Sensing coiled-coil proteins through conformational modulation of energy transfer processes – selective detection of the oncogenic transcription factor c-Jun†
Elena
Pazos
,
Adrián
Jiménez-Balsa
,
José L.
Mascareñas
* and
M. Eugenio
Vázquez
*
Departamento de Química Orgánica y Centro Singular de Investigación en Química Biológica y Materiales Moleculares, Unidad Asociada al CSIC, Universidad de Santiago de Compostela, 15782, Santiago de Compostela, Spain. E-mail: eugenio.vazquez@usc.es; joseluis.mascarenas@usc.es.; Fax: (+34) 981 595 012; Tel: (+34) 881 815 738
First published on 21st July 2011
Abstract
Designed terbium-chelating peptides were used to monitor the formation of leucine zipper associations. The sensing strategy is based on the modulation of the antenna effect from a Trp residue donor and the luminescence increase arising from the folding that takes place upon the formation of the coiled-coil structure. An application in the specific sensing of the oncogenic c-Jun transcription factor is demonstrated.
Introduction
Eukaryotic gene expression is tightly regulated at the transcriptional level by a class of proteins called transcription factors (TFs).1 It is known that the deregulation of many of these TFs is intimately related to aberrant cell proliferation processes2 and, therefore, the development of selective monitoring strategies for these proteins could be of great value in research and diagnostics. In many cases, such as with bZIP transcription factors, their activity involves homo- or heterodimerization processes coupled to folding into α-helical secondary structures.3 We reasoned that such conformational transitions could be used to modulate the sensitization of a lanthanide ion,4,5 exploiting the resulting emission changes as a general strategy for detecting leucine zipper-forming proteins. Herein, we demonstrate the viability of this concept in a model coiled-coil leucine zipper system and extend its application to the specific sensing of the oncoprotein c-Jun, a transcription factor that plays a major role in the development of many tumors.6Results and discussion
Probe design and optimization using a model de novo heterodimeric coiled-coil system
We first studied a proof-of-concept system with the incorporation of a tryptophan antenna and a lanthanide complex at different positions of the external surface of leucine zipper-forming peptides.7 The model peptides were selected on the basis of previously reported de novo heterodimeric coiled-coils featuring two complementary strands designated as the K44 coil (VSALKEK heptad repeat) and the E444 coil (VSALEKE heptad repeat).8 Both strands display complementary electrostatic interactions between the negatively charged Glu and positively charged Lys side chains at positions e and g of the heptads that direct the formation of heterodimeric complexes and select against homodimeric associations. Given the importance of Val and Leu residues at positions a and d for the structural integrity of the leucine zipper,9 we introduced the Tb3+ chelating macrocycle DOTA (1,4,7,10-tetraazacyclododecane-1,4,7,10-tetraacetic acid) to the outer face of one α-helix (position f). Trp antennas were placed at either position i + 4 or i − 4, on the same face of the α-helix (positions b and c), so that in the folded state these residues would be in the vicinity of the metal chelate, therefore hypothetically maximizing the energy transfer efficiency and the luminescence from the metal (Scheme 1).![Top: A helical wheel representation of heterodimeric peptides E444 and K44, indicating as an example the Trp antenna and the DOTA[Tb3+] chelate on the external side of the helix in one of the derivatives (peptide W9W9K44). Positions a and d form the hydrophobic core (shaded grey); positions e and g form complementary electrostatic interactions. Bottom: Peptide sequences of the model heterodimeric leucine zippers and each of the luminescent derivatives studied. Mutations on each sequence: W9W9K44: Ser9 → Trp, Glu13 → ϕ; W17W17K44: Ala17 → Trp, Glu13 → ϕ; W9/W9/1717K4: Ser9 and Ala17 → Trp, Glu13 → ϕ, where ϕ = Dap(DOTA[Tb3+]).](/image/article/2011/SC/c1sc00108f/c1sc00108f-s1.gif) | ||
| Scheme 1 Top: A helical wheel representation of heterodimeric peptides E444 and K44, indicating as an example the Trp antenna and the DOTA[Tb3+] chelate on the external side of the helix in one of the derivatives (peptide W9W9K44). Positions a and d form the hydrophobic core (shaded grey); positions e and g form complementary electrostatic interactions. Bottom: Peptide sequences of the model heterodimeric leucine zippers and each of the luminescent derivatives studied. Mutations on each sequence: W9W9K44: Ser9 → Trp, Glu13 → ϕ; W17W17K44: Ala17 → Trp, Glu13 → ϕ; W9/W9/1717K4: Ser9 and Ala17 → Trp, Glu13 → ϕ, where ϕ = Dap(DOTA[Tb3+]). | ||
Detailed synthetic procedures are given in ESI†. In short, peptides were assembled following standard Fmoc solid phase peptide synthesis protocols10 and the DOTA unit was introduced into the peptide scaffold as the tri-tBu-protected acid through an orthogonally-deprotected 2,3-diaminopropanoic acid (Dap) residue, while the peptides were still attached to the solid support. HPLC-purified peptides were then complexed with TbCl3 in HEPES buffer to give the desired chelates, which were subsequently repurified by reverse-phase HPLC (Scheme 2).
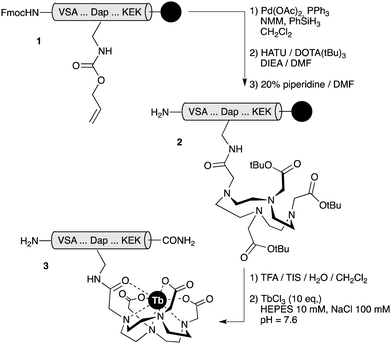 | ||
| Scheme 2 The reaction sequence for the synthesis of therbium-chelated peptides. TIS: triisopropylsilane; NMM: N-methylmorpholine. | ||
Each of the three luminescent peptides, W9W9K44, W17W17K44 and W9/W9/1717K4, were excited at 280 nm and their emission spectra were then compared with those obtained for each peptide in the presence of the complementary sequence E444. As shown in Fig. 1, the relative position of the Trp antenna and the emissive DOTA[Tb3+] chelate has a strong influence on the relative emission of the monomeric/dimeric species. Thus, W9W9K44 shows a moderately intense emission spectrum in the monomeric form, which remains essentially unchanged upon the formation of the W9W9K44/E444 complex. In contrast, peptide W17W17K44 displays a weak emission spectrum as a monomer, but the addition of E444 and the subsequent formation of the W17W17K44/E444 complex results in a large increase in the luminescence emission from the terbium chelate (∼three-fold increase). This increase in the emission from the metal could be fitted to a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 interaction model with a KD of 3.3 ± 0.2 μM. The behavior of W9/W9/1717K4 was intermediate between the other two peptides (see ESI† for details).
1 interaction model with a KD of 3.3 ± 0.2 μM. The behavior of W9/W9/1717K4 was intermediate between the other two peptides (see ESI† for details).
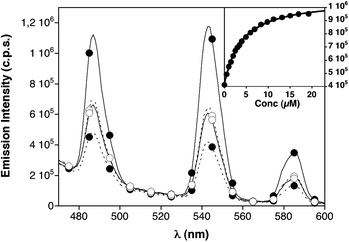 | ||
| Fig. 1 Dashed lines, emission spectra of monomeric peptides W9W9K44 (○), and W17W17K44 (●), 1 μM in HEPES buffer (10 mM HEPES pH 7.6, 100 mM NaCl). Solid lines, emission spectra of their complexes with E444 (19.2 μM). Peptide W9/W9/1717K4 is omitted for clarity (see Fig. S3, ESI†). Inset with the mean emission at 545 nm of 1 μM peptide W17W17K44 in the presence of increasing amounts of the complementary E444peptide and the best fitting curve (each data point in the titration corresponds to an average of three experiments).11 | ||
Circular dichroism experiments are consistent with the formation of an α-helical structure upon addition of the complementary E444peptide and support the expected mechanism of luminescence increase associated with a better relative position of the Trp donor and the DOTA[Tb3+] acceptor in the coiled-coil complexes (see Fig. S11, ESI†).
Application of the sensing strategy to monitor the c-Jun transcription factor
Once we identified the optimum relative positions of the Trp donor and the terbium chelate acceptor that maximizes the emission increase upon dimerization (Trp at i – 4 relative to DOTA[Tb3+]), we sought to translate that information into the appropriate leucine zipper scaffold to specifically recognize the oncogenic transcription factor c-Jun. This protein is an archetypical member of the bZIP family of transcription factors and, as such, is largely unstructured in its monomeric form, but dimerization with c-Fos through a leucine zipper region folds both proteins into stable α helices that specifically recognize the DNA AP1 site (5′–ATGA(c/g)TCAT–3′).12 We thereby designed a c-Fos-based leucine zipper peptide incorporating the Trp and the Dap(DOTA[Tb3+]) residues at equivalent positions to those in which they are in the model W17W17K44peptide (FosWW[Tb], Fig. 2). Additionally, we synthesized a second peptide, A-FosWW[Tb], incorporating a Glu-rich acidic appendage at the N-terminus of the FosWW[Tb] sequence. This negatively-charged extension (EELEKEAEELEQEL) should establish favorable electrostatic interactions with the Arg-rich basic region of c-Jun, resulting in an increased affinity of the sensing peptide (Fig. 2).13![Top: Sequences of the DNA binding domains of c-Fos and c-Jun showing the basic region and leucine zipper regions and the sequences of the designed sensing peptides, where ϕ = Dap(DOTA[Tb3+]). Bottom: a) Dashed lines, emission spectra of 1 μM solutions of monomeric peptides FosWW[Tb] (○) and A-FosWW[Tb] (●) in HEPES buffer (10 mM HEPES pH 7.6, 100 mM NaCl). Solid lines, emission spectra of the complexes of FosWW[Tb] (○) and A-FosWW[Tb] (●) with c-Jun (9.2 μM and 3.8 μM, respectively); control experiment incubating A-FosWW[Tb] with 8 μM of natural c-Fos DNA binding domain (◇); b) titration curves of 1μM solutions of FosWW[Tb] (○) and A-FosWW[Tb] (●) with c-Jun. Emission at 545 nm with the best fit corresponding to a 1 : 1 binding mode (see ESI); c) circular dichroism spectra of c-Jun (▽), A-FosWW[Tb] (▲) and their 1 : 1 complex (●).](/image/article/2011/SC/c1sc00108f/c1sc00108f-f2.gif) | ||
Fig. 2 Top: Sequences of the DNA binding domains of c-Fos and c-Jun showing the basic region and leucine zipper regions and the sequences of the designed sensing peptides, where ϕ = Dap(DOTA[Tb3+]). Bottom: a) Dashed lines, emission spectra of 1 μM solutions of monomeric peptides FosWW[Tb] (○) and A-FosWW[Tb] (●) in HEPES buffer (10 mM HEPES pH 7.6, 100 mM NaCl). Solid lines, emission spectra of the complexes of FosWW[Tb] (○) and A-FosWW[Tb] (●) with c-Jun (9.2 μM and 3.8 μM, respectively); control experiment incubating A-FosWW[Tb] with 8 μM of natural c-Fos DNA binding domain (◇); b) titration curves of 1μM solutions of FosWW[Tb] (○) and A-FosWW[Tb] (●) with c-Jun. Emission at 545 nm with the best fit corresponding to a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 binding mode (see ESI†); c) circular dichroism spectra of c-Jun (▽), A-FosWW[Tb] (▲) and their 1 1 binding mode (see ESI†); c) circular dichroism spectra of c-Jun (▽), A-FosWW[Tb] (▲) and their 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 complex (●). 1 complex (●). | ||
In agreement with the results obtained with the W17W17K44 model peptide, both FosWW[Tb] and A-FosWW[Tb] displayed a large emission increase upon incubation with the DNA binding domain of c-Jun (Fig. 2a).14 As expected, the introduction of the acidic tail resulted in A-FosWW[Tb] binding more strongly to c-Jun (KD = 349 ± 7 nM) than the core leucine zipper peptide FosWW[Tb], which displays a KD of 1.5 ± 0.1 μM (Fig. 2b). The detection limit for c-Jun with the A-FosWW[Tb]probe was found to be approximately 42 nM.15 Control experiments with peptides that do not form stable heterodimeric leucine zippers with A-FosWW[Tb], such as c-Fos, demonstrated that the probe is selective for c-Jun (Fig. 2a). On the other hand, we have found that the luminescent A-FosWW[Tb]/c-Jun complex remains stable after the addition of excess of c-Fos (see Fig. 8, ESI†). The folding of A-FosWW[Tb] into an α-helical structure and formation of a heterodimeric coiled-coil in the presence of c-Jun was further confirmed by circular dichroism spectroscopy, which showed large negative bands at 208 and 222 nm for a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 mixture of A-FosWW[Tb] with c-Jun (Fig. 2c). Interestingly, a significant increase in the terbium luminescence emission was also observed, even when A-FosWW[Tb] was incubated with c-Jun in the presence of a complex biological medium (cell extracts, total protein concentration of ∼65 μg mL−1). In these experiments, we observed a similar binding affinity (mean KD = 630 ± 120 nM) to that in the in vitro studies with purified samples of the protein, therefore demonstrating the specificity and viability of this sensing strategy under demanding conditions. Furthermore, these experiments illustrate the advantages of using lanthanide emission probes, as they allow time-gated experiments that avoid the interference from the intrinsic emission that characterizes complex biological media (see Fig. 3 and see ESI† for a comparison with the results using steady-state emission instrument settings†).
1 mixture of A-FosWW[Tb] with c-Jun (Fig. 2c). Interestingly, a significant increase in the terbium luminescence emission was also observed, even when A-FosWW[Tb] was incubated with c-Jun in the presence of a complex biological medium (cell extracts, total protein concentration of ∼65 μg mL−1). In these experiments, we observed a similar binding affinity (mean KD = 630 ± 120 nM) to that in the in vitro studies with purified samples of the protein, therefore demonstrating the specificity and viability of this sensing strategy under demanding conditions. Furthermore, these experiments illustrate the advantages of using lanthanide emission probes, as they allow time-gated experiments that avoid the interference from the intrinsic emission that characterizes complex biological media (see Fig. 3 and see ESI† for a comparison with the results using steady-state emission instrument settings†).
Conclusions
In summary, we have demonstrated that geometric changes triggered by specific biomolecular recognition events can be exploited for the design of luminescent sensors for bZIP transcription factor proteins. Specifically, the folding of an α-helical peptide upon the formation of heterodimeric coiled-coil complexes allowed the specific detection of the DNA binding domain of the oncogenic transcription factor c-Jun. Additionally, this method might be useful for identifying and screening other heterodimeric coiled-coil proteins. Coiled-coil forming peptides have been previously used to construct biomaterials7a,16 and even reported as in vivo tools for imaging and biological modulation,7b,17 but our work represents the first engineering approach to their use as smart luminescent biosensors. We expect that this approach could be applied in studying biomolecular events coupled to the folding of disordered proteins into α-helical conformations, including other important bZIP transcription factors, such as Myc.![Dashed line: time-gated emission spectra of 1μM solution of monomeric peptide A-FosWW[Tb] in HEPES buffer (10 mM HEPES pH 7.6, 100 mM NaCl). Solid lines: phosphorescence emission spectra of A-FosWW[Tb] with 65 μg mL−1 of protein extracts from cell lysates (●) and the same sample after the addition of 4 equivalents of c-Jun (○).](/image/article/2011/SC/c1sc00108f/c1sc00108f-f3.gif) | ||
| Fig. 3 Dashed line: time-gated emission spectra of 1μM solution of monomeric peptide A-FosWW[Tb] in HEPES buffer (10 mM HEPES pH 7.6, 100 mM NaCl). Solid lines: phosphorescence emission spectra of A-FosWW[Tb] with 65 μg mL−1 of protein extracts from cell lysates (●) and the same sample after the addition of 4 equivalents of c-Jun (○). | ||
Acknowledgements
We thank the support given by the Spanish grants SAF2007-61015, SAF2010-20822-C02, CTQ2009-14431/BQU, Consolider Ingenio 2010 CSD2007-00006 and the Xunta de Galicia INCITE09 209 084PR, GRC2010/12, PGIDIT08CSA-047209PR. We also thank Prof. Anxo Vidal, from the University of Santiago de Compostela, for supplying us with cell lysates, and Prof. Lin Chen, from the University of Southern California, for providing us with recombinant c-Jun for our preliminary experiments. E. P. and A. J. B. thank the Ministry of Education of Spain for their PhD fellowships.Notes and references
- (a) D. S. Latchman in Eukaryotic Transcription Factors, Elsevier, London, 2004, p. 77 Search PubMed; (b) N. M. Luscombe, S. E. Austin, H. M. Berman and J. M. Thornton, GenomeBiology, 2000, 1, 1 CrossRef.
- (a) J. E. Jr. Darnell, Nat. Rev. Cancer, 2002, 2, 740 CrossRef CAS; (b) A. H. Brivanlou and J. E. Jr. Darnell, Science, 2002, 295, 813 CrossRef CAS; (c) P. P. Pandolfi, Oncogene, 2001, 20, 3116 CrossRef CAS.
- (a) H. J. Dyson and P. E. Wright, Nat. Rev. Mol. Cell Biol., 2005, 6, 197 CrossRef CAS; (b) G. M. Verkhivker, Proteins: Struct., Funct., Bioinf., 2005, 58, 706 CrossRef CAS; (c) J. K. Kohler and A. Schepartz, Biochemistry, 2001, 40, 130 CrossRef CAS; (d) K. T. O'Neil, J. D. Shuman, C. Ampe and W. F. DeGrado, Biochemistry, 1991, 30, 9030 CrossRef CAS; (e) M. E. Vázquez, A. M. Caamaño and J. L. Mascareñas, Chem. Soc. Rev., 2003, 32, 338 RSC.
- The choice of a lanthanide metal as a luminescent probe stems from the well-known advantages of these ions for biosensing, due to their stability, long wavelength emission bands and long lifetimes.
- (a) E. Pazos, D. Torrecilla, M. Vázquez-López, L. Castedo, J. L. Mascareñas, A. Vidal and M. E. Vázquez, J. Am. Chem. Soc., 2008, 130, 9652 CrossRef CAS; (b) B. McMahon, P. Mauer, C. P. McCoy, T. C. Lee and T. Gunnlaugsson, J. Am. Chem. Soc., 2009, 131, 17542 CrossRef CAS; (c) T. Gunnlaugsson, A. J. Harte, J. P. Leonard and M. Nieuwenhuyzen, Chem. Commun., 2002, 2134 RSC; (d) J.-C. Bünzli and C. Piguet, Chem. Soc. Rev., 2005, 34, 1048 RSC; (e) E. Pazos, O. Vázquez, J. L. Mascareñas and M. E. Vázquez, Chem. Soc. Rev., 2009, 38, 3348 RSC.
- (a) P. Vaňhara and J. Šmarda, J. Appl. Biomed., 2006, 4, 163 Search PubMed; (b) H. van Dam and M. Castellazzi, Oncogene, 2001, 20, 2453 CrossRef CAS; (c) P. K. Vogt, Oncogene, 2001, 20, 2365 CrossRef CAS.
- (a) J. M. Mason and K. M. Arndt, ChemBioChem, 2004, 5, 170 CrossRef CAS; (b) F. Zhang, K. A. Timm, K. M. Arndt and G. A. Woolley, Angew. Chem., Int. Ed., 2010, 49, 3943 CAS.
- G. De Cresenzo, J. R. Litowsky, R. S. Hodges and M. D. O'Connor-McCourt, Biochemistry, 2003, 42, 1754 CrossRef.
- (a) E. K. O'Shea, K. J. Lumb and P. S. Kim, Curr. Biol., 1993, 3, 658 CrossRef CAS; (b) D. A. D. Parry, R. D. B. Fraser and J. M. Squire, J. Struct. Biol., 2008, 163, 258 CrossRef CAS; (c) P. Burkhard, J. Stetefeld and S. Strelkov, Trends Cell Biol., 2001, 11, 82 CrossRef CAS.
- N. Sewald and H. Jakubke in Peptides: Chemistry and Biology, Wiley-VCH, Weinheim, 2002 Search PubMed.
- Curve fitting analysis was performed using non-linear regression with Mathematica® 7.0.1.0 software (ESI†). (a) M. H. A. Roehrl, J. Y. Wang and G. Wagner, Biochemistry, 2004, 43, 16056 CrossRef CAS; (b) P. Thordarson, Chem. Soc. Rev., 2011, 40, 1305 RSC; (c) M. R. Eftink, Methods Enzymol., 1997, 278, 221 CrossRef CAS; (d) G. Shi, Y. Gong, A. Savchenko, J. G. Zeikus, B. Xiao, X. Ji and H. Yan, Biochim. Biophys. Acta, 2000, 1478, 289 CrossRef CAS.
- (a) Y. Chinenov and T. K. Kerppola, Oncogene, 2001, 20, 2438 CrossRef; (b) D. Bohmann, T. J. Bos, A. Admon, T. Nishimura, P. K. Vogt and R. Tjian, Science, 1987, 238, 1386 CAS; (c) J. N. M. Glover and S. C. Harrison, Nature, 1995, 373, 257 CrossRef CAS; (d) L. Chen, J. N. M. Glover, P. G. Hogan, A. Rao and S. C. Harrison, Nature, 1998, 392, 42 CrossRef CAS; (e) C. Portela, F. Albericio, R. Eritja, L. Castedo and J. L. Mascareñas, ChemBioChem, 2007, 8, 1110 CrossRef CAS.
- (a) M. Olive, D. Krylov, D. R. Echlin, K. Gardner, E. Taparowsky and C. Vinson, J. Biol. Chem., 1997, 272, 18586 CrossRef CAS; (b) D. Krylov, K. Kasai, D. R. Echlin, E. J. Taparowsky, H. Arnheiter and C. Vinson, Proc. Natl. Acad. Sci. U. S. A., 1997, 94, 12274 CrossRef CAS.
- We selected the bZIP domain of human c-Jun, including an extra N-terminal Phe for UV concentration determination. See Fig. 2 for the full amino acid sequence.
- Calculated as described in: D. A. Armbruster and T. Pry, Clin. Biochem. Rev., 2008, 29, S49 Search PubMed.
- (a) B. Apostolovic, M. Danial and H.-A. Klok, Chem. Soc. Rev., 2010, 39, 3541 RSC; (b) G. Grigoryan and A. E. Keating, Curr. Opin. Struct. Biol., 2008, 18, 477 CrossRef CAS.
- (a) Y. Yano, A. Yano, S. Oishi, Y. Sugimoto, G. Tsujimoto, N. Fujii and K. Matsuzaki, ACS Chem. Biol., 2008, 3, 341 CrossRef CAS; (b) H. Tsutsumi, W. Nomura, S. Abe, T. Mino, A. Masuda, N. Ohashi, T. Tanaka, K. Ohba, N. Yamamoto, K. Akiyoshi and H. Tamamura, Angew. Chem., Int. Ed., 2009, 48, 9164 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available: Peptide synthesis and labeling protocols, analytical data and spectroscopic methods. See DOI: 10.1039/c1sc00108f |
| This journal is © The Royal Society of Chemistry 2011 |
