fluorescence before permeabilization) obtained at different GSH/GSSG ratios (3 mM GSH concentration). The arrow in (a) denotes the point at which ΔΨm irreversibly collapsed. (b) Mitochondrial GSH redox status and ΔΨm level in response to decreasing the GSH/GSSG ratio in saponin-permeabilized cardiomyocytes; timing of the depletion of reduced glutathione levels and ΔΨm collapse. Inset, note that the rapid oxidation of the mitochondrial GSB signal occurs ∼30 s before the irreversible collapse of ΔΨm. (c) Montage of a permeabilized cardiomyocyte loaded with the ΔΨm and GSH sensors and sampled at the different GSH/GSSG ratios. Notice the oxidation of the GSH pool and mitochondrial membrane collapse (bottom row). (Modified from Aon, Cortassa, Maack, O'Rourke (2007) J. Biol. Chem.28, 21889–21900). GSSG ratio in saponin-permeabilized cardiomyocytes. Myocytes were resuspended and loaded with TMRM (100 nM) and CM-H2DCFDA (2 μM) for at least 20 min. After loading, the excess dye was washed out, and the cells were briefly superfused with a permeabilizing solution and then continuously perfused with an intracellular solution containing different GSH/GSSG ratios as indicated. ΔΨm, oxidation of the ROS probe, and NADH redox state were simultaneously monitored using two-photon fluorescence excitation. (a) rate of oxidation of the ROS probe (F/F0/unit time), NADH (in % of initial fluorescence before permeabilization), and tetramethyl rhodamine methyl ester, TMRM (ΔΨm in % of initial fluorescence before permeabilization) obtained at different GSH/GSSG ratios (3 mM GSH concentration). The arrow in (a) denotes the point at which ΔΨm irreversibly collapsed. (b) Mitochondrial GSH redox status and ΔΨm level in response to decreasing the GSH/GSSG ratio in saponin-permeabilized cardiomyocytes; timing of the depletion of reduced glutathione levels and ΔΨm collapse. Inset, note that the rapid oxidation of the mitochondrial GSB signal occurs ∼30 s before the irreversible collapse of ΔΨm. (c) Montage of a permeabilized cardiomyocyte loaded with the ΔΨm and GSH sensors and sampled at the different GSH/GSSG ratios. Notice the oxidation of the GSH pool and mitochondrial membrane collapse (bottom row). (Modified from Aon, Cortassa, Maack, O'Rourke (2007) J. Biol. Chem.28, 21889–21900). | ||
A single focused laser flash produced similarly rapid processes within both heart cells and a population of yeast organisms.12,29 A fast localized oxidation of nicotinamide nucleotides and depolarisation of ΔΨm was followed by a rapid migration of both processes as a wave spreading throughout the mitochondrial network in cardiomyocyte and across the epicardium,12,13,18,30,31 and also between mitochondria within and between yeast cells.29 Depletion of cellular GSH and accumulation of ROS closely follow the trigger of the laser flash: all the observed changes follow oscillatory kinetics in the minute domain.
Temporal scales from milliseconds to minutes and hours
Complementary evidence for the omnipresence of oscillatory behaviour comes from studies of the changing respiratory rates of single-cell eukaryotes (yeasts and protists). Almost 40 years ago the respiratory oscillations of a yeast (Schizosaccharomyces pombe)32 and amoeba (Acanthamoeba castellanii)33 as well as 7 other species in synchronous cultures34 were in 1982 eventually assigned with a timekeeping function. The spontaneous self-synchronization of S. cerevisiae in continuous culture provided a more accessible system for biochemical analyses, especially when utilizing the non-invasive potential of membrane inlet mass spectrometry for rapid and continuous monitoring of gases (O2, CO2 and H2S) dissolved in the culture. Automated sampling every 15 s over periods of up to several months, combined with a fast instrument response time gives much higher quality data than that available using an O2electrode.35 Conspicuous periods of 12 h, 40 min and 4 min were revealed (Fig. 6). An explanation for this multioscillatory phenomenon can be deduced from the reactions shown in Fig. 3, but only when changes in mitochondrial activities as energetic and physiological states36,37 are taken into consideration. In isolated mitochondriain vitro, provision of respiratory substrates alone does not produce maximum respiratory rates (i.e. electron transport kinetics). This is because coupled oxidative phosphorylation cannot proceed when ADP (+Pi) is limiting; addition of the nucleotide releases this inhibitory effect and produces an almost instantaneous acceleration of oxygen consumption. This phenomenon of respiratory (or adenine nucleotide) control is responsible for the changing rates of dissolved O2 remaining in the culture medium after the yeasts utilize what they need for their energetic requirements, for biosynthesis of all cellular components during their growth, and for replication by budding. So why should the rapid switching of respiratory states in isolated mitochondria not be the predominant phenomenon in the whole organisms of the culture? Evidently under most circumstances it is the supply of cytoplasmic regenerated ADP that limits mitochondrial dynamics in vivo, and the mitochondria are enslaved to slower reactions of biosynthesis for the recycling of the adenine nucleotide pools. We have suggested that when acting as players on the bigger stage within the whole cell these organelles are dancing to a slower tune played by an orchestra elsewhere.1 The 40 min cycle consists of a complex series of processes that encompasses the entire cellular network, not only of transport and metabolic steps (more than 1200 identified metabolites38 and in reality many times that number), but also a veritable host of transcriptional regulators39 translational controls at the ribosomal machinery, post-transcriptional modifiers, DNA replication and the complexities of the chromosome cycle.39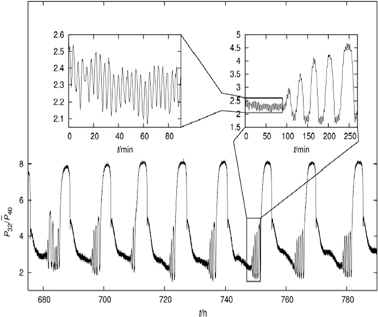 | ||
| Fig. 6 Multi-oscillatory behavior in a self-organized (synchronous) continuous culture of S. cerevisiae. Relative membrane-inlet mass spectrometry (MIMS) signals of the m/z = 32 component corresponding to dissolved O2versus time. Time is given in hours after the start of fermentor continuous operation. Periods of ∼13 h, ∼40 min and ∼4 min can be detected (see the sub-panel). The biological bases for all three oscillatory outputs of the yeast culture have been confirmed by exclusion of the possible influences of variations of aeration or stirring, pulsed medium addition, cycles of NaOH addition and pH variation, or cycles of temperature control. (Reproduced from Aon MA, Roussel MR, Cortassa S, O'Rourke B, Murray DB, et al. (2008) The Scale-Free Dynamics of Eukaryotic Cells. PLoS ONE 3(11): e3624. doi: 10.1371/journal.pone.0003624). | ||
The 40 min period, the synchronisation time-base for the coherent operation of all the parallel processing necessary, is the output of the ultradian clock, a temperature-compensated redox oscillator.40 A spike of H2S produced by sulphite reductase once per 40 min cycle, alongside acetaldehyde and probably other synchronizers, is essential for the auto-dynamic coordination of the yeast population.41 A strange attractor indicating chaotic behaviour is revealed in plots of the concentrations of the three gases (O2, CO2 and H2S).35,42 Deterministic chaos is characteristic of adaptable and robust systems showing very complex behaviour, predictable only in the short-term, but underpinned by a set of simple non-linear kinetic equations.43,44
However, close inspection of the apparently ‘noisy’ output indicates that the highly responsive mass spectrometric monitoring (t1/2 = 2 s) also reveals an oscillator with a 4 min period: this presumably corresponds to the ROS-driven mitochondrial oscillator observed in cardiomyocytes.45 In fact, the presence of the short period oscillations in yeast35 represents evidence of the common redox core shared with cardiomyocytes (Fig. 3). The ability of 4-chlorodiazepam, a mitochondrial benzodiazepine receptor antagonist, to block the oscillations in the minute time domain both in yeast and heart cells14 argues in favor of a common set of redox processes as the mechanistic base of the rhythmicity observed in both systems (Fig. 3), at least in the minute time scale. The long period (12 h) may correspond to a sub-harmonic of the circadian oscillator as the experiments were conducted under natural day-night conditions.
Periodic behaviour was also revealed in the mitochondrial network of cardiac muscle cells at the most fundamental level of their spatio-temporal energetic organization. The ROS-dependent mitochondrial oscillator described in cardiomyocytes exhibits at least two modes of function under physiological conditions or in response to metabolic and oxidative stress. Both modes depend upon network behaviour of mitochondria. In their normal physiological states these organelles behave as a highly integrated network of coupled oscillators (Fig. 7a) with a broad range of frequencies.46 ROS normally weakly couples mitochondria but becomes a strong coupling messenger when under oxidative stress;12,47 the mitochondrial network attains criticality when a threshold level is attained. Then a certain density of mitochondria forms a cluster that spans the whole volume of the cell.13 Under these conditions, the slightest perturbation triggers a cell-wide collapse of ΔΨm, visualized as a wave of depolarization travelling through the cell. There then follows whole-cell synchronized oscillations in NADH, ROS and GSH. This behaviour scales from the mitochondria to the cell by driving cell excitability and to the whole heart in which potentially catastrophic arrhythmias rapidly ensue.17,30,31,48–50 Next-amplitude plots reveal the sharp transition to the patho-physiological state in which an apparently noisy regime gives way to a slow oscillatory pathophysiology in which a high-amplitude period of about 100 s is dominant (Fig. 7b).
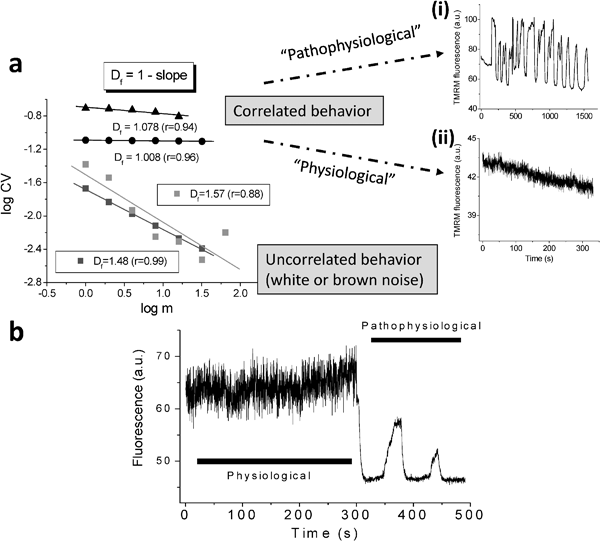 | ||
| Fig. 7 Physiological and pathophysiological behavior of the network of coupled mitochondrial oscillators in cardiac cells. (a) Temporally correlated behavior of the mitochondrial network in cardiac cells. The statistical analysis of the TMRM signal showed that the mitochondrial network of the heart cell functions as a highly correlated network of oscillators. Shown in left panel are the results obtained by Relative Dispersional Analysis (RDA) as a log-log plot of the CV (=SD/mean) of the fluorescence distribution obtained at increasing values of the aggregation parameter, m. This gives a fractal dimension, Df, close to 1.0, either for myocytes showing large (“pathophysiological”) oscillations in ΔΨm (top right panel A) or those under “physiological” conditions (bottom right panel). A completely random process gives Df ∼ 1.5. (b) Before the mitochondrial network reaches criticality, the ΔΨm oscillates at high frequencies and small amplitudes. After criticality, the network behavior evolves into “pathophysiological” behavior characterized by low-frequency, high amplitude oscillations. | ||
Both yeast and cardiomyocyte function on several temporal scales simultaneously; their multi-oscillatory behaviour became evident when investigated by power spectral analysis. Relative dispersional analysis reveals that both exhibit self-similar (fractal) features characteristic of scale-free systems.14,51 These systems have a common feature in that they show a long-term memory in a time series and on all scales share statistically similar characteristics in their frequency-amplitude relationships. Thus these fluctuations are not random excursions but correlated oscillations. Power spectral analysis further confirms these features: the inverse power relationship between amplitude and frequency shows that β (the spectral exponent) has values of 1.8 for cardiomyocytes ΔΨm and 1.95 (i.e. close to that for “colored noise” expected for a chaotic system,52,53 for a similar analysis of yeast respiratory oscillations. Were these time series randomly noisy (“white noise”), β would be 0. These relationships hold over three orders of magnitude (12 h–4 min for yeast and 100 s–220 ms for cardiac muscle cells).14,51,54
The dynamic balance between ROS production and ROS scavenging is one of the underlying basic mechanisms at the origin of oscillations in the redox core (Fig. 3), according to mathematical modelling of the mitochondrial oscillator. Basic network motifs participating in the regulation of this dynamic balance are essentially positive and negative feedbacks. More specifically, the positive feedback is given by mitochondrial respiration and the percentage of the respiratory flux diverted to the free radical superoxide anion, O2˙−; when the latter attains a certain threshold in the matrix facilitates the opening of an inner membrane anion channel (IMAC) which depolarizes the ΔΨm. In order to regenerate the ΔΨm, a negative feedback driven by Cu, Zn SOD on O2˙− closes the IMAC allowing the repolarization of the inner membrane. The computational model of the mitochondrial oscillator is able to produce oscillatory periods ranging from msec to several hours.
Key consequences of disturbed oscillatory behaviour
The pivotal role of mitochondria in the early events leading to necrotic or programmed cell death has been extensively documented.10,11 In cardiomyocytes (Fig. 8) and in the yeast Candida albicans10 (Fig. 9) the importance of the maintenance of cellular redox states can be easily demonstrated. A constituent of garlic (Allium sativum) diallyl disuphide (DADS) at relatively high concentrations (1 mM) when presented to cardiomyocytes leads to accumulation of ROS, depletion of glutathione, NAD(P)H oxidation, depolarization of ΔΨm, and to cell death in a few minutes (Fig. 8 and 10).10,15Dithiothreitol (DTT, a powerful thiol reductant) on the contrary elevates ΔΨm and cellular thiols, whilst preserving cell viability (Fig. 8).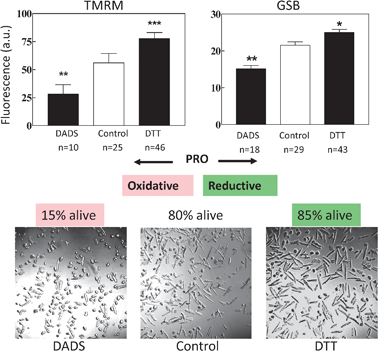 | ||
| Fig. 8 Effect on cell viability and ΔΨm (TMRM) and GSH (GSB) pool of a pro-reductive or a pro-oxidative shift in redox status. (a, b) Cardiomyocytes in the absence (control) or in the presence of 2 mM dithiothreitol (DTT) or 1 mM diallyldisulphide (DADS) were preincubated for 2 h and examined by two-photon microscopy. The pro-oxidative shift in the intracellular thiol pool significantly decreases both the levels of intracellular GSH and ΔΨm with respect to the control (a); in contrast, both variables are increased in the presence of DTT (b). Eighty five percent of cardiomyocytes became non-viable in the presence of DADS (c, left panel) whereas DTT protected them to even slightly, but significantly, higher (85%) values (c, right panel) than the control (80%; c, mid panel) (n = 400; 2 experiments). | ||
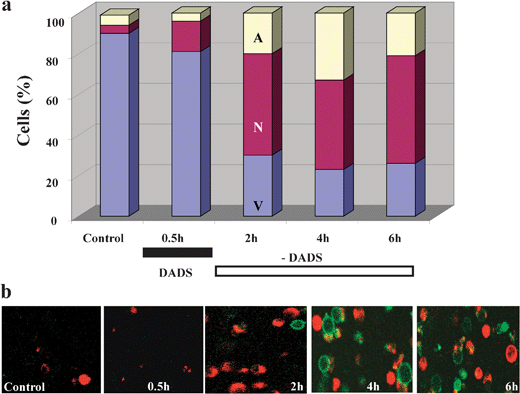 | ||
| Fig. 9 Kinetics of cell death induction in Candida albicans by a short treatment with DADS. (a) Culture-grown C. albicanscells were exposed to 0.5 mM DADS for 30 min at room temperature, while untreated cells remained as control. Cell death was monitored after DADS removal by sampling at the times specified, in which the cells were washed and subjected to protoplasting. Cell death by apoptosis and necrosis was monitored with Alexa Fluor 488 annexin V and propidium iodide on coverslips coated with polylysine, in a thermostated chamber at 30 °C, on the stage of a Nikon E600FN upright microscope. (b) Images of green and red fluorescence were recorded by two photon microscopy. Representative snapshots of the kinetics of appearance of necrotic (red) or apoptotic (green) cells are shown. Key to symbols: A, apoptotic; N, necrotic; V, viable; DADS, diallyl disulphide. (Reproduced from Lemar, Aon, Cortassa, O'Rourke, Muller and Lloyd (2007) Yeast 24, 695–706). | ||
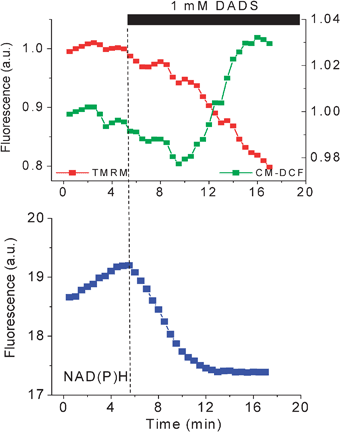 | ||
| Fig. 10 DADS-triggered oxidative stress, ΔΨm depolarization, and cell death. Freshly isolated guinea pig cardiomyocytes were loaded with TMRM and the ROS probe CM-H2DCFDA at 37 °C and imaged by two-photon laser scanning fluorescence microscopy in a perfusion chamber. After exposure to the thiol-oxidizing agent DADS (1 mM), the oxidation of the ROS probe steadily increased after oxidation of the NAD(P)H pool triggering ΔΨm depolarization and sudden cell death. | ||
The apoptotic/necrotic effects of DADS on C.utilis (Fig. 9) can be quantified by the propidium iodide/annexin-fluorescein antibody procedure whereby the permeabilization of plasma membranes is indicated by red fluorescence and the externalization of serine-phospholipid gives a green fluorescence. These processes are measured by the changing distributions of populations of the dying yeasts (Fig. 9a).
In isolated cardiomyocytes perfusion with 1 mM diamide, a powerful glutathione oxidant,15,55 leads to an oscillatory mode that presages oxidation of the autofluorescent nicotinamide nucleotides and collapse of ΔΨm during progressive ROS accumulation.15 By 20–30 min to 2 h cell death was evident (Fig. 5, 8–10). These effects can be clearly demonstrated by poising the redox state of the GSH pool in a series of successively increased oxidation states (Fig. 5). In the perfused guinea pig heart, 30 min exposure to this thiol oxidant produces similar results and the whole organ progresses to an arrhythmic state17 (Fig. 11).
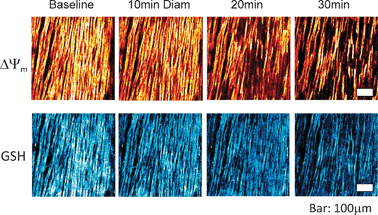 | ||
| Fig. 11 Imaging of ΔΨm (top) and GSB (bottom) in intact guinea pig hearts using 2-photon microscopy after exposure to diamide. Hearts were perfused under normoxic conditions in the presence of 200 μM diamide. Mitochondria remained polarized until approximately 20 min of exposure, at which point Ψm, began to heterogeneously depolarize (top row), and GSB (the adduct marker of GSH, bottom row) became markedly oxidized in parallel with the onset of arrhythmias. Bar is equal to 100 μm. (Modified from Brown, Aon, Frasier, Sloan, Maloney, Anderson, O'Rourke (2010) J. Mol. Cell. Cardiol. 48, 673–679). | ||
Applications of redox state monitoring, pioneered by Britton Chance, have been perfected over 50 years,56 culminating in the in vivo fluorimetric assessment of the effects of cyclosporine A on mitochondrial function during protection against infarction in myocardial ischemia and reperfusion.57
Conclusions and future work: conserved rhythmic functions in yeasts and cardiomyocytes
The core functions described here can be used as early prognostic indicators of patho-physiology.57 Further electronic and optical advances will ensure engineering of portable devices for continuous ambulatory monitoring in health care whilst new translational developments in clinical medicine will take full advantage of the benefits that ensue.Minimally invasive continuous monitoring of mitochondrial functions in yeasts (by membrane inlet mass spectrometry), and in cardiomyocytes by two-photon microscopy, reveal that these two evolutionary-distant systems exhibit strikingly similar oscillatory features. The evidence suggests conservation of an indispensable core metabolic function in the form of intrinsically rhythmic organization involving redox balance. Continuous oscillatory states are observable on multiple timescales, an obligatory requirement for the synchronization and coordination of reactions, events and processes of the coherently-functioning cellular network. Beyond the necessary boundary conditions for adaptable and robust performance, various stress factors (physical, chemical) perturb oscillatory behaviour to signal and initiate the pathways to cell death. Notwithstanding, whether the intrinsic oscillatory dynamics emerging from the redox core exert timekeeping functions that synchronize the spatio-temporal occurrence of processes happening in, e.g. tissue, organ, is still an open theoretical and experimental question. These processes include metabolic-, information-, and signalling-network dynamics which in turn regulate and control essential functions involved in cell growth, division, and differentiation, within tissues, organs, and organisms. This represents a wide research avenue into the future.
Acknowledgements
This work was supported by NIH grants, R21HL106054 (SC), P01HL081427 and R37HL54598 (BOR), and R01-HL091923-01 (MAA).References
- D. Lloyd and D. B. Murray, Trends Biochem. Sci., 2005, 30(7), 373–377 Search PubMed
.
- D. Lloyd, M. A. Aon and S. Cortassa, ScientificWorldJournal, 2001, 1, 133–145 Search PubMed
.
- F. E. Yates, Methods Enzymol., 1992, 210, 636–675 Search PubMed
.
- D. Lloyd and D. B. Murray, FEBS Lett., 2006, 580(12), 2830–2835 Search PubMed
.
- D. Lloyd and D. B. Murray, BioEssays, 2007, 29(5), 465–473 CrossRef CAS
.
- O. H. Petersen and Y. Maruyama, Nature, 1984, 307(5953), 693–696 Search PubMed
.
- D. Lloyd, S. W. Edwards and J. C. Fry, Proc. Natl. Acad. Sci. U. S. A., 1982, 79(12), 3785–3788 Search PubMed
.
- B. M. Sweeney and J. W. Hastings, Cold Spring Harbor symposia on quantitative biology, 1960, 25, 87–104 Search PubMed
.
-
B. C. Goodwin, Temporal Organization in Cells. A Dynamic Theory of Cellular Control Processes, Academic Press, London, 1963 Search PubMed
.
- K. M. Lemar, M. A. Aon, S. Cortassa, B. O'Rourke, C. T. Muller and D. Lloyd, Yeast, 2007, 24(8), 695–706 CrossRef CAS
.
- K. M. Lemar, O. Passa, M. A. Aon, S. Cortassa, C. T. Muller, S. Plummer, B. O'Rourke and D. Lloyd, Microbiology, 2005, 151(Pt 10), 3257–3265 Search PubMed
.
- M. A. Aon, S. Cortassa, E. Marban and B. O'Rourke, J. Biol. Chem., 2003, 278(45), 44735–44744 CrossRef CAS
.
- M. A. Aon, S. Cortassa and B. O'Rourke, Proc. Natl. Acad. Sci. U. S. A., 2004, 101(13), 4447–4452 Search PubMed
.
- M. A. Aon, M. R. Roussel, S. Cortassa, B. O'Rourke, D. B. Murray, M. Beckmann and D. Lloyd, PLoS One, 2008b, 3(11), e3624 Search PubMed
.
- M. A. Aon, S. Cortassa, C. Maack and B. O'Rourke, J. Biol. Chem., 2007a, 282(30), 21889–21900 Search PubMed
.
- D. Lloyd, L. Eshantha, J. Salgado, M. P. Turner and D. B. Murray, FEBS Lett., 2002b, 519(1–3), 41–44 Search PubMed
.
- D. A. Brown, M. A. Aon, C. R. Frasier, R. C. Sloan, A. H. Maloney, E. J. Anderson and B. O'Rourke, J. Mol. Cell. Cardiol., 2010a, 48(4), 673–679 Search PubMed
.
- M. K. Slodzinski, M. A. Aon and B. O'Rourke, J. Mol. Cell. Cardiol., 2008, 45, 650–660 Search PubMed
.
- D. B. Murray, F. A. Engelen, M. Keulers, H. Kuriyama and D. Lloyd, FEBS Lett., 1998, 431(2), 297–299 Search PubMed
.
- D. P. Jones, Methods Enzymol., 2002, 348, 93–112 Search PubMed
.
- L. H. Lash, Chem.-Biol. Interact., 2006, 163(1–2), 54–67 Search PubMed
.
- A. Meister and M. E. Anderson, Annu. Rev. Biochem., 1983, 52, 711–760 CrossRef CAS
.
- D. B. Murray, F. Engelen, D. Lloyd and H. Kuriyama, Microbiology (Reading, England), 1999, 145(Pt 10), 2739–2745 Search PubMed
.
- L. Rapkine, Ann. Physiol. Physicochem., 1931, 7, 382–418 Search PubMed
.
-
D. Lloyd, The Mitochondria of Microorganisms, Academic Press, London, 1974 Search PubMed
.
- N. Beraud, S. Pelloux, Y. Usson, A. V. Kuznetsov, X. Ronot, Y. Tourneur and V. Saks, J. Bioenerg. Biomembr., 2009, 41(2), 195–214 Search PubMed
.
- A. V. Kuznetsov, M. Hermann, V. Saks, P. Hengster and R. Margreiter, Int. J. Biochem. Cell Biol., 2009, 41(10), 1928–1939 Search PubMed
.
- Y. Yaniv, M. Juhaszova, S. Wang, K. W. Fishbein, D. B. Zorov and S. J. Sollott, PLoS One, 2011, 6(7), e21985 Search PubMed
.
- M. A. Aon, S. Cortassa, K. M. Lemar, A. J. Hayes and D. Lloyd, FEBS Lett., 2007b, 581(1), 8–14 Search PubMed
.
- F. G. Akar, M. A. Aon, G. F. Tomaselli and B. O'Rourke, J. Clin. Invest., 2005, 115(12), 3527–3535 Search PubMed
.
- M. A. Aon, S. Cortassa, F. G. Akar, D. A. Brown, L. Zhou and B. O'Rourke, Int. J. Biochem. Cell Biol., 2009, 41(10), 1940–1948 Search PubMed
.
- R. K. Poole and D. Lloyd, The Biochemical Journal, 1973, 136(1), 195–207 Search PubMed
.
- S. W. Edwards and D. Lloyd, FEBS Lett., 1980, 109(1), 21–26 Search PubMed
.
-
D. Lloyd, in Ultradian Rhythms in Life Processes, ed. D. Lloyd and E. L. Rossi, Springer-Verlag, 1992, pp. 5–22 Search PubMed
.
- M. R. Roussel and D. Lloyd, FEBS J., 2007, 274(4), 1011–1018 Search PubMed
.
- B. Chance and G. R. Williams, The Journal of Biological Chemistry, 1956, 17, 65–134 Search PubMed
.
- D. Lloyd, K. M. Lemar, L. E. Salgado, T. M. Gould and D. B. Murray, FEMS Yeast Res., 2003, 3(4), 333–339 Search PubMed
.
- D. B. Murray, M. Beckmann and H. Kitano, Proc. Natl. Acad. Sci. U. S. A., 2007, 104(7), 2241–2246 CrossRef CAS
.
- R. R. Klevecz, J. Bolen, G. Forrest and D. B. Murray, Proc. Natl. Acad. Sci. U. S. A., 2004, 101(5), 1200–1205 Search PubMed
.
- D. B. Murray, S. Roller, H. Kuriyama and D. Lloyd, J. Bacteriol., 2001, 183(24), 7253–7259 Search PubMed
.
- H. Y. Sohn and H. Kuriyama, Yeast, 2001, 18, 125–135 Search PubMed
.
- D. B. Murray and D. Lloyd, BioSystems, 2007, 90, 287–294 Search PubMed
.
- A. L. Lloyd and D. Lloyd, BioSystems, 1993, 29(2–3), 77–85 Search PubMed
.
- A. L. Lloyd and D. Lloyd, Biol. Rhythm Res., 1995, 26, 233–252 Search PubMed
.
- S. Cortassa, M. A. Aon, R. L. Winslow and B. O'Rourke, Biophys. J., 2004, 87(3), 2060–2073 Search PubMed
.
- M. A. Aon, S. Cortassa and B. O'Rourke, Biophys. J., 2006a, 91(11), 4317–4327 Search PubMed
.
- D. B. Zorov, C. R. Filburn, L. O. Klotz, J. L. Zweier and S. J. Sollott, J. Exp. Med., 2000, 192(7), 1001–1014 Search PubMed
.
-
M. A. Aon and S. Cortassa, in Encyclopedia of Complexity and Systems Science, ed. R. Meyers, Springer, New York, 2009 Search PubMed
.
- M. A. Aon, S. Cortassa, F. G. Akar and B. O'Rourke, Biochim. Biophys. Acta, Mol. Basis Dis., 2006b, 1762(2), 232–240 Search PubMed
.
-
M. A. Aon, S. Cortassa and D. Lloyd, in Encyclopedia of Molecular Cell Biology and Molecular Medicine, ed. R. Meyers, Wiley, 2011 Search PubMed
.
- M. A. Aon, S. Cortassa and B. O'Rourke, Adv. Exp. Med. Biol., 2008a, 641, 98–117 Search PubMed
.
-
J. B. Bassingthwaighte, L. S. Liebovitch and B. J. West, Fractal Physiology, Oxford University Press for the American Physiological Society, New York, 1994 Search PubMed
.
-
B. J. West, Physiology, promiscuity and prophecy at The Millennium: A Tale of Tails, World Scientific, Singapore, 1999 Search PubMed
.
-
M. A. Aon, S. Cortassa and B. O'Rourke, in Molecular System Bioenergetics: Energy for Life, ed. V. Saks, Wiley-VCH, 2007, pp. 111–135 Search PubMed
.
- N. S. Kosower and E. M. Kosower, Methods Enzymol., 1995, 251, 123–133 CAS
.
- M. Matsubara, M. Ranji, B. G. Leshnower, M. Noma, S. J. Ratcliffe, B. Chance, R. C. Gorman and J. H. Gorman 3rd, Ann. Thorac. Surg., 2010, 89(5), 1532–1537 Search PubMed
.
- H. N. Xu, S. Nioka, B. Chance and L. Z. Li, Adv. Exp. Med. Biol., 2011, 915, 207–213 Search PubMed
.
Footnote |
| † Dedication: This contribution is dedicated to the memory of Britton Chance, who passed away on November 18th 2010, a mentor and life-long friend of DL, whose many achievements can be viewed at: http://www.med.upenn.edu/biocbiop/chance/reflections.html |
| This journal is © The Royal Society of Chemistry 2012 |
