Bio-inspired materials for parsing matrix physicochemical control of cell migration: A Review
Hyung-Do
Kim
a and
Shelly R.
Peyton
b
aDepartment of Biological Engineering, Massachusetts Institute of Technology, Cambridge, MA
bDepartment of Chemical Engineering, University of Massachusetts, Amherst, MA
First published on 25th October 2011
Abstract
Cell motility is ubiquitous in both normal and pathophysiological processes. It is a complex biophysical response elicited via the integration of diverse extracellular physicochemical cues. The extracellular matrix directs cell motilityvia gradients in morphogens (a.k.a. chemotaxis), adhesive proteins (haptotaxis), and stiffness (durotaxis). Three-dimensional geometrical and proteolytic cues also constitute key regulators of motility. Therefore, cells process a variety of physicochemical signals simultaneously, while making informed decisions about migration viaintracellular processing. Over the last few decades, bioengineers have created and refined natural and synthetic in vitro platforms in an attempt to isolate these extracellular cues and tease out how cells are able to translate this complex array of dynamic biochemical and biophysical features into functional motility. Here, we review how biomaterials have played a key role in the development of these types of model systems, and how recent advances in engineered materials have significantly contributed to our current understanding of the mechanisms of cell migration.
Insight, innovation, integrationIn this review article, we discuss how biomaterials have attempted to recapitulate the complexity of the extracellular matrix in order to mechanistically describe its regulation of cell motility. We show precedent of both bioscientists and engineers in developing materials-based technologies to elucidate the mechanisms of how matrix chemical, mechanical, and topographical features regulate cell motilityin vitro. We review and cite literature at the forefront of emerging technologies, including microfluidics, nanotechnology, and other engineered microenvironments tailored to study complex cell-matrix interactions. The cell biology community has enriched our understanding of the molecular components and their interdependencies involved in generating motility. We urge the biomaterials community to assess the vast space of unanswered, biologically relevant questions that will only be answered with elegant synthetic techniques. In turn, the cell biologists can appreciate the immense opportunities provided by thoughtfully designed materials, wherein carefully controlled microenvironments enable reproducible studies of complex mechanisms. |
Introduction
Human physiology, pathophysiology, and regenerative medicine are each built on the careful orchestration of cellular motile machinery. Embryonic morphogenesis relies on migratory events during gastrulation and neural crest development.1–3 In vertebrate adults, cell motility is a critical part of wound healing and tissue repair.4 Collective motility of epithelial cells constantly renews skin and intestinal tissue. Finally, immune surveillance would be impossible without the unique migratory ability of lymphocytes.5,6 Dysregulation of migratory processes results in disastrous consequences, such as vascular disease, osteoporosis, chronic inflammatory diseases, multiple sclerosis, and even mental retardation. Aberrant cell migration in cancer (metastasis), is the leading cause of cancer-related deaths.7Therefore, understanding cell migration's role in healthy tissue, disease progression, and for tissue engineering, has been of great interest to the scientific community. The elucidation of the signaling pathways underlying the regulation of motility has led to the identification of intracellular components that robustly respond to cues from the microenvironment, such as growth factors, cytokines, and physical cues from the extracellular matrix. Cell migration is a cycle of biophysical processes that are spatio-temporally regulated.8,9 The cycle is initiated by the cell's extension of protrusions, which requires polymerization of actin microfilaments, aided by actin-binding proteins, such as Arp2/3, profilin, cofilin, and Ena/VASP proteins.10 Protrusions at the leading edge of a cell (wide lamellipodia or finger-like filopodia) form stable adhesions to the ECM ligands via transmembrane proteins called integrins.11,12 Protrusions and adhesions, often in conjunction with extracellular soluble gradients, establish an intracellular polarity, and activate a myriad of known and unknown signaling proteins13 including the Rho-family GTPases.14–17 Upon adhesion, active GTPases initiate a cascade of events, and myosin II generates forces by pulling on the actin microfilament network to translocate the polarized cell body.18 The carefully regulated, spatio-temporally controlled activation of this signaling network also leads to the disassembly of adhesions at the trailing edge of the cell, and a net contractile force at the leading edge, allowing the cell to migrate.19 Depending on the cell type, the orchestration of these processes can vary widely. The components of the cell migration cycle: protrusion, attachment, and contraction, can be observed as separate processes in mesenchymal-like cells, such as fibroblasts, whereas in amoeboid-like cells, such as leukocytes, different mechanisms of migration have been proposed,20,21 and individual processes may be less easily discernable.
The resulting cell movement must occur in balance with the physical properties of the extracellular microenvironment. The ECM consists of various filamentous, amorphous, and cross-linking proteins, such as collagens, laminins, fibronectins, glycosaminoglycans, etc., and provides both a physical support and barrier for cell migration.22 Many cells, therefore, have a framework for remodeling the ECM—from cleavage of ECM proteins through various secreted proteases to secretion of ECM proteins—and rely heavily on this framework for productive locomotion.23 In contrast, recent work indicates that leukocytes do not require ECM modifying abilities and employ a proteolysis-independent mechanism to enable fast movement without destruction to the tissue.20,24
The diversity of biophysical processes in cell migration has provided a great opportunity for the field of biomaterials to pose biologically relevant questions via the creative design of well-defined microenvironments. To date, much of the mechanistic research in the field has been generated in a physiologically in appropriate context: tissue culture plastic. Tissues in the human body are not two-dimensional, and have a far more complex physical and chemical microenvironment that dictates cell behavior. Further, two-dimensional studies naturally neglect proteolysis-driven motility and tissue invasion.25 The biosciences community has increasingly sought more physiologically relevant systems, which permit the ability to quantitatively probe various aspects of cell migration in a diverse set of microenvironments (Fig. 1). Over the last twenty years, the biomaterials field has emerged out of a traditional landscape of inert material design into the engineering of bio-instructive, -specific, and -responsive tools (for a historical perspective on biomaterials, we direct you to ref. 26–29). Both tissue-derived and synthetic materials can be rendered biologically sensitive and directive with the incorporation of adhesive matrix factors, signal-initiating growth factors (and growth factor depots), and enzymatic recognition sites (for review see ref. 30). New technology is continually emerging to decipher how physicochemical cues from the extracellular matrix, such as chemical, nanotopographical, mechanical, and enzymatic cues, can feedback and regulate intracellular processes, aiding the study of how the microenvironment regulates cell migration. Cell migration studies using these engineered materials have provided insights into a phenomenon that compels a quantitative approach for establishing relationships between migratory parameters.
In this review, we examine the use of biomaterials in elucidating novel insights into the mechanisms of cell migration, and attempt to place these findings into a physicochemical perspective. We outline our appreciation for the contribution of the current biomaterials literature to the general biochemical and biophysical understanding of cell migration. We specifically focus on the use of biomaterials to control chemotaxis, matrix adhesivity and haptokinesis/-taxis, matrix stiffness and durotaxis, cell polarity, proteolysis, and on the ability of these materials to aid discovery of the key mechanistic parameters cells rely on to make decisions about cell migration in general. For literature describing cell migration relevant to specific physiology and pathophysiology, we refer the audience to other reviews.1–7,9,31
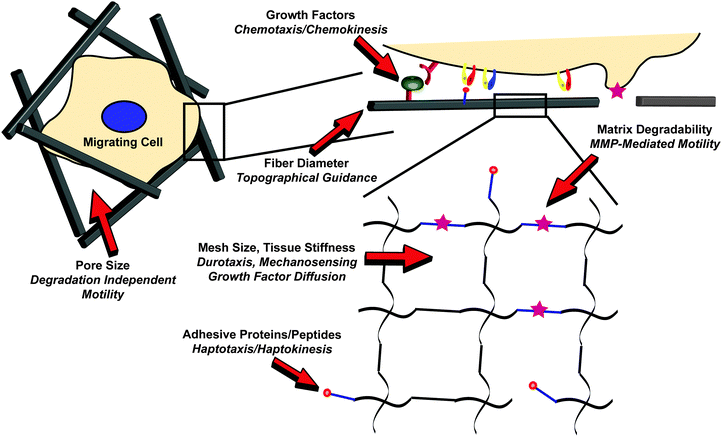 | ||
| Fig. 1 Cell migration processes in a three-dimensional extracellular matrix . In vivo, cell motility is governed by the coordination of multiple extracellular signals, including soluble growth factors, the presentation of insoluble adhesive proteins, and the stiffness and extent of crosslinking of the matrix, among others, across multiple length scales. Reviewed here, biomaterials present the unique opportunity to quantitatively parse each process for a better understanding of the migratory process in a complex microenvironment. Red arrows designate tunable parameters in an engineered biomaterial environment, with the resulting control over cell migration shown in italics. | ||
Chemotaxis
Chemotaxis is the directed migration of cells in response to a soluble chemical signal. Chemotaxis-driven migration is prevalent in many biological events, such as development, the immune response, and wound healing. Interstitial flows induce gradients of chemokines that direct cell migration in lymphatic angiogenesis.32–34 The release of soluble factors from leaky vessels and macrophages is common in inflammation, cardiovascular disease, and cancer, on the one hand initiating a needed immune response, and in the other, resulting in disastrous consequences, such as plaque hardening and metastasis. More recently, it was discovered that a mutual chemotactic gradient established between macrophages and carcinoma cells through paracrine release of EGF and CSF-1 enhances tumor metastasis.35,36In vitro assays to study chemotaxis have been under development for the past three decades, beginning with simple studies of cells migrating on glass surfaces in the presence of localized soluble factors. The Boyden migration chamber was the most notable early advancement in tools development to study cell invasion toward chemoattractive factors,37 where cell invasion is measured through an adhesive porous filter toward a chemoattractive factor. No matrix degradation is required for invasion, but pore sizes of the filter can be varied to assess the effect of steric barrier. The under-agarose assay was the first biomaterials development to add a 3D context to better mimic an in vivo environment for chemotactic studies.38 In this assay, chemoattractants are deposited into a small well within a 3D agarose gel, while a suspension of cells is placed nearby. Cells sense and migrate toward the diffusive gradient of soluble factors through the nanoporous hydrogel. This assay can recapitulate some of the physiological aspects of chemotactic responses important in inflammation. Though quite simple, relevant quantification of diffusion characteristics, as well as visualization of cell motility, can be achieved from this assay.39
Both the under-agarose and Boyden chamber are end-point assays, prohibiting assessment of individual cell kinetics. In addition, because these assays use a single sink of soluble factor, chemoattractants do not form stable gradients, as is often observed in physiological conditions. The development of microfluidic devices to create stable and reproducible gradients of chemical factors has produced chemotaxic microenvironments closer to physiological conditions than ever before achievable. Surprisingly, very basic microfluidic systems have been around and used for short time-point bacterial migration assays for many years.40 More recently, microfluidic platforms have evolved to include 3D gels, overlaid gradients of multiple factors, and separated chambers to analyze cell-cell communication and coordinated motility. Microfluidic platforms are now commercially available to study the migratory response of immune cells to antibodies, shear stresses, and inflammatory cytokines.41
Microfluidic platforms have made quantitative 3D models of real-time chemotaxis possible, both for adherent and suspension cells,42 opening the field for engineers to study complex chemotactic phenomena in biology.43,44 Using a 3D platform that separated cell-seeded and gel-only areas, Chung et al. observed cancer cell migration through 3D materials toward a stable gradient of VEGF,45 recapitulating invasion during metastasis in the presence of leaky vasculature. To investigate coordinated cell–cell communication, this device also allows for sophisticated co-cultures of endothelial cells, smooth muscle cells and cancer cells, as one would find in a well-vascularized tumor microenvironment. The authors observed that cancer cells promoted endothelial migration (to different degrees, depending on the cancer cell subtype), while the presence of smooth muscle cells inhibited endothelial motility. Others have used stable gradients in 3D to investigate metastasis,46,47 and immune responses48,49 as well.
Chemotactic motility in the presence of multiple growth factors is possible with ladder chamber microfluidics, which include multiple compartments, and generate stable diffusion in both 2D and 3D geometries in the absence of shear flow (a common component of other microfluidics platforms, which can influence cell behavior in an uncontrollable fashion).50 With this platform, researchers generated a stable gradient of IL-8, and directed the migration of neutrophils. By being able to tightly control shear forces and chemical gradients independently, these types of systems could lend powerful insight in the study of neutrophils responding to inflammatory cues, as well as cancer cell motility toward leaky vessels in physiologically relevant 3D microenvironments.
The studies discussed thus far have all included diffusion of soluble factors through a media solution or semi-porous 3D milieu. However, growth factors in vivo are commonly found covalently, ionically, or physically connected to the surrounding insoluble fibrous matrix. Wet surface chemistry has seen recent advancements, and has been used to create stable gradients of growth factors tethered to 2D surfaces. The biomaterials community has found chemical methods to tether growth factors to 3D systems without interfering with their bioactivity.51–53 The bulk of studies using growth factor-bound biomaterials has focused on cell differentiation for tissue engineering, with a few exceptions. Recent work by Stefonek-Puccinelli and Masters exploited surface chemistry techniques to overlay gradients of EGF and IGF-1 onto standard tissue culture plates.54 Subtle effects were seen when comparing the growth factors, but they observed a dramatic increase in overall kerotinocyte migration by covalently linking growth factors to the surface of the plates when comparing to the growth factor free control. In addition, they observed a biphasic relationship between cell migration and maximum EGF concentration, with an optimal migration at an intermediate concentration of EGF, which has also been described for chemotaxis with soluble growth factors.55Growth factors can also be encapsulated into degradable polymers for a gradient sustained over multiple weeks. Using this technique, dendritic cells have been coerced to migrate for hundreds of microns.56 In this study, polymer microspheres and cells were embedded in collagen gels, but this technology could be extrapolated into any 3D system, including synthetic biomaterials.
Biomaterials systems have enhanced the study of chemotaxis in a myriad of ways. Unlike glass and plastic substrata, biomaterials have large pore sizes (on the order of nanometres), which allow for diffusion of chemoattractants or flow rates of fluids. Pore sizes are also tunable by varying the crosslinking densities of the biomaterial. The addition of interstitial and vascular flows in controlled environments with microfluidics can be used to study combinatorial effects of chemical and mechanical factors that are physiologically relevant to lymph draining and tumor cell intravasation.57 Finally, chemical techniques to tether growth factors to biomaterials have allowed us to create environments that present soluble factors to cells in a way that growth factor receptors actually encounter them in vivo. Each of these biomaterial advances has allowed us to study cell-matrix interactions in vitro in a manner much more reminiscent of in vivo biology.
Cell adhesion and haptotaxis
In addition to soluble factors, cell motility is also regulated by insoluble adhesive matrix components. Adhesive protein-mediated motility is called haptokinesis, while directed motility by an adhesive gradient is called haptotaxis (Fig. 2). Adhesive domains in the ECM not only serve as a physical mechanism for cell adhesion, but also as complex biochemical regulators of a variety of cellular responses, including motility.58,59 The diversity of ECM composition in various tissues indicates that, like soluble chemokines, levels and distribution of Type I and IV Collagen, fibronectin, and laminins have a profound effect on cell migration responses.60 The dynamic assembly of focal adhesions, a cluster of proteins connecting integrins to the intracellular cytoskeleton, as observed in 2D and 3D cultures, plays an important role in cell migration.58,61,62 Therefore, many efforts have been directed towards quantifying the contribution of adhesions to the overall migratory behavior by controlling the microenvironment in which adhesions are formed.63,64Nearly fifteen years ago, CHO cells and smooth muscle cells were shown to have a biphasic cell migration speed dependence on the concentration of insoluble adhesive proteins passively adsorbed on glass slides.65,66 Simple control of ECM ligand density was achieved via adsorption coating of glass slides with purified natural ECM proteins, ranging from fibronectin, collagen, laminin, or fibrinogen. The observed biphasic dependence could be further tuned by knocking down integrin expression targeting the ECM proteins. Ten years later, in 3D microenvironments, modulation of PtK1 epithelial cell migration under dose-dependent inhibition of myosin67 was consistent with an ECM ligand density-dependent increase in fibroblast cell speed on fibronectin when stimulated with EGF.68 There also existed an optimal fibronectin concentration, at which EGF stimulation resulted in the greatest increase in cell speed. Thus, EGF stimulation, through its activation of various contractility pathways, including ROCK,69 could provide a compensating increase in cell contractility, but could only demonstrate optimal compensation at a particular fibronectin concentration. At very low concentrations of fibronectin, EGF stimulation decreased cell speed, indicating that the balance-disrupting increase in contractility could result in a reduction in motility.68 These studies demonstrated that systematic variation of physicochemical cues is vital to deconvolve the complex biophysical processes involved in cell migration.
The conjugation of adhesion sites into biomaterials has been of great interest to the community in order to optimize cell colonization.64 A variety of polymers have been conjugated with either natural proteins, the IKVAV peptide present in laminin,70 or, most prominently, the RGD peptide, which is present in many ECM proteins, and recognized by multiple integrin subtypes.71 For example, in conjunction with sphingosine-1-phosphate (S1P), researchers at Washington University conjugated varying ratios of synthesized RGD (or cyclic RGD) and poly(ethylene glycol) (PEG)-vinyl sulfone hydrogel precursors.72 They confirmed previous reports that endothelial cell migration on RGD-containing hydrogel depended biphasically on RGD concentration. Importantly, this biphasic curve shifted in the presence of S1P, demonstrating the interplay between soluble and insoluble cues in regulating motility. The biphasic dependence of cell speed has also been demonstrated in numerous 3D constructs including in prostate cancer migration in Matrigel,73 and both fibrosarcoma and smooth muscle cell migration in PEG-based gels.74,75
The combination of cell speed and directional persistence determines the total distance travelled for a single cell.76 The significance of directional persistence during haptokinesis is not fully understood; however, parsing the effects of various migratory phenomena, such as haptokinesis, haptotaxis, chemokinesis, and chemotaxis, will aid in predicting the integration of all these effects during physiologically relevant cell migration. Interestingly, NR6 fibroblasts exhibit directional persistence that is highest at an intermediate Amgel coating concentration.77 However, stimulation with a bath application of EGF decreases directionally persistent migration.77,78 The authors claim that EGF may increase cell speed in metastatic cells, but decrease directional persistence in order to increase the total area probed via randomly directed environment sensing. However, this biphasic directionality response has not been universally reported. In the study on S1P, persistence is only weakly dependent on RGD ligand density, and only subtly responsive to S1P stimulation.72 Therefore, it appears that cell persistent migration is not necessarily correlated with cell migration speed. Future rational biomaterial design will likely require fine-tuning over multiple independent physical parameters to maximize directional cell migration.
A unique area of impact provided by the biomaterials community has been the nano-scale control and presentation of biochemical cues. For example, Spatz et al. initially demonstrated that by allowing di-block copolymer micelles containing gold nanoparticles to form on glass substrates, the authors obtain well-controlled spacing of gold particles.79 Careful variation of the copolymer concentration achieves particle spacing between 28 nm and 110 nm. Subsequent conjugation of gold nanoparticles with thiol-conjugated cyclic RGD peptides resulted in well-defined separation of RGD adhesion sites, in which only one integrin theoretically binds to the adhesion site. This is in sharp contrast to varying bulk ligand density, in which ligand presentation is ultimately stochastic. On surfaces with large RGD spacing, rat fibroblasts are unable to spread properly due to lack of focal adhesion stability.79Fibroblasts on large RGD spacing migrate more quickly, but with less persistence, perhaps due to the lack of cell spreading and its influence on polarity. It has been previously demonstrated that fast turnover in focal adhesions, or the short-lived focal contacts, is correlated with increased cell migration speed.80,81 Spatz et al. also used the gold nanoparticle approach to create well-defined linear gradients of RGD spacing, via controlled retraction of the substrate from the copolymer solution during coating.82 Here, the authors show that cells polarize and exhibit tendency to migrate towards the higher gradient of adhesive ligand. While single RGD peptides were presented in the above studies, others have used controlled substrates to study the effect of integrin clustering on cell motility.68 To do this, RGD was linked to a star-shaped polymer consisting of PEG linkers and conjugated to a PEG hydrogel. Clustered spatial organization of RGD increased NR6 fibroblast migration speeds and robust stress fiber formation compared to unclustered RGDs. This type of controlled spatial presentation of ligands is physiologically relevant, as the in vivo ECM likely presents an unpredicted spatial presentation of integrin binding sites and growth factors.
As stated earlier, physiological migration does not occur on substrates with homogeneously distributed adhesion sites. The cell migration microenvironment consists of varying levels of ECM components with localized gradients. Therefore, great interest exists in understanding the cell's migratory response upon encountering haptotactic cues. There are various biomaterial technologies suited to create molecular gradients, including self-assembled monolayers (SAMs) and PEG hydrogels,63,74,83,84 which can be combined with microfluidic systems85 to enhance local cell motility. SAMs are generally created viaassembly of alkane-thiols onto gold surfaces and subsequent biomolecule conjugation. Migration of bovine aortic endothelial cells on fibronectin gradients is directed distinctly towards increasing adhesion sites.86,87 Not surprisingly, the haptotactic effect can be enhanced by growth factors.88RGD gradients in PEG gels can be created by UV polymerizing RGD-conjugated PEG diacrylate and unconjugated PEG diacrylate at different concentrations,89 or by polymerization of microfluidics-mediated RGD–PEG precursors.90 Both human dermal fibroblasts and mouse embryonic fibroblasts in these studies, respectively, enhanced their migration speeds towards increasing RGD density, and in both cases, migration speed was dependent on the slope of the gradient. Combination of contact printing and SAM can print ligands to different regions to control cell adhesion.91–93 These types of technologies, impossible without the integration of biological insight and biomaterials engineering, show promise in regulating cell migration, both in the context of future rational scaffold design and for mechanistic understanding.
With few exceptions, adhesion to biomaterials has been facilitated with short adhesive peptide sequences, such as RGDs, in lieu of full-length proteins. In fact, RGD, specifically, has been widely used as a generic adhesive sequence, even though certain cell types may not encounter this sequence in vivo as often as other adhesive peptide sequences, such as GFOGER, IKVAV, PHSRN, and many others. Including full-length proteins (with length scales of tens of nanometres or larger) would disrupt the microstructure of a synthetic 3D biomaterial (which have pore sizes on the order of single nanometres).94 However, more effort should be placed on identifying the proper adhesive domain found in the cell's in vivo environment, to better understand physiologically relevant haptotaxis/haptokinesis.
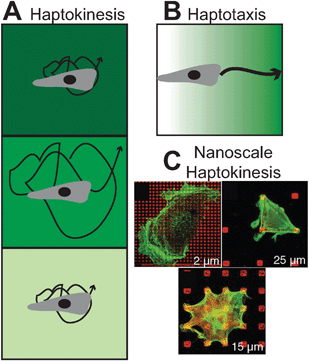 | ||
| Fig. 2 Adhesion-mediated cell migration . In (A), a homogeneous coating of surfaces with insoluble biomolecules at various densities (haptokinesis, shown by increasing background intensity) regulates cell motile speed in a biphasic fashion for many cell types and systems, analogously to chemokinesis. (B) Haptotaxis, on the other hand, is the directed guidance of cell migration by presenting cell adhesive molecules on a gradient, analogously to chemotaxis. (C) On the nanoscale, adhesive biomolecules can be spaced at defined intervals with soft lithography to determine the spatial requirements for focal adhesion formation to generate functional motility (reprinted with permission from ref. 178). | ||
Matrix mechanics and durotaxis
It is now widely accepted that mechanical forces from the extracellular matrix play a large role in directing tissue morphogenesis and progenitor cell lineage commitment, where mechanical forces can be transferred between cells through cadherins and the intermediate filaments, or between cells and the matrix through focal adhesions and the actin cytoskeleton. A subset of mature tissues experience dynamic mechanical loading in vivo, such as in the cardiovascular and musculoskeletal systems, and these mechanical forces are thought to be critical for the maintenance of proper smooth muscle and bone cell phenotype.6,19,23,95,96 Also, endothelial cell function is regulated by the constant shear stresses conferred to them in cardiovascular and pulmonary tissues. In fact, when cells from these tissues are removed from the body and cultured on tissue culture plastic, they markedly lose proper differentiation marker expression, and convert to a synthetic phenotype. Mechanical forces (both static and dynamic) from the ECM in vitro are known to influence endothelial cell behavior (for a review, see ref. 97), stem cell lineage commitment,98 both smooth and skeletal muscle cell plasticity,99–101osteoblast phenotype,102 as well as many other cell and tissue behaviors (for a review, see ref. 103).The first evidence that the static mechanical properties of the ECM could be translated into functional changes in migratory phenotype came from Yu-Li Wang's lab over a decade ago. They created engineered substrates from polyacrylamide (PAA) hydrogels, more traditionally known for their tunable mesh sizes, and used in molecular biology for protein separation.104 Although disputed by some,105 these substrates were reportedly the first of their kind to demonstrate independent control over substrate stiffness and the concentration of adhesive ligand presented at the surface. These tunable mechanical properties were exploited to create a 2D substrate that contained an interface between a compliant and a stiff surface, with the integrin-binding collagen protein covalently linked to the surface to facilitate cell adhesion. When 3T3 fibroblast migration was tracked using time-lapse microscopy, cells initially adhered on the soft gel were found to preferentially migrate onto the stiffer surface. Conversely, cells initially adhered to the stiffer surface would preferentially stay on the stiffer substrate (Fig. 3A–B). This first evidence of stiffness-directed cell motility is now widely known as mechanotaxis, or durotaxis.
In a more quantitative study, human aortic smooth muscle cell migration was compared on uniform PAA substrates of different stiffnesses.106 Similar to variation of adhesivity, cells migrated fastest on substrates of intermediate stiffness and exhibited a biphasic migration speed as a function of static matrix stiffness (Fig. 3C). This experiment corroborates that force balance between ECM tension and cytoskeletal contractility is critical in generating motility. Indeed, the stiffness value at which this maximum migration speed occurred depended on the concentration of adhesive protein presented at the surface. This biphasic phenomenon has also been seen with both osteoblasts107 and neutrophils,108 but not necessarily with all cell types,109 suggesting a differential role of contractility in cell migration across cell types. Nevertheless, these studies provide a mechanistic foundation for the durotactic behavior observed in Yu-Li Wang's study.
The development of stable gradients of stiffness within 2D substrates has been a powerful advancement in the study of durotaxis phenomena. PAA substrates can be created with a gradient in stiffness with a simple, yet elegant technique of combining photo-masking with photo-sensitive polymerization.110 On these surfaces with gradients in stiffness, vascular smooth muscle cells (SMCs) migrated radially from soft-to-stiff regions of the hydrogel. Using a sophisticated microfluidics approach, Burdick et al. created gradients in both the concentration of adhesive peptide (RGDS) and PEG crosslinker content.111 Though not fully explored within this study, systems such as these could be used to study the cross talk of adhesive and stiffness cues in the directed migration of cells in 2D. Expanding on this microfluidics approach, Zaari et al. created gradients in stiffness ranging from ∼1 to 40 kPa on a single PAA gel within a 3mm total distance.112 In agreement with this lab's previous work, an increase in SMC attachment, total spread area, and F-actin fiber definition was observed with increasing substrate stiffness along the gradient. It was not clear from this study, however, if cells were able to migrate preferentially toward the stiff region of the gel, or if the increase in cell number on the stiffer regions was due to an increase in attachment. Isenberg et al. followed up this study using the same microfluidic platform to discern whether or not a gradient in stiffness could direct SMC migration.113 They found that SMCs were responsive to the stiffness cue, and that the strength of the gradient controlled how responsive cells were to the stiffness cue given. Presumably, if a stiffness gradient is too gradual, changes in stiffness will not occur within the length scale of a cell. Unlike soluble growth factors, this stiffness cue is static, and a cell may have to encounter the cue probabilistically during its random walk for it to have an effect.
Mechanosensing in 3D model systems is just beginning to be investigated. To parse the roles of 3D matrix mechanics and cell motility, a number of natural biopolymers have been widely embraced, especially Type I Collagen114 and Matrigel. These systems offer an extreme ease of use, as they can be commercially purchased, contain natural cell-adhesive domains, are enzymatically degradable, and can be made to span a small range of stiffnesses. Using a nested collagen matrix technique pioneered by Fred Grinnell, Miron-Mendoza et al. showed that the stiffness of the outer cell-free matrix increased the ability of human foreskin fibroblasts to migrate.96 When the outer matrix was soft, cell migration slowed, and it was possible to visualize collagen fibers moving under cell tractional forces. They hypothesized that motile cells use tractional forces to pull on collagen fibrils to move forward, so when the outer matrix is restrained by internal crosslinks, cell tractional forces can be completely transduced into cell motility. In lieu of increasing crosslinker content, cell migration was also maximized by constraining the outer gel onto the walls of the polystyrene tissue culture plate.
Using Matrigel as a model system, Zaman et al. reproduced the biphasic dependence of migration speed on material stiffness in human prostate carcinoma cells (Fig. 3D).73 Interestingly in this 3D environment, the relationship between cell migration speed, matrix stiffness, and matrix adhesion did show the same curve shift as was previously shown on 2D substrates.106 As the 3D environment was softened, cell migration speed was maximized in conditions with lower adhesivity. At first glance, this result contradicts previous work on 2D substrata. However, many parameters require a closer look in this Matrigel study, including the difference in cell types studied (highly invasive prostate carcinoma cellsvs. the generally less-motile primary, fully differentiated SMCs), and the difference in the stiffness ranges tested. The authors make many suggestions as well, including the added viscoelastic resistance of a 3D matrix of entangled fibers affects the ability of cells to polarize, which may account for this difference in balance of adhesive and mechanical traction forces, forcing cells to alter their morphology to squeeze through pores. The authors compared these 3D relationships between stiffness, adhesion, and proteolysis in a parallel computational model.115 However, the lack of control of physicochemical properties in this and other protein-based 3D systems may produce convoluting factors. Matrigel concentration is increased to stiffen the resulting 3D gel, which also alters the adhesive ligand concentration, pore diameters, diffusion of growth factors, and the availability of enzymatically-sensitive peptide domains.
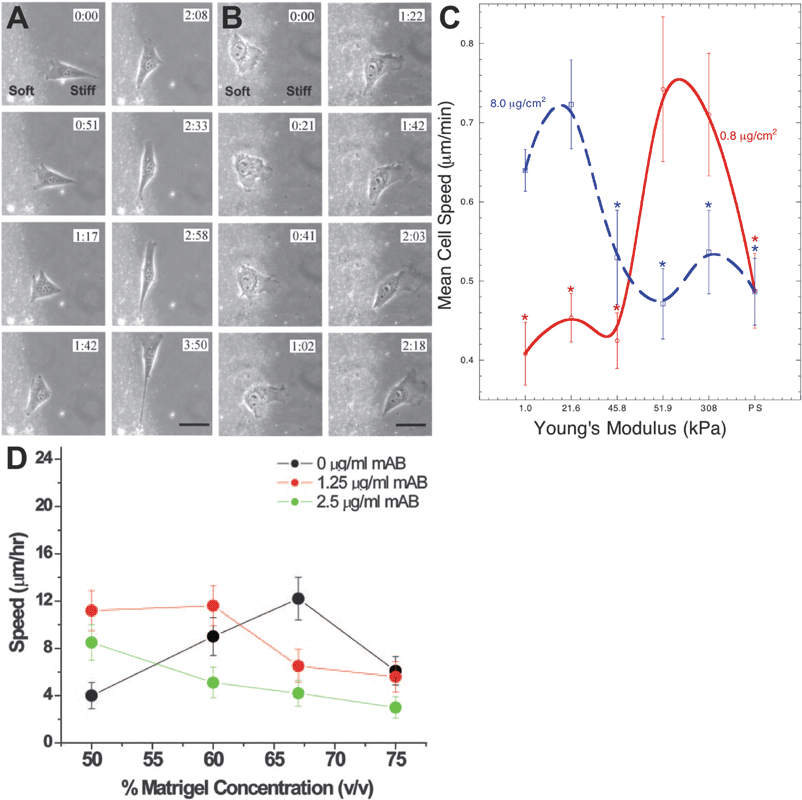 | ||
| Fig. 3 The stiffness of the matrix regulates cell motility . PAA substrates were used to show the first evidence that the stiffness of an underlying substratum could direct cell motility (A–B). Migrating cells starting on a stiff (A) or soft (B) substrate would come to an interface between soft and stiff substrates and preferentially migrate on stiffer substrates (reprinted from ref. 104 with permission from Elsevier). On PAA substrates spanning a range of stiffnesses, smooth muscle cells were shown to have a biphasic dependence on substrate stiffness (C). Cell motility was maximized on substrates of intermediate stiffness, which further depended on the density of fibronectin coupled to the PAA surface (reprinted from ref. 106). This biphasic relationship between matrix stiffness and cell motility was later shown in prostate cancer cells in 3D Matrigel environments (D,73 Copyright (2006) National Academy of Sciences, USA). | ||
One challenge ahead is to elucidate the mechanism of force transduction. Canonically, it is theorized that mechanical signals from the ECM are transmitted to the internal structure of the cellvia integrins (for review, see ref. 116). Several studies have pointed to integrins as providing a link between mechanical changes in the matrix and cell migratory phenotypes.73,117,118 Further, focal adhesions are known to be sensitive to substrate mechanical properties.119,120 However, it is still not completely understood how integrins are able to transfer mechanical information between the cell and the ECM. One theory is that cells are constantly probing their mechanical environment and exerting tension viamyosin contraction along F-actin filaments.121,122 A motor-clutch mechanism may explain how maximum migration speeds have been observed on substrates of intermediate stiffness, as intracellular tension increases with increasing substrate stiffness up to a point of frictional slippage.122 The mechanisms behind which cells are able to respond to mechanical cues in 3D are less clear, as natural biopolymer systems (such as Type I Collagen and Matrigel), are unable to separate stiffness cues from proteolytic and adhesive cues. Current synthetic gel systems achieve this goal, but still have problems of convoluting stiffness and porosity/diffusion parameters. New technologies are on the verge of achieving this goal, including macroporous synthetic scaffolds,123 and 3D systems with gradients in elasticity.124
Given the diversity of mechanical environments in vivo, the ability to engineer substrates that contain tunable static and dynamic mechanical properties is extremely useful. Further, the loss of these naturally occurring mechanical forces with tissue culture plastic dishes may be a key factor in why cells undergo unnatural changes in their phenotype in culture. Ideally, biomaterial tools will include tight control over all these physicochemical features, such as adhesivity, microarchitecture, and stiffness, so that engineers and biologists are able to study cell motility in more physiologically relevant environments, yet more reliably, reproducibly, and cost effectively than in animal models.
Proteolysis
There is increasing interest in studying migration in 3D environments, with the awareness that the vast majority of migratory microenvironments in vivo are composed of cells surrounded by matrix in all dimensions.22,125,126 The most researched 3D motility phenomenon is ECM proteolysis, specifically proteolysis mediated by members of the matrix metalloproteinase (MMP) family.127–129Cell migration in 3D ECM is complicated by the existence of steric barriers, created by the integration of a diverse profile of insoluble scaffolding proteins, such as fibrous, porous Type I Collagen networks and the denser, amorphous laminin–fibronectin networks.59 To overcome steric barriers, cells possess an arsenal of MMPs, as well as proteolysis-independent motility mechanisms.21 The secretion of MMPs and their inhibitors, is, like all cellular processes, a tightly regulated process stimulated by a variety of physicochemical cues.127 Biophysically, matrix remodeling results in local and global changes in ligand density, matrix stiffness, and scaffold geometry, which feedback to regulate cell motility. Therefore, proteolysis-mediated cell migration poses an interesting feedback mechanism that still requires elucidating for a variety of purposes, including understanding tumor invasion and the rational design of tissue engineering constructs. Controlled microenvironments provide an opportunity to deconvolve the complexity involved in proteolysis-mediated cell migration.As an extension to the previous computational model addressing elasticity and adhesivity in 3D cell migration,115 Zaman et al. incorporated the effects of matrix porosity and MMP activity in a lattice Monte-Carlo model.130 Not surprisingly, the model predicted a non-linear dependence of cell speed on ligand density. However, due to the advantages of the modeling technique, it also predicted that directional persistence depended biphasically on ligand density. In their model, persistent migration is enhanced in the presence of MMP activity; however, the increase is less dramatic if the model incorporates the cell's ability to deform the matrix and decrease local steric hindrance. While mesenchymal-type migration in physiology involves both matrix deformation and proteolysis,129 this study conceptualizes the effects of distinct matrix remodeling processes involved during 3D cell migration.
Experimentally, in one of the earliest controlled 3D motility studies, RGD peptides were conjugated into Type I Collagen matrices, which are naturally degradable.131 Mouse melanoma cells moving in these matrices exhibited a biphasic dependence of directional persistence on RGD concentration. Unlike the previously mentioned studies, increasing adhesivity by incorporating RGD into these matrices does not convolute other parameters, such as proteolysis and matrix stiffness. However, it is unclear how long soluble RGD persists in such a 3D matrix without additional chemical coupling. In agreement with the 2D studies discussed in the Haptotaxis section, RGD incorporation only affected cell persistence, and not cell speed. Further, as in Wacker et al.,72 the addition of a soluble factor enhanced sensitivity to RGD. Secondly, in a study utilizing a novel engineered PEG-based hydrogel containing MMP degradation sites, fibroblasts enhanced MMP release upon TNF-α stimulation, which increased the motility persistence length.132 In the first study, gradually modulating cellular MMP activity via pharmacological inhibition, EGF-enhanced human glioblastoma cell migration in 3D collagen matrices was compared to 2D collagen-coated substrata.133 Surprisingly, EGF stimulation increased directional persistence in 3D collagen, but decreased persistence on 2D collagen surfaces. By modulating MMP activity and matrix degradation with a broad MMP inhibitor, authors showed that the EGF-enhanced increase in directional persistence arises due to MMP activity, further corroborating hypotheses generated in the above computational model.130 Interestingly, cell speed varied minimally with matrix degradation, which was consistent with the minimal dependence of cell speed on bulk collagen concentration. Collagenase itself has been shown to exhibit a persistent proteolysis along collagen fibrils,134 perhaps laying the molecular foundation for the high dependence of directional persistence on MMP-mediated ECM degradation.
Most of the quantitative parametric studies involving proteolytic migration have been gathered on biopolymer matrices.96,135 While physiologically relevant, they have the distinct disadvantage of convoluting matrix parameters, i.e. the number of available proteolysis sites is related to ligand density, matrix porosity, and bulk matrix stiffness. In response to this, a burgeoning number of studies have begun using synthetic materials, equipped with adhesive ligands and enzymatically-sensitive crosslinks.136 PEG hydrogels have been particularly popular in this regard for their versatile chemistry and quantitative control. One of the first of such hydrogels incorporated integrin-binding peptides, the MT-1 MMP-sensitive peptide sequence GPQGIWGQ, and a plasmin-sensitive sequence YKNRDvia multi-arm PEG monomers.136,137 Subsequent studies have utilized step-growth polymerization of PEG monomers138 or different functional end groups75 to incorporate both adhesion and MMP-sensitive sequences, or even full length proteins.100 Further, collagenase-sensitive substrates have been incorporated into PEG hydrogels, allowing live visualization of collagenase activity during cell migration.139 PEG hydrogels also have the ability to present a variety of receptor-binding biomolecules, with cell-demanded release. In collaboration between the Langer and Hubbell groups, an MMP-responsive PEG hydrogel was used to encapsulate thymosin β4, which induces vascular cell survival and upregulation of vascularization genes and MMP secretion, mimicking, perhaps, the release of matrix-associated growth factors in natural ECMs. Lastly, a recent report describes the use peptide-based hydrogels containing MMP2 recognition sites and RGD.140 Parameterizing cell migration using synthetic ECMs is still scarce. However, due to the variety of ECM proteins with specificities for a diverse set of proteases, such studies would undoubtedly provide interesting insights into other mechanisms for regulating cell migration through presentation of diverse physicochemical cues.
Fueled by innovations in microscopy in the last ten years, interest in 3D cell migration has identified non-proteolytic forms of cell migration. The amoeboid mode of migration was first observed in fast-moving lymphocytes, and verified in vitro when fibrosarcoma cells were observed to undergo dramatic changes in morphology to allow for continued movement in 3D Type I Collagen gels in the presence of a cocktail of MMP and protease inhibitors. This seminal study provided a putative explanation for the poor success of MMP inhibitors as metastasis-targeted chemotherapy.141 This mode of migration is integrin-independent, and utilizes a squeezing motion based on expansive actin network-based protrusive flowing and myosin-dependent contraction, which is required to pass the narrow gaps of ECM pores.20,24 While biologists agree with this mechanism for lymphocytes, the mesenchymal-amoeboid transition of fibrosarcomas is currently disputed, due to the convoluting nature of pepsin-treated Type I Collagen. Ensuing studies in native Type I Collagen, which retain the natural crosslinks existent in vivo, were not able to replicate this MMP-independent motility.142 These studies reiterate the importance of providing disease-relevant physicochemical cues. Synthetic PEG hydrogels incorporating both RGD and MMP-sensitive peptides, with inherent pore sizes orders of magnitude smaller than the smallest cell protrusion, have demonstrated that fibrosarcoma cells exhibited rounded and contraction-dependent migration, which is only weakly dependent on integrins.75 While no proteolysis-specific measurements were performed, the authors suggest that fibrosarcoma cellsin vivo are more rounded than they appear in traditional in vitro studies, and synthetic ECMs could provide a unique framework to examine these proposed mechanisms of motility and morphology.
Microarchitecture
Several groups have highlighted the relevance of studying migration in 3D models due to the striking differences drawn between cells in 2D versus 3D matrices.143,144 Evidence suggests that both cell morphology and adhesion structure change dramatically between 2D and 3D cultures,62 perhaps due to the difference in structure between a monolayer of adhesive proteins on a flat surface versus the fibrous presentation of adhesive sites in 3D. In a study using the same matrix protein, research has shown that migration speeds on 2D collagen-coated surfaces do not correlate with speeds in 3D collagen gels, and that protrusion activity is controlled by distinct focal adhesion proteins (p130Cas in 2D and zyxin in 3D) in the geometrically distinct microenvironments.145From an engineering perspective, although interesting, this study and others attempting to decipher the role of geometry on cell behavior separately from other biophysical factors are limited by the fact that these parameters are convoluted in natural biopolymers, such as Type I Collagen gels. This is highlighted when comparing the work of others in these gels, wherein the mode of matrix polymerization had profound affects on migratory phenotype (Fig. 4).133,146,147 Work by Kim et al. observed that glioblastoma cells were insensitive to changes in 3D collagen stiffness and adhesivity.133 However, Harley et al. showed that cell motility was dependent on the stiffness of collagen matrices that were formed using freeze-drying techniques.146 This freeze-drying technique of matrix formation created pore sizes that were in excess of 100 μm in diameter, so it is likely that cells in these matrices were experiencing a quasi-1D migratory microenvironment, rather than true 3D migration.
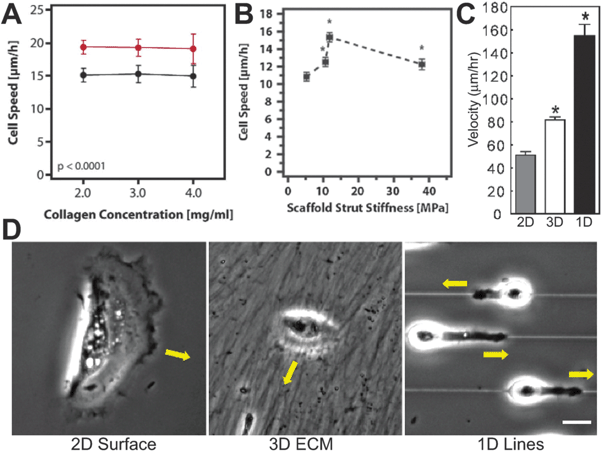 | ||
| Fig. 4 Fiber Length-Scale Dictates Geometric Migratory Microenvironment. In (A) Kim et al., observed no dependence of glioblastoma motility on 3D collagen gel stiffness (reprinted from ref. 133 with permission from the American Society for Cell Biology). In (B) Harley et al., used collagen gels with pore sizes 1–2 orders of magnitude larger than in Kim et al. (reprinted from ref. 146 with permission from Elsevier). They observed cell speeds that had a biphasic dependence on collagen fiber stiffness. Pore sizes in this microenvironment are an order of magnitude larger than the length-scale of the cell, so collagen fibers appear as 1D lines. This may explain the discrepancy in observed motility between (A) and (B). In (C–D) Doyle et al., used micropatterned PDMS substrata and cell-generated 3D ECMs to show that cell migration is fastest along 1D printed lines and slowest on a non-fibrillar, flat 2D surface (©2009 Rockefeller University Press. Originally published in ref. 147). | ||
To assess the effects of dimensional geometry on motility, Doyle and colleagues compared motility on a fibrillar cell-derived 3D matrix, flat PDMS model substrates, and PDMS printed lines.147 They found that cell speeds along the PDMS lines (“1D mode”) mimicked speeds along matrix fibers (“3D mode”), and that both of these morphologies led to speeds that were much faster than cells on uniform 2D surfaces. Work by Liu et al. implies that there is a minimal fiber diameter of 1 μm to cause cells to polarize along the fiber.148Cell motile phenotype on fibers less than 1 μm in diameter mimicked those on 2D surfaces. Most interestingly, the dependence of migration on ligand density, myosin, and microtubules was different between 1D and 2D migration, with 1D relationships most mimicking the 3D environment.147
A recent study by Ochsner et al. has attempted to separate geometric influences from adhesive and stiffness effects by micropatterning different-shaped wells in PDMS substrata.149 Their evidence suggests that actin filaments contribute to cytoskeletal tension, matrix remodeling, and metabolism differently on 2D surfaces versus 3D microenvironments. In fact, their study concluded that the geometric microenvironment determined the extent to which cells were sensitive to alterations in matrix stiffness by regulating cytoskeletal tension. Macroporous scaffolds made from inverse-opal processing are additional biomaterial tools capable of providing a nondegradable 3D environment with independently tunable adhesivity, stiffness, and pore diameters.150,151 Generally made with a PEG background, these scaffolds are inherently nondegradable, allowing for long-term cell tracking in a static system. Although not fibrillar, nor necessarily representative of in vivo tissues, these types of scaffolds are excellent model systems appropriate for parsing the relationships between cell motility and biophysical cues in 3D. Their macroporosity allows them to be overlaid with other degradable materials as well.152 Recently, these PEG-based macroporous scaffolds were used to determine the effects of pore diameters on mesenchymal stem cell (MSC) motility.153 Surprisingly, MSCs were observed to migrate in a non-intuitive fashion, where maximum displacement occurred in an intermediate pore diameter that was smaller than the spherical cell diameter, and maximum displacement did not correlate with maximum observed cell speeds. Cell speed had a biphasic dependence on scaffold adhesivity, but only in environments that had very large pore sizes, and likely were a quasi-2D environment.
Thus far, we have discussed 3D synthetic systems that, although are more than ninety percent water, consist of a nanoporous mesh. These mesh sizes, resulting from crosslinking between polymer chains, are orders of magnitude smaller than the smallest cellular processes, and do not resemble the microarchitecture of an in vivo fibrous matrix (Fig. 1). To create synthetic scaffolds that more closely mimic the native architecture and the nano- or micro-topology of the ECM, researchers have applied electrospinning techniques. Electrospinning has been in use for more than seven decades, but has only recently been employed to create synthetic tissue environments. Electrospinning can be used with a variety of polymer systems, and can create fibers with diameters ranging from 100 nm–10 μm, which spans the length scale of many natural fibers, such as collagens, chitosan, fibrin, chitin, and fibrinogen (for review, see ref. 154). Thus far, research with electrospun fibers has been focused on in vivo applications for tissue regeneration, but an electrospun polymer system could be used to study cell motility mechanisms in a synthetic polymer system with control of fiber architecture.
Electrospinning results in a heterogeneous distribution of fiber diameters and pore sizes, which is not unlike the in vivo environment. For directed migration, and predictable cell infiltration into a tissue-engineered device, however, fiber dimensions and arrangement can be tightly controlled with rapid prototyping techniques. Rapid prototyping is a general term for computer-aided design of scaffolds, and encompasses many different techniques, including fused deposition, 3D printing™, selective laser sintering, melt-dissolution, and cryogenic prototyping (for reviews specific to rapid prototyping, see ref. 155–157. Rapid prototyping of scaffolds is popular in tissue engineering, since it allows for precise structural design, but work on using this for basic mechanistic understandings of cell migration has not been explored. Basic connections have been made between increased pore sizes of rapid prototyped scaffolds and increased cell infiltration,158,159 but there is no mechanistic understanding between these scaffold geometries and cell migration processes. Given the tight control for fiber design allowed by these methods, this could be a very interesting new area for mechanistic studies of nano-topographical control of cytoskeletal and focal adhesion assembly in fibrous materials.
Self-assembled peptide gels (SAPGs) also form fibrillar matrices, and are able to mimic the geometrical features of in vivocollagen fibers. SAPGs are designed from natural amino acids and undergo spontaneous assembly into nanofibers, approximately 10 nm in diameter. The pore sizes of SAPGs are on the order of hundreds of nanometres, and as many cells' smallest effective protrusions are on the order of microns (excluding immune cells and some adaptive cancer cells), cell-mediated degradation is required for productive locomotion. Although the length scale features of these SAPGs are comparable to natural biopolymers, the motility of cells within them is markedly reduced in comparison.160 Addition of osteogenic growth factor peptides into SAPGs can increase osteoblast motility alongside proliferation and expression of differentiation-specific markers.161 Much smaller length scales have also been shown to be important for regulating cell responses to materials. Non-biological TiO2 nanotubes have shown that cells are able to sense material properties on the order of nanometres, and have helped parse the relationship between protein spacing and cell migrationin an inherently non-fouling environment. Focal adhesion formation and cytoskeletal assembly appears to be directly regulated by the nano-length scale and presentation of these nanotubes,162,163 presumably mediated by adhesive matrix protein presentation.
Overall, it's clear from these studies that the architecture of the microenvironment plays a large role in directing cell motility. In vivo, varied microenvironments exist, including relatively constant 2D, planar basal laminas, largely porous, and potentiallyquasi-2D on the length scale of a cell, trabecular bone, fibrous and dense connective tissue, etc. Certain subtypes of cell populations dominate these environments, and these microarchitectures likely regulate the motility of these cellsin vivo. Studies in which the microarchitecture of the 2D or 3D substrate can be controlled in vitro are therefore extremely relevant for the proper understanding and manipulation of the migratory behavior.
Tool development
Despite the many studies discussed within this review, the vast majority of cell motility studies are conducted on traditional glass or plastic surfaces. One of the reasons for the hesitation of the biological community to embrace biomaterial systems is the chemistry expertise needed (or perceived to be needed) to create biomaterial model systems versus the traditional platforms. Many scientists prefer naturally derived biopolymers, such as Type I Collagen and Matrigel, because although they have obvious reproducibility limitations, they are very easy to create, and their fibrous nature represents the in vivo microenvironment much better than the nonporous or macroporous 3D hydrogels described here. Certain technological and cost limitations also exist in imaging technologies to transfer mechanistic studies, canonically performed on thin, flat, and optically clear cover slips into more realistic 3D microenvironments. This section describes some of the more recent advances both within and outside the biomaterials community to overcome these hurdles.Numerous biomaterial systems developed by the community have been created for tissue engineering purposes, but also show extensive promise as model substrates to study mechanisms of cell motility. For instance, because of their modular peptide design, protein-engineered biomaterials can mimic various properties of the natural ECM while maintaining the versatility to include non-naturally occurring binding sites for some synthetic “plug and play” control.164 Though touted for their potential impact in the field of tissue regeneration, due to their natural bioresorbability and biofunctionality, these materials could also be useful for more basic mechanistic questions about cell-microenvironment relationships due to the inherent molecular-level design control.
Confocal and two-photon lasers have taken advantage of UV-mediated polymerization of hydrogels to create 3D micropatterned materials (Fig. 5A). Using this sophisticated form of photolithography (two-photon laser scanning, or TPLS), Lee et al. made 3D gels with precise control over the location of cell-adhesive RGDS in a degradable environment.165 In doing so, they were able to guide the adhesion, and therefore migration, of fibroblasts in 3D. These types of 3D micropatterned gels may lead to advances in guided tissue regeneration. Micropatterning of 3D natural biopolymers has already been realized in Type I Collagen gels, originally described by Nelson et al. (Fig. 5C).166 Simple stamping techniques were applied to these 3D gels to form reproducible arrays of rectangular cultures. This geometrical conformation of tissue has allowed for very interesting studies of how mechanotaxis and morphogen gradients regulate cell invasion in three dimensions.167,168
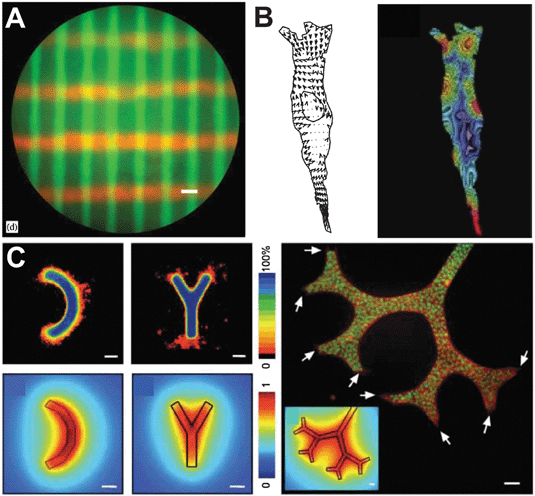 | ||
| Fig. 5 Emerging tools for studying matrix control of cell motility. The combination of light sensitive polymerization initiators and confocal microscopy has made 3D patterning of hydrogels possible ((A) reprinted from ref. 179 with permission from Elsevier). Embedding and tracking the displacement of fluorescent beads in compliant substrates can be exploited to map tractional forces of migrating cells ((B) reprinted from ref. 180 with permission from Elsevier). Patterning in 3D matrices allows for formation of microtissues to study cell migration from tissue-like structures ((C) reprinted with permission from ref. 166). | ||
As discussed in the durotaxis and haptotaxis sections, the ability of cells to exert tractional forces on the surrounding matrix directly affects their ability to migrate. Measurements of cell tractional forces, either through the cytoskeleton, or the matrix itself, is critical in observing this exerted force. Traction force microscopy has been a powerful tool in the past two decades in observing these tractional dynamics (Fig. 5B, for review, see ref. 18). Substrate deformations can be visualized by incorporating fluorescent beads in the hydrogel matrix. Displacement of the beads is translated to tractional forces using continuum mechanics.169 Traction force microscopy has been used to calculate the ability of cells to sense and exert stress in two dimensions viaPAA substrates,170–172 and to describe cancer mechanosensing.173,174 More recently, thin film arrays have been developed to measure force exertion by cells without the need for embedded beads.175 Analogous to the widely published post-arrays, the displacement of the thin films viacell-generated tension can be measured by unsophisticated optical microscopy.176
As the number of interesting convoluting physicochemical parameters increases, one lingering limitation of biomaterials-based migration studies is the lack of high-throughput technology. The ability to screen chemotherapeutics in 96-well formats, to test a wide variety of variables and combinations at once, is common in pharmaceutical companies. This type of format has been recently adapted for migration studies, to quantify migration and signaling in response to growth factors and inhibitors.177 Though not yet available, adapting this type of technology to a biomaterials system would allow one to study these combinatorial agents in an engineered environment with control of stiffness, mesh sizes, adhesive background, etc.
Future outlook
While in vivo studies have provided the importance of cell migration in the context of physiology and disease, in vitro studies have been critical in providing mechanistic insights into the migratory process. The cell biology community has indeed enriched our understanding of the molecular components and their interdependencies involved in generating motility. However, the particular complexity of the locomotive process discussed in this review calls for an integrative approach for a comprehensive and predictive understanding of cell migration. Studies of cell motility using biomaterials systems have demonstrated the power of natural and synthetic materials in contributing to the mechanistic regulation of relevant phenomenological parameters, such as cell speed and directional persistence. Unfortunately, synthetic biomaterials systems have not yet gained sufficient traction in the biosciences community, often due to a mismatch between the pertinent biological questions and the physiological relevance of the materials. With the recent efforts for fostering interdisciplinary work, we posit that this gap will soon be bridged. We urge the biomaterials community to assess the vast space of unanswered, biologically relevant questions that will only be answered with elegant synthetic techniques. In turn, the cell biologists can appreciate the immense opportunities provided by thoughtfully designed materials, wherein carefully controlled microenvironments enable reproducible studies of complex mechanisms.References
- K. L. Maschhoff and H. S. Baldwin, Molecular determinants of neural crest migration, Am. J. Med. Genet., 2000, 97(4), 280–288 CrossRef CAS.
- R. Keller, Cell migration during gastrulation, Curr. Opin. Cell Biol., 2005, 17(5), 533–541 CrossRef CAS.
- A. Aman and T. Piotrowski, Cell migration during morphogenesis, Dev. Biol., 2010, 341(1), 20–33 CrossRef CAS.
- L. Braiman-Wiksman, et al., Novel insights into wound healing sequence of events, Toxicol. Pathol., 2007, 35(6), 767–779 CrossRef.
- F. Sánchez-Madrid and M. A. d. Pozo, Leukocyte polarization in cell migration and immune interactions, EMBO J., 1999, 18(3), 501–511 CrossRef.
- A. J. el Haj, et al., Cellular responses to mechanical loading in vitro, J. Bone Miner. Res., 1990, 5(9), 923–32 CrossRef CAS.
- J. Yang and R. A. Weinberg, Epithelial-mesenchymal transition: at the crossroads of development and tumor metastasis, Dev. Cell, 2008, 14(6), 818–829 CrossRef CAS.
- D. A. Lauffenburger and A. F. Horwitz, Cell migration: a physically integrated molecular process, Cell, 1996, 84, 359–69 CrossRef CAS.
- A. J. Ridley, et al., Cell migration: integrating signals from front to back, Science, 2003, 302(5651), 1704–1709 CrossRef CAS.
- L. M. Machesky, Lamellipodia and filopodia in metastasis and invasion, FEBS Lett., 2008, 582(14), 2102–2111 CrossRef CAS.
- D. J. Webb, T. Parsons and A. F. Horwitz, Adhesion assembly, disassembly and turnover in migrating cells—over and over and over again, Nat. Cell Biol., 2002, 4, E97–E100 CrossRef CAS.
- M. A. Wozniak, et al., Focal adhesion regulation of cell behavior, Biochim. Biophys. Acta, Mol. Cell Res., 2004, 1692(2–3), 103–19 CrossRef CAS.
- S. Etienne-Manneville, Polarity proteins in migration and invasion, Oncogene, 2008, 27(55), 6970–6980 CrossRef CAS.
- S. Etienne-Manneville and A. Hall, Rho GTPases in cell biology, Nature, 2002, 420(6916), 629–35 CrossRef CAS.
- A. Hall, Rho GTPases and the actin cytoskeleton, Science, 1998, 279(5350), 509–14 CrossRef CAS.
- O. Pertz, et al., Spatiotemporal dynamics of RhoA activity in migrating cells, Nature, 2006, 440(7087), 1069–72 CrossRef CAS.
- T. Wittmann and C. M. Waterman-Storer, Cell motility: can Rho GTPases and microtubules point the way?, J. Cell Sci., 2001, 114(Pt 21), 3795–803 CAS.
- C. T. Mierke, et al., Contractile forces in tumor cell migration, Eur. J. Cell Biol., 2008, 87(8–9), 669–676 CrossRef CAS.
- T. M. Skerry, et al., Early strain-related changes in enzyme activity in osteocytes following bone loading in vivo, J. Bone Miner. Res., 1989, 4(5), 783–8 CrossRef CAS.
- T. Lammermann, et al., Rapid leukocyte migration by integrin-independent flowing and squeezing, Nature, 2008, 453(7191), 51–5 CrossRef.
- K. Wolf, et al., Compensation mechanism in tumor cell migration: mesenchymal-amoeboid transition after blocking of pericellular proteolysis, J. Cell Biol., 2003, 160(2), 267–277 CrossRef CAS.
- S. Even-Ram and K. M. Yamada, Cell migration in 3D matrix, Curr. Opin. Cell Biol., 2005, 17(5), 524–32 CrossRef CAS.
- S. Lehoux and A. Tedgui, Signal transduction of mechanical stresses in the vascular wall, Hypertension, 1998, 32(2), 338–45 CAS.
- K. Wolf, et al., Amoeboid shape change and contact guidance: T-lymphocyte crawling through fibrillar collagen is independent of matrix remodeling by MMPs and other proteases, Blood, 2003, 102(9), 3262–3269 CrossRef CAS.
- R. v. Horssen and T. L. M. t. Hagen, Crossing barriers: the new dimension of 2D cell migration assays, J. Cell. Physiol., 2011, 226(1), 288–290 CrossRef.
- M. Gonen-Wadmany, L. Oss-Ronen and D. Seliktar, Protein-polymer conjugates for forming photopolymerizable biomimetic hydrogels for tissue engineering, Biomaterials, 2007, 28(26), 3876–86 CrossRef CAS.
- A. G. Mikos, et al., Engineering complex tissues, Tissue Eng., 2006, 12(12), 3307–39 CrossRef CAS.
- J. Kopecek, Hydrogel biomaterials: a smart future?, Biomaterials, 2007, 28(34), 5185–92 CrossRef CAS.
- N. Huebsch and D. J. Mooney, Inspiration and application in the evolution of biomaterials, Nature, 2009, 462(7272), 426–32 CrossRef CAS.
- J. A. Hubbell, Bioactive biomaterials, Curr. Opin. Biotechnol., 1999, 10(2), 123–9 CrossRef CAS.
- A. Locascio and M. A. Nieto, Cell movements during vertebrate development: integrated tissue behaviour versus individual cell migration, Curr. Opin. Genet. Dev., 2001, 11(4), 464–469 CrossRef CAS.
- J. M. Rutkowski and M. A. Swartz, A driving force for change: interstitial flow as a morphoregulator, Trends Cell Biol., 2007, 17(1), 44–50 CrossRef CAS.
- M. E. Fleury, K. C. Boardman and M. A. Swartz, Autologous morphogen gradients by subtle interstitial flow and matrix interactions, Biophys. J., 2006, 91(1), 113–21 CrossRef CAS.
- C. L. Helm, et al., Synergy between interstitial flow and VEGF directs capillary morphogenesis in vitro through a gradient amplification mechanism, Proc. Natl. Acad. Sci. U. S. A., 2005, 102(44), 15779–84 CrossRef CAS.
- D. Kedrin, et al., Cell motility and cytoskeletal regulation in invasion and metastasis, J. Mammary Gland Biol. Neoplasia, 2007, 12(2–3), 143–52 CrossRef.
- J. Condeelis and J. W. Pollard, Macrophages: obligate partners for tumor cell migration, invasion, and metastasis, Cell, 2006, 124(2), 263–6 CrossRef CAS.
- S. Boyden, The chemotactic effect of mixtures of antibody and antigen on polymorphonuclear leucocytes, J. Exp. Med., 1962, 115, 453–66 CrossRef CAS.
- R. D. Nelson, P. G. Quie and R. L. Simmons, Chemotaxis under agarose: a new and simple method for measuring chemotaxis and spontaneous migration of human polymorphonuclear leukocytes and monocytes, Journal of immunology, 1975, 115(6), 1650–6 CAS.
- C. Rothman and D. Lauffenburger, Analysis of the linear under-agarose leukocyte chemotaxis assay, Ann. Biomed. Eng., 1983, 11(5), 451–77 CrossRef CAS.
- R. M. Ford, et al., Measurement of bacterial random motility and chemotaxis coefficients: I. Stopped-flow diffusion chamber assay, Biotechnol. Bioeng., 1991, 37(7), 647–60 CrossRef CAS.
- S. Toetsch, et al., The evolution of chemotaxis assays from static models to physiologically relevant platforms, Integr. Biol., 2009, 1(2), 170–81 RSC.
- S. Y. Cheng, et al., A hydrogel-based microfluidic device for the studies of directed cell migration, Lab Chip, 2007, 7(6), 763–9 RSC.
- S. Kim, H. J. Kim and N. L. Jeon, Biological applications of microfluidic gradient devices, Integrative Biology: Quantitative Biosciences from Nano to Macro, 2010, 2(11–12), 584–603 CAS.
- S. Toetsch, et al., The evolution of chemotaxis assays from static models to physiologically relevant platforms, Integrative Biology: Quantitative Biosciences from Nano to Macro, 2009, 1(2), 170–81 CAS.
- S. Chung, et al., Cell migration into scaffolds under co-culture conditions in a microfluidic platform, Lab Chip, 2009, 9(2), 269–75 RSC.
- D. J. Beebe, et al., A platform for assessing chemotactic migration within a spatiotemporally defined 3D microenvironment, Lab Chip, 2008, 8(9), 1507–1515 RSC.
- R. D. Kamm, W. J. Polacheck and J. L. Charest, Interstitial flow influences direction of tumor cell migration through competing mechanisms, Proc. Natl. Acad. Sci. U. S. A., 2011, 108(27), 11115–11120 CrossRef.
- F. Lin and E. C. Butcher, T cell chemotaxis in a simple microfluidic device, Lab Chip, 2006, 6(11), 1462–1469 RSC.
- M. A. Swartz, et al., Dendritic cell chemotaxis in 3D under defined chemokine gradients reveals differential response to ligands CCL21 and CCL19, Proc. Natl. Acad. Sci. U. S. A., 2011, 108(14), 5614–5619 CrossRef.
- W. Saadi, et al., Generation of stable concentration gradients in 2D and 3D environments using a microfluidic ladder chamber, Biomed. Microdevices, 2007, 9(5), 627–35 CrossRef.
- M. O. Platt, et al., Sustained epidermal growth factor receptor levels and activation by tethered ligand binding enhances osteogenic differentiation of multi-potent marrow stromal cells, J. Cell. Physiol., 2009, 221(2), 306–17 CrossRef CAS.
- J. A. Hubbell, Matrix-bound growth factors in tissue repair, Swiss Med Wkly, 2007, 137(Suppl 155), 72S–76S CAS.
- V. H. Fan, et al., Tethered epidermal growth factor provides a survival advantage to mesenchymal stem cells, Stem Cells, 2007, 25(5), 1241–51 CrossRef CAS.
- T. J. Stefonek-Puccinelli and K. S. Masters, Co-immobilization of gradient-patterned growth factors for directed cell migration, Ann. Biomed. Eng., 2008, 36(12), 2121–33 CrossRef.
- U. Philippar, et al., A Mena invasion isoform potentiates EGF-induced carcinoma cell invasion and metastasis, Dev. Cell, 2008, 15(6), 813–28 CrossRef CAS.
- D. J. Irvine, et al., Directed cell migration via chemoattractants released from degradable microspheres, Biomaterials, 2005, 26(24), 5048–5063 CrossRef.
- C. P. Ng and M. A. Swartz, Fibroblast alignment under interstitial fluid flow using a novel 3-D tissue culture model, Am. J. Physiol. Heart Circ. Physiol., 2003, 284(5), H1771–7 CAS.
- B. Geiger, et al., Transmembrane crosstalk between the extracellular matrix-cytoskeleton crosstalk, Nat. Rev. Mol. Cell Biol., 2001, 2(11), 793–805 CrossRef CAS.
- B. Alberts, Molecular biology of the cell, Book, 2002 Search PubMed.
- S. B. Carter, Haptotaxis and the mechanism of cell motility, Nature, 1967, 213(5073), 256–60 CrossRef CAS.
- K. E. Kubow and A. R. Horwitz, Reducing background fluorescence reveals adhesions in 3D matrices, Nat. Cell Biol., 2011, 13(1), 3–5 CrossRef CAS ; author reply 5-7.
- E. Cukierman, et al., Taking cell-matrix adhesions to the third dimension, Science, 2001, 294(5547), 1708–12 CrossRef CAS.
- M. N. Yousaf, Model substrates for studies of cell mobility, Curr. Opin. Chem. Biol., 2009, 13(5–6), 697–704 CrossRef CAS.
- A. Seidi, et al., Gradient biomaterials for soft-to-hard interface tissue engineering, Acta Biomaterialia, 2011 Search PubMed.
- S. P. Palecek, et al., Integrin-ligand binding properties govern cell migration speed through cell-substratum adhesiveness, Nature, 1997, 385(6616), 537–40 CrossRef CAS.
- P. A. DiMilla, et al., Maximal migration of human smooth muscle cells on fibronectin and type IV collagen occurs at an intermediate attachment strength, J. Cell Biol., 1993, 122(3), 729–37 CrossRef CAS.
- S. L. Gupton and C. M. Waterman-Storer, Spatiotemporal feedback between actomyosin and focal-adhesion systems optimizes rapid cell migration, Cell, 2006, 125(7), 1361–1374 CrossRef CAS.
- G. Maheshwari, et al., Cell adhesion and motility depend on nanoscale RGD clustering, J. Cell Sci., 2000, 113(Pt 10), 1677–1686 CAS.
- A. Wells, et al., Growth factor-induced cell motility in tumor invasion, Acta Oncol., 2002, 41(2), 124–130 CrossRef CAS.
- K. Tashiro, et al., A synthetic peptide containing the IKVAV sequence from the A chain of laminin mediates cell attachment, migration, and neurite outgrowth, The Journal of Biological Chemistry, 1989, 264(27), 16174–16182 CAS.
- E. Ruoslahti, RGD and other recognition sequences for integrins, Annu. Rev. Cell Dev. Biol., 1996, 12, 697–715 CrossRef CAS.
- B. K. Wacker, et al., Endothelial cell migration on RGD-peptide-containing PEG hydrogels in the presence of sphingosine 1-phosphate, Biophys. J., 2008, 94(1), 273–285 CrossRef CAS.
- M. H. Zaman, et al., Migration of tumor cells in 3D matrices is governed by matrix stiffness along with cell-matrix adhesion and proteolysis, Proc. Natl. Acad. Sci. U. S. A., 2006, 103(29), 10889–94 CrossRef CAS.
- B. K. Mann and J. L. West, Cell adhesion peptides alter smooth muscle cell adhesion, proliferation, migration, and matrix protein synthesis on modified surfaces and in polymer scaffolds, J. Biomed. Mater. Res., 2002, 60(1), 86–93 CrossRef CAS.
- M. P. Schwartz, et al., A synthetic strategy for mimicking the extracellular matrix provides new insight about tumor cell migration, Integr. Biol., 2010, 2(1), 32–40 RSC.
- R. J. Petrie, A. D. Doyle and K. M. Yamada, Random versus directionally persistent cell migration, Nat. Rev. Mol. Cell Biol., 2009, 10(8), 538–49 CrossRef CAS.
- M. F. Ware, A. Wells and D. A. Lauffenburger, Epidermal growth factor alters fibroblast migration speed and directional persistence reciprocally and in a matrix-dependent manner, J. Cell Sci., 1998, 111(Pt 16), 2423–32 CAS.
- B. D. Harms, et al., Directional persistence of EGF-induced cell migration is associated with stabilization of lamellipodial protrusions, Biophys. J., 2005, 88(2), 1479–88 CrossRef CAS.
- E. A. Cavalcanti-Adam, et al., Cell spreading and focal adhesion dynamics are regulated by spacing of integrin ligands, Biophys. J., 2007, 92(8), 2964–2974 CrossRef CAS.
- A. Glading, et al., Epidermal growth factor activates m-calpain (calpain II), at least in part, by extracellular signal-regulated kinase-mediated phosphorylation, Mol. Cell. Biol., 2004, 24(6), 2499–2512 CrossRef CAS.
- T. Takino, et al., Membrane-type 1 matrix metalloproteinase modulates focal adhesion stability and cell migration, Exp. Cell Res., 2006, 312(8), 1381–1389 CrossRef CAS.
- M. Arnold, et al., Induction of cell polarization and migration by a gradient of nanoscale variations in adhesive ligand spacing, Nano Lett., 2008, 8(7), 2063–2069 CrossRef CAS.
- A. Pulsipher and M. N. Yousaf, Surface chemistry and cell biological tools for the analysis of cell adhesion and migration, ChemBioChem, 2010, 11(6), 745–753, 730 CrossRef CAS.
- A. S. Gobin and J. L. West, Effects of epidermal growth factor on fibroblast migration through biomimetic hydrogels, Biotechnol. Prog., 2003, 19(6), 1781–1785 CrossRef CAS.
- B. M. Lamb, N. P. Westcott and M. N. Yousaf, Microfluidic lithography to create dynamic gradient SAM surfaces for spatio-temporal control of directed cell migration, ChemBioChem, 2008, 9(16), 2628–2632 CrossRef CAS.
- J. T. Smith, J. T. Elkin and W. M. Reichert, Directed cell migration on fibronectin gradients: effect of gradient slope, Exp. Cell Res., 2006, 312(13), 2424–2432 CrossRef CAS.
- J. T. Smith, et al., Measurement of cell migration on surface-bound fibronectin gradients, Langmuir: The ACS Journal of Surfaces and colloids, 2004, 20(19), 8279–8286 CAS.
- J. T. Smith, D. H. Kim and W. M. Reichert, Haptotactic gradients for directed cell migration: stimulation and inhibition using soluble factors, Comb. Chem. High Throughput Screening, 2009, 12(6), 598–603 CrossRef CAS.
- S. A. DeLong, A. S. Gobin and J. L. West, Covalent immobilization of RGDS on hydrogel surfaces to direct cell alignment and migration, J. Controlled Release, 2005, 109(1–3), 139–148 CrossRef CAS.
- D. Guarnieri, et al., Covalently immobilized RGD gradient on PEG hydrogel scaffold influences cell migration parameters, Acta Biomater., 2010, 6(7), 2532–2539 CrossRef CAS.
- E. W. L. Chan and M. N. Yousaf, A photo-electroactive surface strategy for immobilizing ligands in patterns and gradients for studies of cell polarization, Mol. BioSyst., 2008, 4(7), 746–753 RSC.
- E.-J. Lee, E. W. L. Chan and M. N. Yousaf, Spatio-temporal control of cell coculture interactions on surfaces, ChemBioChem, 2009, 10(10), 1648–1653 CrossRef CAS.
- B. M. Lamb, S. Park and M. N. Yousaf, Microfluidic permeation printing of self-assembled monolayer gradients on surfaces for chemoselective ligand immobilization applied to cell adhesion and polarization, Langmuir: The ACS Journal of Surfaces and Colloids, 2010, 26(15), 12817–12823 CAS.
- M. Gonen-Wadmany, R. Goldshmid and D. Seliktar, Biological and mechanical implications of PEGylating proteins into hydrogel biomaterials, Biomaterials, 2011, 32(26), 6025–33 CAS.
- B. Williams, Mechanical influences on vascular smooth muscle cell function, J. Hypertens., 1998, 16(12 Pt 2), 1921–9 CrossRef CAS.
- A. Uchida, et al., The effect of mechanical stress on cultured growth cartilage cells, Connect. Tissue Res., 1988, 17(4), 305–11 CrossRef CAS.
- J. P. Califano and C. A. Reinhart-King, Exogenous and endogenous force regulation of endothelial cell behavior, J. Biomech., 43(1), 79–86 CrossRef.
- A. J. Engler, et al., Matrix elasticity directs stem cell lineage specification, Cell, 2006, 126(4), 677–89 CrossRef CAS.
- A. J. Engler, et al., Myotubes differentiate optimally on substrates with tissue-like stiffness: pathological implications for soft or stiff microenvironments, J. Cell Biol., 2004, 166(6), 877–87 CrossRef CAS.
- S. R. Peyton, et al., The effects of matrix stiffness and RhoA on the phenotypic plasticity of smooth muscle cells in a 3-D biosynthetic hydrogel system, Biomaterials, 2008, 29(17), 2597–607 CrossRef CAS.
- S. R. Peyton, et al., The use of poly(ethylene glycol) hydrogels to investigate the impact of ECM chemistry and mechanics on smooth muscle cells, Biomaterials, 2006, 27(28), 4881–93 CrossRef CAS.
- C. B. Khatiwala, et al., ECM compliance regulates osteogenesis by influencing MAPK signaling downstream of RhoA and ROCK, J. Bone Miner. Res., 2009, 24(5), 886–98 CrossRef CAS.
- S. R. Peyton, et al., The emergence of ECM mechanics and cytoskeletal tension as important regulators of cell function, Cell Biochem. Biophys., 2007, 47(2), 300–20 CrossRef CAS.
- C. M. Lo, et al., Cell movement is guided by the rigidity of the substrate, Biophys. J., 2000, 79(1), 144–52 CrossRef CAS.
- M. T. Thompson, et al., Biochemical functionalization of polymeric cell substrata can alter mechanical compliance, Biomacromolecules, 2006, 7(6), 1990–1995 CrossRef CAS.
- S. R. Peyton and A. J. Putnam, Extracellular matrix rigidity governs smooth muscle cell motility in a biphasic fashion, J. Cell. Physiol., 2005, 204(1), 198–209 CrossRef CAS.
- C. B. Khatiwala, S. R. Peyton and A. J. Putnam, Intrinsic mechanical properties of the extracellular matrix affect the behavior of pre-osteoblastic MC3T3-E1 cells, Am. J. Physiol.: Cell Physiol., 2006, 290(6), C1640–50 CrossRef CAS.
- K. M. Stroka and H. Aranda-Espinoza, Neutrophils display biphasic relationship between migration and substrate stiffness, Cell Motil. Cytoskeleton, 2009, 66(6), 328–41 CrossRef CAS.
- T. A. Ulrich, E. M. de Juan Pardo and S. Kumar, The mechanical rigidity of the extracellular matrix regulates the structure, motility, and proliferation of glioma cells, Cancer Res., 2009, 69(10), 4167–74 CrossRef CAS.
- J. Y. Wong, et al., Directed movement of vascular smooth muscle cells on gradient-compliant hydrogels, Langmuir, 2003, 19(5), 1908–1913 CrossRef CAS.
- J. A. Burdick, A. Khademhosseini and R. Langer, Fabrication of gradient hydrogels using a microfluidics/photopolymerization process, Langmuir, 2004, 20(13), 5153–5156 CrossRef CAS.
- N. Zaari, et al., Photopolymerization in microfluidic gradient generators: Microscale control of substrate compliance to manipulate cell response, Adv. Mater., 2004, 16(23–24), 2133–+ CrossRef CAS.
- B. C. Isenberg, et al., Vascular Smooth Muscle Cell Durotaxis Depends on Substrate Stiffness Gradient Strength, Biophys. J., 2009, 97(5), 1313–1322 CrossRef CAS.
- F. Grinnell and W. M. Petroll, Cell Motility and Mechanics in Three-Dimensional Collagen Matrices, Annu. Rev. Cell Dev. Biol Search PubMed.
- M. H. Zaman, et al., Computational model for cell migration in three-dimensional matrices, Biophys. J., 2005, 89(2), 1389–97 CrossRef CAS.
- E. L. Baker and M. H. Zaman, The biomechanical integrin, J. Biomech., 43(1), 38–44 CrossRef.
- J. Shih and R. Keller, Patterns of cell motility in the organizer and dorsal mesoderm of Xenopus laevis, Development, 1992, 116(4), 915–30 CAS.
- S. Chien, et al., Molecular basis of mechanical modulation of endothelial cell migration, Front. Biosci., 2005, 10, 1985–2000 CrossRef CAS.
- T. Shemesh, et al., Focal adhesions as mechanosensors: a physical mechanism, Proc. Natl. Acad. Sci. U. S. A., 2005, 102(35), 12383–8 CrossRef CAS.
- B. Geiger and A. Bershadsky, Exploring the neighborhood: adhesion-coupled cell mechanosensors, Cell, 2002, 110(2), 139–42 CrossRef CAS.
- T. Mitchison and M. Kirschner, Cytoskeletal dynamics and nerve growth, Neuron, 1988, 1(9), 761–72 CrossRef CAS.
- C. E. Chan and D. J. Odde, Traction dynamics of filopodia on compliant substrates, Science, 2008, 322(5908), 1687–91 CrossRef CAS.
- A. N. Stachowiak, et al., Bioactive hydrogels with an ordered cellular structure combine interconnected macroporosity and robust mechanical properties, Adv. Mater., 2005, 17(4), 399–403 CrossRef CAS.
- S. Nemir, H. N. Hayenga and J. L. West, PEGDA hydrogels with patterned elasticity: Novel tools for the study of cell response to substrate rigidity, Biotechnol. Bioeng., 105(3), 636–44 CrossRef CAS.
- L. G. Griffith and M. A. Swartz, Capturing complex 3D tissue physiology in vitro, Nat. Rev. Mol. Cell Biol., 2006, 7(3), 211–24 CrossRef CAS.
- K. S. Smalley, M. Lioni and M. Herlyn, Life isn't flat: taking cancer biology to the next dimension, In Vitro Cell. Dev. Biol.: Anim., 2006, 42(8–9), 242–7 CrossRef CAS.
- I. Stamenkovic, Matrix metalloproteinases in tumor invasion and metastasis, Semin. Cancer Biol., 2000, 10(6), 415–433 CrossRef CAS.
- P. Friedl and K. Wolf, Proteolytic interstitial cell migration: a five-step process, Cancer metastasis reviews, 2009, 28(1–2), 129–135 Search PubMed.
- K. Wolf and P. Friedl, Molecular mechanisms of cancer cell invasion and plasticity, Br. J. Dermatol., 2006, 154(Suppl 1), 11–15 CrossRef CAS.
- M. H. Zaman, P. Matsudaira and D. A. Lauffenburger, Understanding effects of matrix protease and matrix organization on directional persistence and translational speed in three-dimensional cell migration, Ann. Biomed. Eng., 2007, 35(1), 91–100 CrossRef.
- B. T. Burgess, J. L. Myles and R. B. Dickinson, Quantitative analysis of adhesion-mediated cell migration in three-dimensional gels of RGD-grafted collagen, Ann. Biomed. Eng., 2000, 28(1), 110–118 CrossRef CAS.
- G. P. Raeber, M. P. Lutolf and J. A. Hubbell, Molecularly engineered PEG hydrogels: a novel model system for proteolytically mediated cell migration, Biophys. J., 2005, 89(2), 1374–88 CrossRef CAS.
- H.-D. Kim, et al., Epidermal Growth Factor-induced Enhancement of Glioblastoma Cell Migration in 3D Arises from an Intrinsic Increase in Speed But an Extrinsic Matrix- and Proteolysis-dependent Increase in Persistence, Mol. Biol. Cell, 2008, 19(10), 4249–4259 CrossRef CAS.
- S. Saffarian, et al., Interstitial collagenase is a Brownian ratchet driven by proteolysis of collagen, Science, 2004, 306(5693), 108–111 CrossRef CAS.
- C. M. Ghajar, et al., A novel three-dimensional model to quantify metastatic melanoma invasion, Mol. Cancer Ther., 2007, 6(2), 552–61 CrossRef CAS.
- M. P. Lutolf and J. A. Hubbell, Synthetic biomaterials as instructive extracellular microenvironments for morphogenesis in tissue engineering, Nat. Biotechnol., 2005, 23(1), 47–55 CrossRef CAS.
- G. P. Raeber, M. P. Lutolf and J. A. Hubbell, Mechanisms of 3-D migration and matrix remodeling of fibroblasts within artificial ECMs, Acta Biomater., 2007, 3(5), 615–629 CrossRef CAS.
- J. S. Miller, et al., Bioactive hydrogels made from step-growth derived PEG-peptide macromers, Biomaterials, 2010, 31(13), 3736–3743 CrossRef CAS.
- S.-H. Lee, et al., Poly(ethylene glycol) hydrogels conjugated with a collagenase-sensitive fluorogenic substrate to visualize collagenase activity during three-dimensional cell migration, Biomaterials, 2007, 28(20), 3163–3170 CrossRef CAS.
- K. M. Galler, et al., Self-assembling multidomain peptide hydrogels: designed susceptibility to enzymatic cleavage allows enhanced cell migration and spreading, J. Am. Chem. Soc., 2010, 132(9), 3217–3223 CrossRef CAS.
- L. M. Coussens, B. Fingleton and L. M. Matrisian, Matrix metalloproteinase inhibitors and cancer: trials and tribulations, Science, 2002, 295(5564), 2387–2392 CrossRef CAS.
- F. Sabeh, R. Shimizu-Hirota and S. J. Weiss, Protease-dependent versus-independent cancer cell invasion programs: three-dimensional amoeboid movement revisited, J. Cell Biol., 2009, 185(1), 11–19 CrossRef CAS.
- P. Friedl, K. S. Zanker and E. B. Brocker, Cell migration strategies in 3-D extracellular matrix: differences in morphology, cell matrix interactions, and integrin function, Microsc. Res. Tech., 1998, 43(5), 369–78 CrossRef CAS.
- G. G. Martins and J. Kolega, Endothelial cell protrusion and migration in three-dimensional collagen matrices, Cell Motil. Cytoskeleton, 2006, 63(2), 101–15 CrossRef.
- S. I. Fraley, et al., A distinctive role for focal adhesion proteins in three-dimensional cell motility, Nat. Cell Biol Search PubMed.
- B. A. Harley, et al., Microarchitecture of three-dimensional scaffolds influences cell migration behavior via junction interactions, Biophys. J., 2008, 95(8), 4013–24 CrossRef CAS.
- A. D. Doyle, et al., One-dimensional topography underlies three-dimensional fibrillar cell migration, J. Cell Biol., 2009, 184(4), 481–90 CrossRef CAS.
- Y. Liu, et al., Control of cell migration in two and three dimensions using substrate morphology, Exp. Cell Res., 2009, 315(15), 2544–57 CrossRef CAS.
- M. Ochsner, et al., Dimensionality controls cytoskeleton assembly and metabolism of fibroblast cells in response to rigidity and shape, PLoS One, 2010, 5(3), e9445 Search PubMed.
- J. da Silva, et al., The cavity-to-cavity migration of leukaemic cells through 3D honey-combed hydrogels with adjustable internal dimension and stiffness, Biomaterials, 2010, 31(8), 2201–2208 CrossRef CAS.
- A. N. Stachowiak, et al., Bioactive hydrogels with an ordered cellular structure combine interconnected macroporosity and robust mechanical properties, Adv. Mater., 2005, 17(4), 399–+ CrossRef CAS.
- A. N. Stachowiak and D. J. Irvine, Inverse opal hydrogel-collagen composite scaffolds as a supportive microenvironment for immune cell migration, J. Biomed. Mater. Res., Part A, 2008, 85A(3), 815–828 CrossRef CAS.
- S. R. Peyton, et al., Marrow-Derived stem cell motility in 3D synthetic scaffold is governed by geometry along with adhesivity and stiffness, Biotechnol. Bioeng., 2011 Search PubMed.
- A. Martins, et al., Electrospun nanostructured scaffolds for tissue engineering applications, Nanomedicine, 2007, 2(6), 929–42 CrossRef.
- D. B. Wolfe, D. Qin and G. M. Whitesides, Rapid prototyping of microstructures by soft lithography for biotechnology, Methods Mol. Biol., 2010, 583, 81–107 CAS.
- M. G. Li, X. Y. Tian and X. B. Chen, A brief review of dispensing-based rapid prototyping techniques in tissue scaffold fabrication: role of modeling on scaffold properties prediction, Biofabrication, 2009, 1(3), 032001 CrossRef CAS.
- S. M. Peltola, et al., A review of rapid prototyping techniques for tissue engineering purposes, Ann. Med., 2008, 40(4), 268–80 CrossRef CAS.
- T. C. Lim, K. S. Chian and K. F. Leong, Cryogenic prototyping of chitosan scaffolds with controlled micro and macro architecture and their effect on in vivo neo-vascularization and cellular infiltration, Journal of biomedical materials research. Part A, 2010, 94(4), 1303–11 Search PubMed.
- S. Park, et al., 3D polycaprolactone scaffolds with controlled pore structure using a rapid prototyping system, J. Mater. Sci.: Mater. Med., 2009, 20(1), 229–34 CrossRef CAS.
- K. Mi, et al., Influence of a self-assembling peptide, RADA16, compared with collagen I and Matrigel on the malignant phenotype of human breast-cancer cells in 3D cultures and in vivo, Macromol. Biosci., 2009, 9(5), 437–43 CrossRef CAS.
- A. Horii, et al., Biological designer self-assembling peptide nanofiber scaffolds significantly enhance osteoblast proliferation, differentiation and 3-D migration, PLoS One, 2007, 2(2), e190 Search PubMed.
- J. Park, et al., TiO2 Nanotube Surfaces: 15 nm—An Optimal Length Scale of Surface Topography for Cell Adhesion and Differentiation, Small, 2009, 5(6), 666–671 CrossRef CAS.
- K. S. Brammer, et al., Enhanced cellular mobility guided by TiO2 nanotube surfaces, Nano Lett., 2008, 8(3), 786–93 CrossRef CAS.
- D. Sengupta and S. C. Heilshorn, Protein-Engineered Biomaterials: Highly Tunable Tissue Engineering Scaffolds, Tissue Eng., Part B: Rev., 2010, 16(3), 285–293 CrossRef CAS.
- S. H. Lee, J. J. Moon and J. L. West, Three-dimensional micropatterning of bioactive hydrogels via two-photon laser scanning photolithography for guided 3D cell migration, Biomaterials, 2008, 29(20), 2962–8 CrossRef CAS.
- C. M. Nelson, et al., Tissue geometry determines sites of mammary branching morphogenesis in organotypic cultures, Science, 2006, 314(5797), 298–300 CrossRef CAS.
- N. Gjorevski and C. M. Nelson, Endogenous patterns of mechanical stress are required for branching morphogenesis, Integr. Biol., 2(9), 424–34 RSC.
- A. L. Pavlovich, S. Manivannan and C. M. Nelson, Adipose Stroma Induces Branching Morphogenesis of Engineered Epithelial Tubules, Tissue Eng Part A Search PubMed.
- R. J. Pelham, Jr. and Y. Wang, Cell locomotion and focal adhesions are regulated by substrate flexibility, Proc. Natl. Acad. Sci. U. S. A., 1997, 94(25), 13661–5 CrossRef.
- B. Sabass, et al., High resolution traction force microscopy based on experimental and computational advances, Biophys. J., 2008, 94(1), 207–20 CrossRef CAS.
- S. Sen, A. J. Engler and D. E. Discher, Matrix Strains Induced by Cells: Computing How Far Cells Can Feel, Cell. Mol. Bioeng., 2009, 2(1), 39–48 CrossRef.
- J. P. Califano and C. A. Reinhart-King, Exogenous and endogenous force regulation of endothelial cell behavior, J. Biomech., 2010, 43(1), 79–86 CrossRef.
- C. T. Mierke, et al., Contractile forces in tumor cell migration, Eur. J. Cell Biol., 2008, 87(8–9), 669–76 CrossRef CAS.
- C. Raupach, et al., Stress fluctuations and motion of cytoskeletal-bound markers, Phys. Rev. E: Stat., Nonlinear, Soft Matter Phys., 2007, 76(1 Pt 1), 011918 CrossRef.
- J. A. Zimberlin, P. Wadsworth and A. J. Crosby, Living microlens arrays, Cell Motil. Cytoskeleton, 2008, 65(9), 762–7 CrossRef.
- G. Miquelard-Garnier, et al., Polymer microlenses for quantifying cell sheet mechanics, Soft Matter, 2010, 6(2), 398–403 RSC.
- E. J. Joslin, et al., EGF-receptor-mediated mammary epithelial cell migration is driven by sustained ERK signaling from autocrine stimulation, J. Cell Sci., 2007, 120(Pt 20), 3688–99 CrossRef CAS.
- D. Lehnert, et al., Cell behaviour on micropatterned substrata: limits of extracellular matrix geometry for spreading and adhesion, J. Cell Sci., 2004, 117(Pt 1), 41–52 CrossRef CAS.
- M. S. Hahn, et al., Photolithographic patterning of polyethylene glycol hydrogels, Biomaterials, 2006, 27(12), 2519–24 CrossRef CAS.
- K. A. Beningo and Y. L. Wang, Flexible substrata for the detection of cellular traction forces, Trends Cell Biol., 2002, 12(2), 79–84 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2012 |
