A sensitive graphene oxide–DNA based sensing platform for fluorescence “turn-on” detection of bleomycin†
Feng
Li
ab,
Yan
Feng
b,
Can
Zhao
b,
Peng
Li
b and
Bo
Tang
*a
aCollege of Chemistry, Chemical Engineering and Materials Science, Key Laboratory of Molecular and Nano Probes, Ministry of Education, Shandong Normal University, Jinan 250014, China. E-mail: tangb@sdnu.edu.cn; Fax: +86-531-86180017
bCollege of Chemistry and Molecular Engineering, Qingdao University of Science and Technology, Qingdao 266042, China
First published on 3rd November 2011
Abstract
A sensitive and selective fluorescent sensing platform for bleomycin (BLM) was developed based on BLM-induced DNA strand scission and the difference in affinity of graphene oxide for single-stranded DNA containing different numbers of bases in length.
Cancer is a generic term for a large group of diseases which result from the metastases of abnormal cells to any part of the body. It accounted for 7.6 million deaths (around 13% of all deaths) in 2008 and the deaths from cancer worldwide are projected to continue rising, with an estimated 11 million deaths in 2030.1 The usage of antitumor drugs is a major protocol in curing the disease or considerably prolonging life while improving the patient's quality of life. Among them, bleomycins (BLMs), a family of naturally occurring glycopeptide-derived antibiotics, have been widely used as an available chemotherapeutic agent against certain cancers, including Hodgkin's disease, non-Hodgkin lymphoma, testicular cancer and malignant pleural effusions.2
The therapeutic activity of the BLM is generally believed to correlate with its ability to bind to and degrade DNA with the combination of oxygen and a metal ion in a low oxidation state.3 Meanwhile, BLM is less toxic to the human body due to its specific advantages of low myelosuppression and low immunosuppression.4 However, the remarkable dose-limiting side effect of BLM is the potential for pneumonitis and lung fibrosis.5Fever, rigors and skin toxicity may occur as well. To achieve the best therapeutic effect and weaken the toxicity of BLM, in the past decades, researchers have developed numerous reliable and sensitive methods for BLM detection, including high-performance liquid chromatography (HPLC), radioimmunoassay (RIA), enzyme immunoassay, microbiological assay and fluorescent methods.6 Recently, our group established a simple colorimetric sensing platform for trace BLM detection with gold nanoparticles as the sensing element.7BLM was found to induce the aggregation of gold nanoparticles, revealing a visible red-to-blue color change. With the in-depth investigation into the interaction between BLM and DNA, DNA has become a powerful tool in BLM sensing. For example, Yin et al. proposed an electrochemical detection assay for BLM based on BLM-induced DNA strand scission.8 However, less attention has been paid to applying this specific reaction in BLM detection via other protocols, such as fluorescent methods. Owing to the features of high sensitivity, facile operation, on-site and real-time detection, fluorescent sensors have been extensively explored in various applications.9
Graphene oxide (GO), a water-soluble derivative of graphene, is of great importance because of its unique characteristics such as good dispersibility and facile surface functionality.10 It is able to adsorb single-stranded DNA (ssDNA) via non-covalent π-stacking interactions between the hexagonal cells of graphene and the ring structure of nucleobases, while double-stranded DNA (dsDNA) cannot be adsorbed due to its more rigid structure.11 Based on the affinity difference, the GO–DNA complex has emerged as a novel bioassay platform for DNA and protein detection, metal ion sensing, etc.12 Very recently, Zhao et al. made a detailed investigation on the difference in affinity of GO for ssDNA containing different numbers of bases in length and proved that short ssDNA had weaker affinity to GO than long ssDNA.13 Besides the above unique properties, GO is a fluorescence (FL) superquencher due to its long-range nanoscale energy transfer property.14
Herein, a simple and sensitive GO–DNA complex-based sensing platform was constructed for FL “turn-on” detection of BLM. FAM-labeled long ssDNA (F-ssDNA) undergoes an irreversible cleavage event under the oxidative effect of BLM with Fe(II) as a cofactor, releasing the FAM-linked DNA fragment from the GO surface due to the weakened affinity, thus resulting in the restoration of FL of the sensing system. To the best of our knowledge, it is the first attempt to apply the specific reaction in BLM detection using a fluorescent method.
Scheme 1 depicts GO–DNA complex-based fluorescent detection of BLMviaDNA strand scission. The 16-mer F-ssDNA oligonucleotide bound strongly to a GO sheet to form a self-assembled complex via π-stacking interactions, thus GO significantly quenched the FL of FAM due to the close proximity to GO. In the presence of both a metal ion in a low oxidation state and oxygen, BLM is capable of recognizing and noncovalently binding to specific sequences in DNA and then inducing strand scission in both ssDNA and dsDNA, with Fe(II) being the most extensively studied and most active.15 The generally observed selectivity is for scission at pyrimidines (T or C) located at the 3′-position with respect to a guanine (G).16 It is generally accepted that BLM·Fe(II) binds with oxygen to form BLM·Fe(III)OOH which is responsible for abstraction of a hydrogen from the C4′-position of the deoxyribose sugar of DNA. As a result, upon addition of BLM·Fe(II), the 16-mer F-ssDNA underwent an irreversible strand scission at the scission site indicated by the black arrow, releasing a 3-mer FAM-linked oligonucleotide fragment from the GO surface, which was responsible for FL restoration of the sensing system.
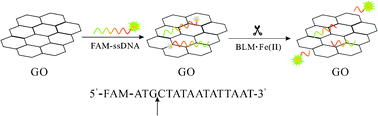 | ||
| Scheme 1 Schematic illustration of GO–DNA complex-based fluorescent detection of BLMviaDNA strand scission. | ||
In order to demonstrate the feasibility of our strategy, BLM·Fe(II)-induced FL response of the GO–DNA complex was recorded. As shown in Fig. 1, in the absence of GO, the F-ssDNA exhibits strong FL emission that can be attributed to the presence of the fluorescein-based dye (a). However, the addition of GO results in significant quenching of the FL emission (b) due to the long-range energy transfer between GO and the fluorescein. Upon incubation with BLM·Fe(II), one can observe a significant FL enhancement of the GO–DNA complex, indicating the occurrence of BLM·Fe(II)-induced strand scission in the F-ssDNA. The released 3-mer FAM-linked ssDNA is too short to be adsorbed on the GO, resulting in the FL restoration of FAM. It is worthy to point out that BLM·Fe(II) scarcely influences the FL of the free F-ssDNA in the absence of GO. A similar phenomenon also has been observed in a Fenton-like reaction regulated oxidative DNA damage investigation, using the GO–DNA complex as the sensing platform as well.17
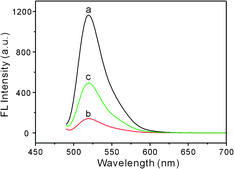 | ||
| Fig. 1 FL emission spectra of F-ssDNA (40 nM) under different conditions: (a) F-ssDNA, (b) F-ssDNA + 20 μg mL−1 GO and (c) F-ssDNA + 20 μg mL−1 GO + 1.0 μM BLM·Fe(II). | ||
To achieve the best sensing performance, the concentration of GO was optimized, with the concentration of F-ssDNA being fixed at 40 nM (Fig. S1 in ESI†). The FL intensity change (F0 − F)/F0 increases gradually with increasing the concentration of GO from 0 to 12 μg mL−1 and then changes sharply with further increasing GO up to 16 μg mL−1, finally gives a slight increase when it is above 18 μg mL−1. F0 and F are the FL intensity of F-ssDNA at 520 nm in the absence and presence of GO, respectively. The FL of F-ssDNA is efficiently quenched by 20 μg mL−1 GO, suggesting the formation of the GO–DNA complex. Since a transitional metal ion is usually required during the strand scission process, an experiment was performed to investigate the effect of different metal ions on DNA scission (Fig. S2 in ESI†). The BLM samples were prepared by mixing 500 nM BLM with different metal ions in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 molar ratio. Obviously, Fe(II) is the most active metal ion in combination with BLM for pronounced FL restoration. Moreover, it is reported that BLM·Fe(II) is significantly less toxic than other metal ions coordinated BLM complexes at therapeutically effective concentrations in vivo.18 A control experiment in which the GO–DNA complex was treated only with Fe(II) was performed and the negligible FL enhancement demonstrated the necessity of BLM.
1 molar ratio. Obviously, Fe(II) is the most active metal ion in combination with BLM for pronounced FL restoration. Moreover, it is reported that BLM·Fe(II) is significantly less toxic than other metal ions coordinated BLM complexes at therapeutically effective concentrations in vivo.18 A control experiment in which the GO–DNA complex was treated only with Fe(II) was performed and the negligible FL enhancement demonstrated the necessity of BLM.
Fig. 2 shows the FL spectra of the GO–DNA complex upon incubation with different concentrations of BLM·Fe(II). The FL intensity increases dramatically with increasing the concentration of BLM·Fe(II) from 0.5 nM to 10 μM, suggesting that the GO–DNA complex is effective for FL “turn-on” detection of BLMvia BLM·Fe(II)-induced DNA strand scission. An abnormal phenomenon is observed that the FL intensity decreases when the concentration of BLM·Fe(II) is above 10 μM (data not shown). It is probable that the accumulation of Fe(III) after oxidation of Fe(II) by oxygen potentially inhibits the degradation of DNA in the system. The inset indicates that the FL intensity change (F/F0 − 1) of the sensing system varies linearly over the concentration of BLM from 5.0 nM to 1.0 μM, and a detection limit of 0.2 nM was estimated based on 3σ, where F0 and F are the FL intensities of the GO–DNA complex before and after addition of BLM·Fe(II), respectively. The linear concentration range is wider than those of some HPLC and RIA-based analysis.6a,b The detection limit is lower than those of the aforementioned traditional methods,6,7 and is comparable to that of the electrochemical assay.8
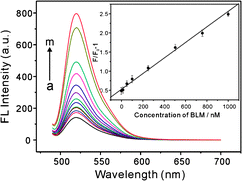 | ||
Fig. 2
FL
spectra of GO–DNA complex upon incubation with different concentrations of BLM·Fe(II): (a) 0, (b) 0.5 nM, (c) 1.0 nM, (d) 5.0 nM, (e) 10 nM, (f) 50 nM, (g) 100 nM, (h) 250 nM, (i) 500 nM, (j) 750 nM, (k) 1000 nM, (l) 5000 nM and (m) 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 nM. (inset) FL intensity ratio of the GO–DNA complex with {(F/F0 − 1)} plotted against the concentration of BLM·Fe(II). 000 nM. (inset) FL intensity ratio of the GO–DNA complex with {(F/F0 − 1)} plotted against the concentration of BLM·Fe(II). | ||
To further test the effect of BLM·Fe(II) on the restoration of the FL of the GO–DNA complex, the kinetic behavior of the reaction between BLM·Fe(II) and DNA was examined. Fig. 3 shows the time-dependent FL responses of the GO–DNA complex upon addition of different concentrations of BLM·Fe(II). Obviously, BLM·Fe(II) induces dramatic FL enhancement at the early stages of the reaction and reaches a plateau in less than 5 min, revealing a fast reaction kinetics. Moreover, the reaction is almost complete within 30 s with BLM·Fe(II) at high concentrations. Thus the FL measurements were performed after a 5 min incubation of the GO–DNA complex with BLM·Fe(II). Moreover, no obvious FL increase is obtained in the absence of BLM·Fe(II), indicating that the FL enhancement is indeed induced by BLM·Fe(II).
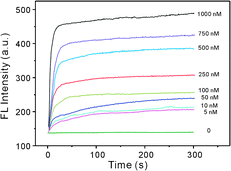 | ||
| Fig. 3 Kinetics study for FL changes of the GO–DNA complex upon incubation with various concentrations of BLM·Fe(II). | ||
The selectivity of the fluorescent biosensor for BLM was evaluated by monitoring the FL responses of the GO–DNA complex upon addition of a mixture of BLM·Fe(II) and some coexisting substances, including hydrogen peroxide (H2O2), urea, dopamine (DA), ascorbate (AA), glucose, cysteine (Cys), and human serum albumin (HSA), all of which are kept at a concentration of 500 nM (see Fig. S3 in ESI†). All the coexisting substances cause little FL intensity changes of the sensing system, indicating that they have no significant effect on the conformational change or cleavage of F-ssDNA. Although it has been reported that some reducing agents, such as AA and H2O2, are capable of acting with BLM to induce efficient degradation of DNA, on a molar basis, Fe(II) is far more effective than the reducing agents to degrade DNA.19 Thus it is reasonable to achieve the above results. The selectivity of the biosensor for BLM is satisfactory. To investigate the analytical capability of the BLM fluorescent biosensor, three fresh serum samples supplied by the Affiliated Hospital of Qingdao University were 2-fold diluted by PBS and then pre-doped with 100 nM BLM·Fe(II) before mixing with the GO–DNA complex, respectively. The recovery was tested by the standard addition method (see Table S1 in ESI†). The recoveries for the determination of BLM are in the range from 96.0% to 103%, indicating that the proposed fluorescent biosensor for BLM can be used in real sample analysis.
In conclusion, on the basis of BLM-induced DNA strand scission and the difference in affinity of GO for ssDNA containing different numbers of bases in length, a FL “turn-on” biosensor was developed for BLM detection. F-ssDNA was adsorbed on the surface of GO via π-stacking interactions, resulting in the quenching of FL due to energy transfer between F-ssDNA and GO. Upon addition of BLM·Fe(II), F-ssDNA underwent an irreversible cleavage event, releasing the shortened FAM-linked DNA fragment from the GO surface for the restoration of FL. BLM-induced DNA strand scission was complete within 5 min, which contributed to the sensitive detection of BLM. Moreover, some coexisting substances in body fluids had negligible influence on BLM detection, and the recoveries of serum samples analysis were satisfactory.
This work was supported by the National Natural Science Funds for Distinguished Young Scholar (No. 20725518), the National Key Natural Science Foundation of China (No. 21035003), the National Natural Science Foundation of China (No. 21175076), and the Science Foundation of China Postdoctor (Nos. 20100471568, 201104648).
Notes and references
- http://www.who.int/mediacentre/factsheets/fs297/en/index.html .
- (a) H. Umezawa, in Anticancer Agents Based on Natural Product Models, ed. J. M. Cassady and J. D. Douros, Academic Press, New York, 1980, p. 147 Search PubMed; (b) S. M. Hecht, in Cancer Chemotherapeutic Agents, ed. W. O. Foye, American Chemical Society, Washington D.C., 1995, p. 369 Search PubMed; (c) J. S. Lazo and B. A. Chabner, in Cancer Chemotherapy and Biotherapy; Principles and Practice, ed. B. A. Chabner and D. L. Longo, Lippincott-Raven Publishers, Philadelphia, 1996, 2nd edn, p. 379 Search PubMed; (d) U. Galm, M. H. Hager, S. G. Van Lanen, J. Ju, J. S. Thorson and B. Shen, Chem. Rev., 2005, 105, 739 CrossRef CAS.
- (a) S. M. Hecht, Acc. Chem. Res., 1986, 19, 383 CrossRef CAS; (b) J. Stubbe and J. W. Kozarich, Chem. Rev., 1987, 87, 1107 CrossRef CAS.
- G. Tisman, V. Herbert, L. T. Go and L. Brenner, Blood, 1973, 41, 721 CAS.
- J. Hay, S. Shahzeidi and G. Laurent, Arch. Toxicol., 1991, 65, 81 CrossRef CAS.
- (a) Q. M. Chen, Y. Shen and Q. W. Zhang, J. Tianjin Med. Univ., 2000, 6, 156 Search PubMed; (b) J. Yan, J. Zhu and C. S. Li, China Pharm., 2003, 12, 40 Search PubMed; (c) K. Fujiwara, M. Isobe, H. Saikusa, H. Nakamura, T. Kitagawa and S. Takahashi, Cancer Treat. Rep., 1983, 67, 363 CAS; (d) T. Onuma, J. F. Holland, H. Masuda, J. A. Waligunda and G. A. Goldberg, Cancer, 1974, 33, 1230 CrossRef CAS; (e) J. T. Liu, Z. F. Liu, X. L. Hu, L. Kong and S. P. Liu, Luminescence, 2008, 23, 1–6 CrossRef CAS.
- F. Li, Y. Feng, C. Zhao and B. Tang, Biosens. Bioelectron., 2011, 26, 4628 CrossRef CAS.
- B. C. Yin, D. Wu and B. C. Ye, Anal. Chem., 2010, 82, 8272 CrossRef CAS.
- (a) C. W. Liu, C. C. Huang and H. T. Chang, Anal. Chem., 2009, 81, 2383 CrossRef CAS; (b) N. Nagraj, J. W. Liu, S. Sterling, J. Wu and Y. Lu, Chem. Commun., 2009, 4103 RSC; (c) H. H. Wang, L. Xue and H. Jiang, Org. Lett., 2011, 13, 3844 CrossRef CAS.
- Z. Liu, J. T. Robinson, X. M. Sun and H. J. Dai, J. Am. Chem. Soc., 2008, 130, 10876 CrossRef CAS.
- (a) N. Mohanty and V. Berry, Nano Lett., 2008, 8, 4469 CrossRef CAS; (b) C. H. Lu, H. H. Yang, C. L. Zhu, X. Chen and G. N. Chen, Angew. Chem., Int. Ed., 2009, 48, 785.
- (a) L. Tang, Y. Wang and J. Li, ACS Nano, 2011, 5, 3817 CrossRef CAS; (b) H. F. Dong, W. C. Gao, F. Yan, H. X. Ji and H. X. Ju, Anal. Chem., 2010, 82, 5511 CrossRef CAS; (c) Y. Q. Wen, F. F. Xing, S. J. He, S. P. Song, L. H. Wang, Y. T. Long, D. Li and C. H. Fan, Chem. Commun., 2010, 46, 2596 RSC.
- X. H. Zhao, R. M. Kong, X. B. Zhang, H. M. Meng, W. N. Liu, W. H. Tan, G. L. Shen and R. Q. Yu, Anal. Chem., 2011, 83, 5062 CrossRef CAS.
- R. S. Swathi and K. L. Sebastian, J. Chem. Phys., 2008, 129, 054703 CrossRef CAS.
- M. Takeshita, A. P. Grollman, E. Ohtsubo and H. Ohtsubo, Proc. Natl. Acad. Sci. U. S. A., 1978, 75, 7983.
- (a) L. F. Povirk, Y. H. Han and R. J. Steighner, Biochemistry, 1989, 28, 5808 CrossRef CAS; (b) Q. Ma, Y. Akiyama, Z. Xu, K. Konishi and S. M. Hecht, J. Am. Chem. Soc., 2009, 131, 2013 CrossRef CAS.
- M. Liu, Q. Zhang, H. M. Zhao, S. Chen, H. T. Yu, Y. B. Zhang and X. Quan, Chem. Commun., 2011, 47, 4084 RSC.
- E. A. Rao, L. A. Saryan, W. E. Antholine and D. H. Petering, J. Med. Chem., 1980, 23, 1310 CAS.
- E. A. Sausville, J. Peisach and S. R. Horwitz, Biochemistry, 1978, 17, 2470.
Footnote |
| † Electronic supplementary information (ESI) available: Detailed experimental procedures and results. See DOI: 10.1039/c1cc15694b |
| This journal is © The Royal Society of Chemistry 2012 |
