Glutathione (GSH)-decorated magnetic nanoparticles for binding glutathione-S-transferase (GST) fusion protein and manipulating live cells†
Yue
Pan‡
a,
Marcus J. C.
Long‡
b,
Xinming
Li
a,
Junfeng
Shi
a,
Lizbeth
Hedstrom
ac and
Bing
Xu
*a
aDepartment of Chemistry, Brandeis University, 415 South Street, Waltham, MA, USA. E-mail: bxu@brandeis.edu; Fax: +7817362516; Tel: +781-736-5201
bGraduate Program in Biochemistry and Biophysics, Brandeis University, 415 South Street, Waltham, MA, USA
cDepartment of Biology, Brandeis University, 415 South Street, Waltham, MA 02454, USA
First published on 3rd March 2011
Abstract
Iron oxide-based magnetic nanoparticles (MNP) surface-decorated with glutathione (GSH) via a dopamine anchor bind to human α1-glutathione S-transferase (GST) with high affinity and specificity and are able to separate GST fusion proteins from cell lysates. Both the purified GST and the protein of interest (POI) preserve their innate properties. The conjugate of MNP and the GST fusion protein also enables magnetic manipulation of cells.
Introduction
This article reports the use of glutathione (GSH)-decorated iron oxide nanoparticles as a general agent to separate a protein fused with human α1-glutathione S-transferase (GST) and to manipulate live cells. Because their properties and functions differ drastically from their bulk counterparts, nanoscale structures (e.g., nanoparticles, nanowires, nanofibers, and nanotubes) have emerged as a novel class of materials and are being intensively explored for many biological applications.1,2 Particularly, their high surface to volume ratio, surface tailorability, improved dispersity, and capacity for carrying multiple functions open many new possibilities for their applications in biomedical research. Among a variety of nanomaterials, magnetic nanoparticles (MNPs)2–4 are unique because they respond to a magnetic field and may facilitate probing specific functions of bioactive molecules in localized domains or compartments of live cells.5 To achieve this attractive goal, one has to develop an effective and selective strategy to attach the protein(s) of interest (POIs) to the magnetic nanoparticles.The development of various protein tags not only has led to successful separation of recombinant proteins by conventional affinity chromatography,6 but also provided an opportunity for using magnetic nanoparticles to bind the POIs with high specificity. For example, magnetic nanoparticles decorated with the complex of nickel and nitrilotriacetic acid (Ni-NTA) have shown high specificity and capacity for binding and separating histidine-tagged proteins.7–9 The cytotoxicity associated with nickel ions,10 however, excludes the application of Ni-NTA based magnetic nanoparticles on live cells for long duration. The biocompatibility of iron oxide nanoparticles, GSH, and GST makes GSH-decorated iron oxide magnetic nanoparticles an ideal candidate for binding and manipulating GST fusion proteins. Moreover, the ligand–receptor interaction between GSH and GST may allow the POIs to adopt a specific orientation on MNPs, an attractive feature for studying protein–protein interactions. Despite these merits, GSH-decorated magnetic nanoparticles have been elusive until this work. Using dopamine as the anchor7,11 to link glutathione to iron oxide nanoparticles, we demonstrate here the first example of magnetic nanoparticles to bind GST fusion proteins.
Though the specificity/reversibility of the GST/GSH interaction has successfully resulted several types of GST-decorated, commercial microbeads for many years, those microparticles still suffer from limitations such as non-specific protein absorption, low capacity, and the requirement of excessive washing.9,12 Unlike in the case of microbeads, the target proteins cover the surface of the nanoparticles effectively and quickly, thus reducing the overall unoccupied surface area for nonspecific absorption of proteins and achieving high specificity. Indeed, the nanoparticles (6) exhibit very low non-specific binding and essentially have no dead volume, thus leading to ultra-high purity of eluted protein with the need for only a single wash, which could prove decisive if dealing with very unstable proteins. In addition, the low ionic strength binding/washing conditions should preserve even weakly associated proteins with the POI, thus making these nanoparticles ideal for proteomic work. Moreover, we show that these functionalized nanoparticles are biocompatible (essentially no cell death after incubation for 24 h) and that they associate strongly with or enter cells, which facilitates the manipulation of live cells. These results suggest that the use of GSH-decorated nanoparticles promises applications beyond simple protein separation.
Results and discussion
Scheme 1 illustrates the general strategy for the separation of GST fusion proteins from a cell lysate with MNP-GSH and the further applications. The well-established ligand–receptor interaction between GSH to GST ensures that MNP-GSH selectively binds to GST-POI (in this work, POI = enhanced green fluorescent protein (GFP)),13 which forms the conjugate of MNP-GSH∼GST-POI (∼ for denoting ligand–receptor interaction). The use of magnetic separation yields pure MNP-GSH∼GST-POI, which produces GST-POI upon elution, affords POI via enzymatic cleavage, or renders cells responsive to magnetic attraction. After the separation, both GST and the fusion protein preserve their own properties (in this case, the catalytic activity of GST and the green fluorescence of the GFP fused to GST). Our results demonstrate that these nanoparticles, which exhibit high affinity and specificity to GST fusion proteins, not only allow a simple procedure to separate recombinant proteins fused to GST, but also provide a versatile, cell-compatible platform for manipulating proteins or cells by a magnetic force.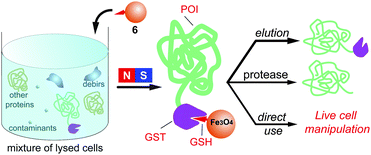 | ||
| Scheme 1 The GSH-decorated iron oxide nanoparticles selectively bind to GST fusion proteins from a cell lysate and their down-stream applications. | ||
To execute the processes outlined in Scheme 1, we first made the key component, MNP-GSH (6). As shown in Scheme 2, the simple coupling of dopamine (1) with maleimide (2) in acetic acid affords 3 (40% yield), which reacts with GSH in PBS buffer solution (pH 7.4) to generate 5 in 80% yield. Then, iron oxide nanoparticles14 react with 5 as a dispersion in CHCl3. The addition of hexane to the dispersion produces brown precipitates, which disperse in water (pH 9.0) to yield 6 (i.e., MNP-GSH). Having the average diameter (6 nm) similar to those of as-prepared iron oxide nanoparticles, the dispersed 6 moves to the wall of the Eppendorf vial upon the magnetic attraction created by a commercial rack (Fig. 1) with surface magnetic field about 4000 G.7UV-Vis spectrum of 6 displays a maximum at about 270 nm (see Fig. S1 in supporting information†) that originates from the aromatic ring of 5, indicating the attachment of 5 to the MNP. Besides, the IR spectra (see Fig. S2 in supporting information†) of 6 exhibits broad peaks around 3300 cm−1 and a sharp peak at about 1695 cm−1, which belong to a carboxylic acid group, indicating the presence of 5 on the MNP.
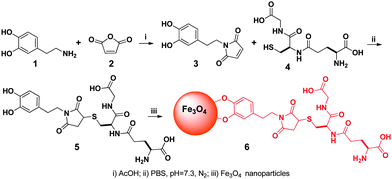 | ||
| Scheme 2 The synthetic route of the glutathione-decorated magnetic nanoparticles (for clarity, surfactants on the nanoparticle are not shown). | ||
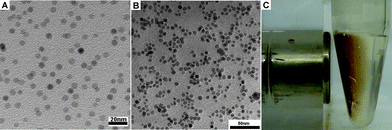 | ||
| Fig. 1 Transmission electron micrograph (TEM) images of (A) as-prepared iron oxide nanoparticles and (B) 6. (C) Optical image of 6 attracted by a magnet. | ||
In addition, weight analysis gives the estimation of about 48 molecules of 5 on each iron oxide nanoparticle (see supporting information†). The GSH-decorated iron oxide nanoparticles are stable and can be stored in PBS at 4 °C for over 2 months before usage.
After its characterization, we tested the affinity of 6 towards pure GST (see the SDS/PAGE analysis (Fig. S4) in supporting information†). After being incubated with 6 for 2 h in 50 mM PBS at pH 7.6, the amount of GST in the solution decreases by about 40% (from 470 ng (lane 12, Fig. S4†) to 290 ng (lane 11, Fig. S4†)), suggesting the binding of GST to 6. The washing solutions (PBS buffer, 50 mM phosphate, 75 mM NaCl, and pH 7.6) for the nanoparticles (i.e., 6 incubated with GST solution) show that the amounts of GST in three consecutive washes drop from around 130 ng (lane 8, Fig. S4†) to 80 ng (lane 6, Fig. S4†). While the use of low concentration GSH (30 mM) to elute the nanoparticles gives 40 ng GST (lane 10, Fig. S4†), the use of high concentration GSH (100 mM) liberates 130 ng GST (lane 9, Fig. S4†). These results clearly indicate the specific binding of GST to 6, which warrants the use of 6 to separate GST or GST fusion protein from the lysates of cells.
We optimized the procedure for using 6 to separate GST or GST fusion proteins, which consists of three steps: (1) incubating 6 (340 μg mL−1) with the lysate of E. coli that overexpresses the GST-POI for 2 h in 50 mM phosphate buffer at pH 6.6; (2) using a magnet to attract the incubated 6 to the wall of the vial and washing the nanoparticles with a buffer (50 mM phosphate, 75 mM NaCl, and pH 7.6) at 4 °C; and (3) eluting the nanoparticles by a GSH solution (in a Tris buffer, pH 8.0) at room temperature. As shown in Fig. 2A, the elution of the nanoparticles by GSH solution gives only GST band (lane 1). In addition, the protein band (lane 3) of the lysate (after incubation) shows that the amount of GST is about 10% relative to the GST in the initial lysate (lane 4), but the amounts of other proteins change little. These results confirm that 6 selectively binds to GST. Quantitative analysis (see Figs. S5 and S7 in supporting information†) indicates the binding capacity of 6 to GST to be 60 μg GST protein mg−1.
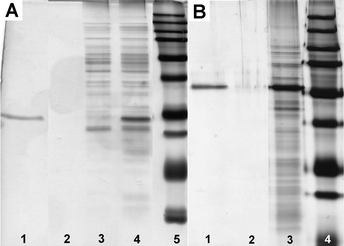 | ||
| Fig. 2 (A) SDS/PAGE analysis (by silver staining15) of the binding of 6 to the GST in an E. coli lysate (1/3 of total volume in each lane): lane 1, elution by GSH (see Fig. S5 in supporting information†); lane 2, 1st wash; lane 3, lysate after the removal of 6; lane 4, cell lysate; lane 5, molecular weight marker. (B) SDS/PAGE analysis (by silver staining) of the binding of 6 to the GFP-TEV-HA-GST in an E. coli lysate (lane 1, 1/10 of total volume; the other lanes, 1/3 of total volume in each lane): lane 1, elution by GSH (see Fig. S8 in supporting information†); lane 2, 1st wash; lane 3, cell lysate; lane 4, molecular weight marker. All washes were carried using 50 mM sodium phosphate buffer at pH 7.6. | ||
Considering that a medium length linker (e.g., 16 amino acid residues) would reduce domain–domain interference between GST and POI, we designed a linker consisting of both a TEV protease (a highly specific cysteine protease from the tobacco etch virus) recognition sequence to allow liberation of the POI under mild conditions, and an HA sequence (an epitope derived from hemagglutinin for which highly specific/sensitive antibodies are available) to facilitate the confirmation of the eluted proteinvia Western blot. We used the optimized procedure to isolate the fusion protein, GFP-TEV-HA-GST (see supporting information†), overexpressed in an E. coli lysate. As shown in Fig. 2B, the pre-elution wash by PBS buffer hardly removes any proteins (lane 2), and the elution by a GSH solution (300 mM) results in only one strong band (lane 1) of the GFP-TEV-HA-GST. These results indicate that 6 selectively binds to the GFP-TEV-HA-GST fusion protein from the cell lysate. In addition, according to Fig. 2B and Figs. S8 and S9 (see supporting information†), there is 100 μg of GFP-TEV-HA-GST on 1 mg of 6. Furthermore, we used a Western blot (see Fig S10 in supporting information†) to confirm that the eluted protein contains the HA tag, proving that it is GFP-TEV-HA-GST.
Beside elution, we used TEV protease to treat the conjugate of 6 and GFP-TEV-HA-GST. SDS/PAGE analysis (see Fig. S11 and 12 in supporting information†) shows the liberation of POI and confirms that GST remains on the nanoparticles. This result suggests that the action of the proper protease can successfully liberate the POI from the conjugate of 6 and the fusion proteins. In addition, 6 is reusable after elution (see Fig. S13 in supporting information†), indicating that the surface anchored GSH is stable. In a control experiment, we used 6 to bind E. colidihydrofolate reductase (DHFR)-GFP fusion protein and hardly observed any fluorescence from the nanoparticles (see Fig. S14 in supporting information†), further supporting that 6 exhibits little non-specific binding to other proteins.
After being separated, both the GST and the POI preserve their innate properties: the conjugate of GFP-TEV-HA-GST and 6 exhibits green fluorescence (Fig. 3A), and the eluted GFP-TEV-HA-GST protein is enzymatically active (Fig. 3B). We also incubated the conjugate of 6 and GFP-TEV-HA-GST with HeLa cells overnight, then observed the cell behavior in a trypan blue solution, which acts as a negative stain for live cells (i.e., colorless cells are viable). As shown in Fig. 3C and D, more than 95% of the total population of the cells is viable. Some colorless cells migrate in the trypan blue solution upon applying a small magnet on one side of the culture well. This result suggests that the HeLa cells become “magnetized” after the cells uptake the conjugate of 6 and GFP-TEV-HA-GST. Thus, the conjugate of 6 and GST fusion proteins permits the direct manipulation of cellvia a magnet.
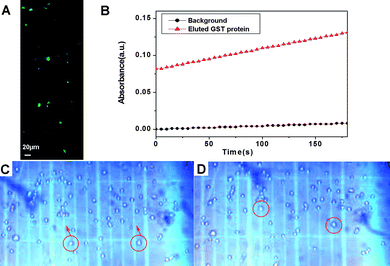 | ||
| Fig. 3 (A) Fluorescent image of the conjugate of 6 and GFP-TEV-HA-GST; (B) GST activity (assay conditions: 50mM Tris/NaCl, pH 8.0, 2 mM GSH, 2 mM chlorodinitrobenzene) indicated by the increase of UV-Vis absorption at 340 nm. (C, D) Optical images (10×) of the magnet-guided migration of HeLa cells in trypan blue solution: (C) t = 0 s and (D) t = 1 s. The migrating cells are white, indicating that they remain viable in the presence of the conjugate of 6 and GFP-TEV-HA-GST. The magnet (not shown) was placed on the upper sides of (C), (D). | ||
To verify that cells uptake the conjugate of 6 and GFP-TEV-HA-GST, we added the conjugate 6 and GFP-TEV-HA-GST to the COS cell together with 40 μg mL−1Rhodamine B isothiocyanate (RITC)-dextran and incubated for 12 h. After the removal of extracellular RITC-dextran by several washes, we found that RITC-dextran overlaps with the distribution of the conjugate of 6 and GFP-TEV-HA-GST (green), suggesting the conjugates are inside the cells (see Fig. S15 in supporting information†).
Conclusions
In conclusion, we demonstrated that GSH-decorated magnetic nanoparticles selectively bind to GST fusion protein from cell lysates without altering the function of the protein of interest. Although the mechanism of the uptake of the conjugate of 6 and GFP-TEV-HA-GST has yet to be elucidated, this work has provided design principles and significant insights for decorating magnetic nanoparticles to manipulate proteins and cells. This approach should be applicable to other ligand and receptor pairs, which ultimately will lead to a general and comprehensive platform for studying biological interactions and biological system using a magnetic force.Acknowledgements
The authors acknowledge the financial support from start-up grant from Brandeis University. YP thanks the assistance of Brandeis EM facility. JFS thanks the Scholarship from Chinese Scholarship Council (2008638092)Notes and references
- W. C. W. Chan and S. M. Nie, Science, 1998, 281, 2016 CrossRef CAS; G. M. Whitesides, Nat. Biotechnol., 2003, 21, 1161 CrossRef CAS; Y. N. Xia, P. D. Yang, Y. G. Sun, Y. Y. Wu, B. Mayers, B. Gates, Y. D. Yin, F. Kim and Y. Q. Yan, Adv. Mater., 2003, 15, 353 CrossRef CAS; Y. W. Jun, J. W. Seo and A. Cheon, Acc. Chem. Res., 2008, 41, 179 CrossRef CAS; A. Bajaj, S. Rana, O. R. Miranda, J. C. Yawe, D. J. Jerry, U. H. F. Bunz and V. M. Rotello, Chem. Sci., 2010, 1, 134 RSC; S. Bi, H. Zhou and S. Zhang, Chem. Sci., 2010, 1, 681 RSC; S. Chen, L. J. Wang, S. L. Duce, S. Brown, S. Lee, A. Melzer, S. A. Cuschieri and P. Andre, J. Am. Chem. Soc., 2010, 132, 15022 CrossRef CAS.
- T. Hyeon, Chem. Commun., 2003, 927 RSC; S. H. Sun, Adv. Mater., 2006, 18, 393 CrossRef CAS.
- S. A. Majetich, J. O. Artman, M. E. McHenry, N. T. Nuhfer and S. W. Staley, Phys. Rev. B: Condens. Matter, 1993, 48, 16845 CrossRef CAS; S. A. Majetich and Y. Jin, Science, 1999, 284, 470 CrossRef CAS; S. H. Sun, C. B. Murray, D. Weller, L. Folks and A. Moser, Science, 2000, 287, 1989 CrossRef CAS; J. Ge, Y. Hu, T. Zhang and Y. Yin, J. Am. Chem. Soc., 2007, 129, 8974 CrossRef CAS; U. Jeong, X. W. Teng, Y. Wang, H. Yang and Y. N. Xia, Adv. Mater., 2007, 19, 33 CrossRef CAS; J. P. Ge and Y. D. Yin, J. Mater. Chem., 2008, 18, 5041 RSC; J. Lim, A. Eggeman, F. Lanni, R. D. Tilton and S. A. Majetich, Adv. Mater., 2008, 20, 1721 CrossRef CAS; J. P. Ge, L. He, J. Goebl and Y. D. Yin, J. Am. Chem. Soc., 2009, 131, 3484 CrossRef CAS; J. Park, E. Lee, N. M. Hwang, M. S. Kang, S. C. Kim, Y. Hwang, J. G. Park, H. J. Noh, J. Y. Kini, J. H. Park and T. Hyeon, Angew. Chem., Int. Ed., 2005, 44, 2872 CrossRef CAS.
- J. H. Gao, H. W. Gu and B. Xu, Acc. Chem. Res., 2009, 42, 1097 CrossRef CAS.
- J. Gao, W. Zhang, P. Huang, B. Zhang, X. Zhang and B. Xu, J. Am. Chem. Soc., 2008, 130, 3710 CrossRef CAS.
- F. M. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, K. Struhl, L. M. Albright, D. M. Coen and A. Varki, ed., Current protocols in molecular biology, John Wiley & Sons, New York, 2003 Search PubMed.
- C. J. Xu, K. M. Xu, H. W. Gu, H. Liu, R. K. Zheng, X. X. Zhang, Z. H. Guo and B. Xu, J. Am. Chem. Soc., 2004, 126, 9938 CrossRef CAS.
- C. J. Xu, K. M. Xu, H. W. Gu, X. F. Zhong, Z. H. Guo, R. K. Zheng, X. X. Zhang and B. Xu, J. Am. Chem. Soc., 2004, 126, 3392 CrossRef CAS; K. S. Lee and I. S. Lee, Chem. Commun., 2008, 709 RSC.
- I. S. Lee, N. Lee, J. Park, B. H. Kim, Y. W. Yi, T. Kim, T. K. Kim, I. H. Lee, S. R. Paik and T. Hyeon, J. Am. Chem. Soc., 2006, 128, 10658 CrossRef CAS.
- X. Lue, X. Bao, Y. Huang, Y. Qu, H. Lu and Z. Lu, Biomater., 2009, 30, 141 CrossRef CAS.
- L. Wang, Z. M. Yang, J. H. Gao, K. M. Xu, H. W. Gu, B. Zhang, X. X. Zhang and B. Xu, J. Am. Chem. Soc., 2006, 128, 13358 CrossRef CAS; H. Lee, S. M. Dellatore, W. M. Miller and P. B. Messersmith, Science, 2007, 318, 426 CrossRef CAS; J. Xie, K. Chen, H. Y. Lee, C. J. Xu, A. R. Hsu, S. Peng, X. Y. Chen and S. H. Sun, J. Am. Chem. Soc., 2008, 130, 7542 CrossRef CAS; K. L. Young, C. J. Xu, J. Xie and S. H. Sun, J. Mater. Chem., 2009, 19, 6400 RSC.
- J. Kim, Y. Piao, N. Lee, Y. I. Park, I. H. Lee, J. H. Lee, S. R. Paik and T. Hyeon, Adv. Mater., 2010, 22, 57 CrossRef CAS.
- R. Y. Tsien, Annu. Rev. Biochem., 1998, 67, 509 CrossRef CAS.
- S. H. Sun, H. Zeng, D. B. Robinson, S. Raoux, P. M. Rice, S. X. Wang and G. X. Li, J. Am. Chem. Soc., 2004, 126, 273 CrossRef CAS.
- M. Chevallet, S. Luche and T. Rabilloud, Nat. Protoc., 2006, 1, 1852 CrossRef CAS.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c1sc00030f |
| ‡ Equal contribution from Yue Pan and Marcus J. C. Long. |
| This journal is © The Royal Society of Chemistry 2011 |
