Lipid-coated nanocapillaries for DNA sensing†
Silvia
Hernández-Ainsa
a,
Christoph
Muus
b,
Nicholas A. W.
Bell
a,
Lorenz J.
Steinbock
ac,
Vivek V.
Thacker
a and
Ulrich F.
Keyser
*a
aCavendish Laboratory University of Cambridge, Department of Physics, JJ Thomson Avenue, Cambridge, CB3 0HE, UK. E-mail: ufk20@cam.ac.uk; Fax: +44 (0)1223 337000; Tel: +44 (0)1223337272
bUniversity of Heidelberg, Institute of Pharmacy and Molecular Biotechnology, Im Neuenheimer Feld 364, 69120 Heidelberg, Germany
cLaboratory of Nanoscale Biology, Institute of Bioengineering, School of Engineering, EPFL, 1015 Lausanne, Switzerland
First published on 5th November 2012
Abstract
We report a simple and efficient way to accomplish the chemical modification of glass nanopores by means of lipid self-assembly. Lipid coating improves the success rate of these glass nanopores as biosensors to detect λ-DNA.
Controlling the internal architecture inside non-biological nanopores is one of the major challenges in the development of more specific, robust and longer lasting biosensing devices. Although some elegant approaches such as the combination of hybrid biological and solid-state nanopores1 or the use of DNA platforms2,3 have been recently reported, chemical modification has been by far the most common way to customize man-made nanopores.4 Different chemical coating strategies have been used involving both covalent5,6 and electrostatic7,8 surface-chemistry. Molecular self-assembly provides an alternative route to simple and versatile coating. By means of a rationalized molecular design, supramolecular architectures with a required organization can be achieved.9 Of particular interest are self-assembled structures inspired by biological systems such as lipid bilayers.10 Biological membranes form the physical barrier between the interior of cells and their extracellular environments and play an important role in cellular structure and function. Lipid coating has been successfully employed by Mayer and colleagues in silicon nitride nanopores to create biosensors capable of detecting several proteins, including the amyloid-beta (Ab) peptide implicated in Alzheimer's disease.11,12 However, fabrication of silicon nitride nanopores is challenging and requires the use of a transmission electron microscope to ablate the surface. Nanopores from pulled glass capillaries represent a good alternative due to their lower cost (less than £1) and fast preparation time (less than 1 min).13,14 Inspired by these results, we adapted this lipid-coating procedure to nanopores constructed by using pulled-glass capillaries. In addition to the successful assembly of lipid bilayers on our glass nanopores, we show that these nanopores can be used to detect linear double-stranded λ-DNA by resistive-pulse sensing. Lipid coating is expected to yield nanopores with much better controlled surface properties and should prevent non-specific adsorption on the nanopore surface as shown earlier,11 thus increasing the success in λ-DNA detection as demonstrated in this paper.
Measurements were carried out in quartz nanocapillaries (made from quartz glass capillaries, Hilgenberg, Germany) assembled in polydimethylsiloxane (PDMS) cells fabricated following a previously described procedure.15 Briefly, a laser assisted pipette puller (Sutter P-2000) was used to draw down the diameter of quartz capillaries to a few tens of nanometers. The nanocapillary was glued into a PDMS mould connecting two fluid reservoirs. After the assembled device was plasma cleaned for 10 minutes, 150 mM KCl solution (10 mM HEPES, pH = 7.5) was added immediately to both reservoirs. Finally, the assembled cells were placed under vacuum to remove air bubbles in the capillaries. Silver wires (200 μm diameter, Advent) were chlorinated (Ag/AgCl) and inserted in both reservoirs. The nanocapillaries were tested for low noise and ionic current vs. voltage (IV) curves were recorded to estimate the diameter (see ESI†). Ionic current measurements were performed using an Axopatch 200B (Axon Instruments, USA) amplifier in voltage-clamp mode. All signals were digitized with an NI-PCIe-6251card (National Instruments, USA) and with Axoscope 10.2 software (Molecular Devices). Custom written LabVIEW (LabVIEW 8.6, National Instruments) programs were used to record and process the data. Data analysis was performed in Origin 8.5.
Small unilamellar vesicles (SUVs) were prepared by sonication of 2-oleoyl-1-palmitoyl-sn-glycero-3-phosphocholine (POPC) lipids in a 150 mM KCl (10 mM HEPES (pH = 7.5)) solution and further extruded through 30 nm polycarbonate membranes (Nuclepore Track-Etched Membranes, Whatman) according to the protocols of Avanti Polar Lipids, Inc.16 The mean diameter of the extruded POPC SUVs was 53 ± 12 nm, measured via dynamic light scattering (Malvern Zetasizer Nano ZS). The obtained mean SUV diameter was found to be appropriate to prevent the formation of spanned bilayers onto the nanopores.17
Lipid coating was performed by exposing the tip of the nanocapillary to 100 μL of the aqueous suspension of POPC small unilamellar vesicles (SUVs) for 5 min. This procedure allows SUVs to rupture and spontaneously spread onto the nanopore walls (Fig. 1a).18 Subsequently, the cell is immersed in a large volume of fresh buffer for 5–10 min to remove the excess of SUVs. Fresh buffer is added to both reservoirs before recording the ionic current of the lipid-coated nanopore. After lipid-coating, we always observe a reduction of the nanopore diameter as indicated by the lower slope detected in the IV curve of the coated pore compared to the bare nanopore (Fig. 1b). Taking into account the bilayer thickness of POPC lipids (3.7 ± 0.1 nm)19 and the thickness of the water layer on the quartz surface (1.0 ± 0.1 nm)20 a decrease of the nanopore diameter of 9.4 ± 0.4 nm is expected. The experimental value of the decrease in diameter obtained by fitting the distribution with a Gaussian function yields 12.5 ± 5.0 nm, which is in good agreement with the calculated diameter considering the variance in the diameter of the glass capillaries (Fig. 1b). Successful coating leads to changes in the ionic current power spectral density, showing an enhancement in the 1/f type low-frequency noise upon coating of the nanopore (see ESI†). This feature has been previously reported and is connected with the dynamic fluctuations produced by the coating.21
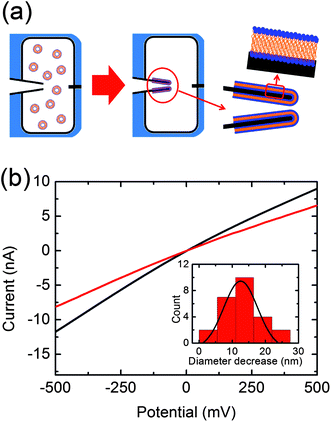 | ||
| Fig. 1 (a) Schematic representation of the lipid-coating process. (b) Current voltage curves recorded before (black) and after (red) lipid-coating. The inserted histogram represents the decrease of the nanopore diameter after lipid-coating in 25 nanocapillaries. A peak at 12.5 ± 5.0 nm is obtained by fitting the distribution with a Gaussian function. | ||
We used fluorescence microscopy to demonstrate successful spreading of the lipids coating the nanocapillary walls. POPC vesicles were prepared containing 1% mol of 1,2-diphytanoyl-sn-glycero-3-phosphoethanolamine-N-(7-nitro-2,1,3-benzoxadiazol-4-yl) (ammonium salt) (NBD-DPhPE) (λabs max = 460 nm). After performing the same coating procedure as previously described, nanocapillaries were imaged under a fluorescence microscope (λexc = 458 nm, λem = 535 nm). We observed almost uniform fluorescence on the nanocapillaries, indicating a successful coating (see ESI†).
After establishing a working coating protocol, we investigated the performance of these nanopores for resistive-pulse sensing. Linear double-stranded λ-DNA (48.5 kbp) is established as the gold standard to prove robust single-molecule sensing. When positive voltages are applied to the electrode placed in the back reservoir, negatively charged λ-DNA is expected to translocate into the nanocapillary due to electrophoretic force. We first perform control measurements by applying +500 mV in a DNA-free solution (500 mM KCl, 10 mM HEPES (pH = 7.5)) to show the voltage-stability of these lipid-coated pores as well as the absence of either λ-DNA or possible SUV translocation22 (upper panel, Fig. 2a). Upon addition of λ-DNA (1 nM) to the reservoir containing the lipid-coated nanopore, ionic current blockade events are observed (lower panel, Fig. 2a), a clear indication of λ-DNA translocations.15,23 Analyses of mean current blockage and duration are shown in Fig. 2b. Mean peak amplitude and duration values were calculated by fitting the corresponding histograms with a Gaussian peak function, yielding −76 pA and 1.4 ms respectively. As previously reported, higher event currents are observed for shorter durations.15,24
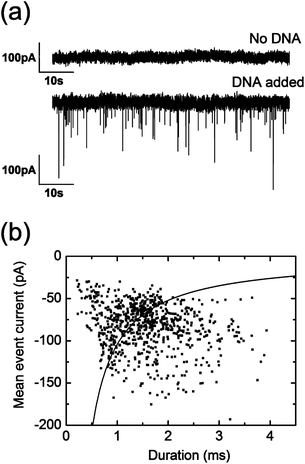 | ||
| Fig. 2 (a) Current trace of a lipid-coated pore when a potential of 500 mV is applied in the absence (upper panel) or presence (lower panel) of λ-DNA. (b) Scatter plot of the duration and mean peak amplitude of the 774 recorded λ-DNA events in the same lipid-coated pore described in (a) measured at 500 mM KCl (10 mM HEPES (pH = 7.5)) upon applying a potential of 500 mV. Black line fit is calculated as stated in ref. 24. | ||
Different λ-DNA folding states during translocation can be detected by using bare glass nanopores.15 Importantly, DNA folding can also be detected in our lipid-coated nanopores. Hence, when plotting the histogram of the whole current trace during λ-DNA translocations, several peaks can be observed (see ESI†), corresponding to different λ-DNA folding states.15,24
The most interesting characteristic of the lipid coating is its ability to decrease the non-specific interactions between the biomolecules and nanocapillary walls. The inherent variability between nanocapillaries due to surface differences can be eliminated with lipid coating, thus enhancing the robustness of sensing. Remarkably, our experiments show that the ratio of successful detection of λ-DNA is higher in lipid-coated nanopores (40%) when compared with non-coated ones (13%) (see ESI†). This increase in success rate is directly related to the better defined surface of the lipid-coated nanopores in comparison with the bare glass nanopores. One of the main reasons for the low success rate of detection in bare glass nanopores is the variable negative surface charge of quartz glass,25,26 which can produce electro-osmotic flow opposite to the direction of DNA translocation through the nanopore and prevent DNA from entering the sensing region.27
In summary, we have chemically modified glass nanocapillaries by means of lipid self-assembly. Both the decrease in the nanopore diameter and the increase in the 1/f-type low-frequency noise evidence the success of our lipid coating process. We have shown that lipid-coated nanopores can be used as single-molecule biosensors for efficient detection of λ-DNA. Lipid coating significantly reduces surface charge differences of glass nanopores and, as a consequence, single-molecule sensing success and robustness are more reproducible. Our work shows that lipid-coating significantly improves the characteristics of nanocapillaries, allowing for the design of specific nanopores with lipids bearing recognition sites with the aim to control molecular transport.28
Acknowledgements
SHA and UFK acknowledge support from an ERC starting grant. CM thanks the DAAD (German Academic Exchange Service) for a scholarship. NAWB was supported by the EPSRC. LJS was supported by an Emmy Noether grant from the Deutsche Forschungsgemeinschaft (DFG). VVT gratefully acknowledges funding from the Cambridge Commonwealth Trust and the Jawaharlal Nehru Memorial Trust. The authors thank Joanne L. Gornall for help with the SUV preparation.Notes and references
- A. R. Hall, A. Scott, D. Rotem, K. K. Mehta, H. Bayley and C. Dekker, Nat. Nanotechnol., 2010, 5, 874 CrossRef CAS.
- N. A. W. Bell, C. R. Engst, M. Ablay, G. Divitini, C. Ducati, T. Liedl and U. F. Keyser, Nano Lett., 2012, 12, 512 CrossRef CAS.
- R. Wei, T. G. Martin, U. Rant and H. Dietz, Angew. Chem., Int. Ed., 2012, 51, 1 CrossRef.
- M. Wanunu and A. Meller, Nano Lett., 2007, 7, 1580 CrossRef CAS.
- Y. Fu, H. Tokuhisa and L. A. Baker, Chem. Commun., 2009, 4877 RSC.
- B. Yameen, M. Ali, R. Neumann, W. Ensinger, W. Knoll and O. Azzaroni, Chem. Commun., 2010, 46, 1908 RSC.
- M. Ali, B. Yameen, J. Cervera, P. Ramírez, R. Neumann, W. Ensinger, W. Knoll and O. Azzaroni, J. Am. Chem. Soc., 2010, 132, 8338 CrossRef CAS.
- S. Umehara, N. Pourmand, C. D. Webb, R. W. Davis, K. Yasuda and M. Karhanek, Nano Lett., 2006, 6, 2486 CrossRef CAS.
- M. Muñoz-Úbeda, S. K. Misra, A. L. Barrán-Berdón, C. Aicart-Ramos, M. B. Sierra, J. Biswas, P. Kondaiah, E. Junquera, S. Bhattacharya and E. Aicart, J. Am. Chem. Soc., 2011, 133, 18014 CrossRef.
- J. L. Gornall, K. R. Mahendran, O. J. Pambos, L. J. Steinbock, O. Otto, C. Chimerel, M. Winterhalter and U. F. Keyser, Nano Lett., 2011, 11, 3334 CrossRef CAS.
- E. C. Yusko, J. M. Johnson, S. Majd, P. Prangkio, R. C. Rollings, J. Li, J. Yang and M. Mayer, Nat. Nanotechnol., 2011, 6, 253 CrossRef CAS.
- E. C. Yusko, P. Prangkio, D. Sept, R. C. Rollings, J. Li and M. Mayer, ACS Nano, 2012, 6, 5909 CrossRef CAS.
- C. A. Morris, A. K. Friedman and L. A. Baker, Analyst, 2010, 135, 2190 RSC.
- A. Bruckbauer, L. Ying, A. M. Rothery, D. Zhou, A. I. Shevchuk, C. Abell, Y. E. Korchev and D. Klenerman, J. Am. Chem. Soc., 2002, 124, 8810 CrossRef CAS.
- L. J. Steinbock, O. Otto, C. Chimerel, J. L. Gornall and U. F. Keyser, Nano Lett., 2010, 10, 2493 CrossRef CAS.
- http://www.avantilipids.com .
- P. S. Cremer and S. G. Boxer, Langmuir, 2011, 27, 10920 CrossRef.
- P. S. Cremer and S. G. Boxer, J. Phys. Chem. B, 1999, 103, 2554 CrossRef CAS.
- B. A. Lewis and D. M. Engelman, J. Mol. Biol., 1983, 166, 211 CrossRef CAS.
- T. J. Zwang, W. R. Fletcher, T. J. Lane and M. S. Johal, Langmuir, 2010, 26, 4598 CrossRef CAS.
- S. W. Kowalczyk, L. Kapinos, T. R. Blosser, T. Magalhães, P. van Nies, R. Y. H. Lim and C. Dekker, Nat. Nanotechnol., 2011, 6, 433 CrossRef CAS.
- D. A. Holden, J. J. Watkins and H. S. White, Langmuir, 2012, 28, 7572 CrossRef CAS.
- C. Dekker, Nat. Nanotechnol., 2007, 2, 209 CrossRef CAS.
- J. Li, M. Gershow, D. Stein, E. Brandin and J. A. Golovchenko, Nat. Mater., 2003, 2, 611 CrossRef CAS.
- Z. Siwy, E. Heins, C. C. Harrell, P. Kohli and C. R. Martin, J. Am. Chem. Soc., 2004, 126, 10850 CrossRef CAS.
- W. J. Lan, D. A. Holden and H. S. White, J. Am. Chem. Soc., 2011, 133, 13300 CrossRef CAS.
- U. F. Keyser, S. van Dorp and S. G. Lemay, Chem. Soc. Rev., 2010, 39, 939 RSC.
- U. F. Keyser, J. R. Soc., Interface, 2011, 8, 1369 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available: Details of experimental procedures, graphs and images. See DOI: 10.1039/c2an36397f |
| This journal is © The Royal Society of Chemistry 2013 |
