The anti-tumor antibiotic PD 113,271 binds to microtubule-associated protein 1B (MAP1B)
Toshifumi
Takeuchi†
a,
Takahiko
Imai†
a,
Kazutomo
Ishi
a,
Takeki
Saitoh
a,
Kouji
Kuramochi
b and
Fumio
Sugawara
*a
aApplied Biological Science, Tokyo University of Science (RIKADAI), Chiba, 278-8510, Japan
bKyoto Prefectural University, Graduate School of Life and Environmental Science, Kyoto, 602-8522, Japan
First published on 14th September 2011
Abstract
PD 113,271 and fostriecin are structurally related phosphate monoesters with potent cytotoxicity against several tumor cell lines. Although there have been several biological studies on fostriecin such as that on protein phosphatase (PP) 2A inhibition and its mode of action, little is known about PD 113,271. In the present study, we identified microtubule-associated protein (MAP) 1B as a binding protein of PD 113,271 using phage display biopanning. Phages that expressed the highly basic region of MAP1B bound to the immobilized PD 113,271. In addition, PD 113,271 bound to MAP1B in the lysate of human liver cells. Moreover, we identified that PD 113,271 directly interfered with MAP1B binding to microtubules and induced a morphological abnormality in the distribution of MAP1B in cells. These results showed that PD 113,271 binds to the tubulin binding domain of MAP1B and may be useful for studying MAP1B function.
Introduction
PD 113,271 is a cytotoxic phosphate monoester of Streptomyces pulveraceus (Fig. 1).1–6 It exhibits excellent cytotoxic activity against mouse leukemia (L1210; IC50 = 1.8 μM) and human ileocecal carcinoma (HCT-8; IC50 = 9.0 μM) in vitro and anti-tumor activity against L1210 and P388 leukemia in vivo.7–9 It has been reported that PD 113,271 inhibits the enzyme topoisomerase II with a minimum inhibitory concentration (MIC) of 12.5 μM in an in vitro assay.10,11 Fostriecin1–6 (parent compound of PD 113,271), cytostatin12,13 and phoslactomycins14,15 comprising the fostriecin family share common structures such as α,β-unsaturated-δ-lactones, phosphate monoesters, and 9,11-diols, and commonly inhibit protein phosphatase (PP) 2A16–19 (Fig. 1). Several synthetic and biological studies on the fostriecin family revealed that these common structures play an important role in PP2A inhibition.20–30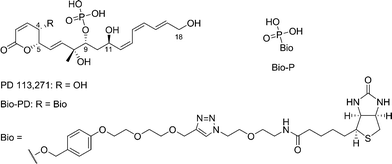 | ||
| Fig. 1 Structures of PD 113,271, biotin-labeled PD 113,271 (bio-PD), and biotin-labeled phosphate monoester (bio-P). | ||
These compounds have been reported to have many biological activities of fostriecin such as in vivo and in vitro anti-tumor activity (L1210, IC50 = 0.46 μM; HTC-8, IC50 = 5.1 μM, respectively),7–9 inhibition of topoisomerase II (MIC, 50 μM),10,11 selective inhibition of PP2A (IC50 = 1.5 nM),16,17 and interruption of the cellular mitotic entry checkpoint through PP2A inhibition.12,13,31–34 However, the relationship between fostriecin's activity against topoisomerase II or PP2A and potent cytotoxicity is not well understood.35 Based on the similarity between PD 113,271 and fostriecin, PD 113,271 could also inhibit PP2A. However, there have been few reports of biological studies on PD 113,271. Therefore, we attempted to screen a PD 113,271-binding protein using phage display biopanning36–38 that is widely used for target identification. In the present study, we have shown that PD 113,271 bound to the highly basic region of microtubule-associated protein (MAP) 1B, a domain known to bind to tubulin,39in vitro and in a cell lysate. Moreover, PD 113,271 inhibited the binding activity of MAP1B to tubulin. Furthermore, we investigated that PD 113,271 induced a morphological abnormality in the distribution of MAP1B in pheochromocytoma (PC12) cells. To our knowledge, the present study is the first to show that PD 113,271 binds to MAP1B.
Results
Selection of phages associated with PD 113,271
To identify PD 113,271-binding proteins, phage display biopanning was performed using biotin-labeled PD 113,271 (bio-PD; Fig. 1).29 Our previous study had shown that this bio-PD probe had an inhibitory activity against proliferation of mouse leukemia cells (L1210, IC50 = 58 μM), and covalently bound the PP2A catalytic subunit (PP2Ac),29 suggesting that PP2Ac-expressing phages should covalently bind to bio-PD immobilized on wells. Therefore, PP2Ac-expressing phages should not be selected by phage display biopanning. Before biopanning, we confirmed the affinity for the bio-PD immobilized on wells using a PP2Ac-expressing phage clone and observed that PP2Ac-expressing phages were not eluted from the well (data not shown), indicating that these phages covalently bound to the PD 113,271 immobilized on the well. Thus, we decided that bio-PD could be used to identify PD 113,271-binding proteins.A phage library displaying human liver cancer cell line cDNA was screened by affinity selection using bio-PD. During 5 rounds of selection, the elution phage titer increased from 9.0 × 105 (PFU mL−1) in the first round to 2.9 × 107 (PFU mL−1) in the fifth round (Fig. 2), suggesting that PD 113,271-bound phages should be efficiently enriched after 5 rounds of selection. Next, we randomly selected 16 clones from the phage particles eluted in the fifth round and sequenced their inserted cDNA (Table 1). A PP2Ac-expressing phage clone was not obtained, as expected. Fourteen–sixteenths of the phages displayed a part of MAP1B (Table 1, clones #1, #2, and #3), and three–sixteenths displayed ribosomal L1 domain-containing protein (RSL1D1) (Table 1, clone #4).
| Phage clone | Sequencea | Frequencyb | Protein name | E value |
|---|---|---|---|---|
| a Amino acid sequences were determined from the sequences of the inserted DNA. b Frequency is the number of sequences among the bound phage clones. c A partial sequence is shown. The following sequence was not analyzed. d Sequences similar to the corresponding protein are underlined. | ||||
| 1 | NS![[S with combining low line]](https://www.rsc.org/images/entities/char_0053_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[E with combining low line]](https://www.rsc.org/images/entities/char_0045_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[V with combining low line]](https://www.rsc.org/images/entities/char_0056_0332.gif) ![[M with combining low line]](https://www.rsc.org/images/entities/char_004d_0332.gif) ![[V with combining low line]](https://www.rsc.org/images/entities/char_0056_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[D with combining low line]](https://www.rsc.org/images/entities/char_0044_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[P with combining low line]](https://www.rsc.org/images/entities/char_0050_0332.gif) ![[V with combining low line]](https://www.rsc.org/images/entities/char_0056_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif) ![[E with combining low line]](https://www.rsc.org/images/entities/char_0045_0332.gif) ![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[P with combining low line]](https://www.rsc.org/images/entities/char_0050_0332.gif) ![[S with combining low line]](https://www.rsc.org/images/entities/char_0053_0332.gif) ![[V with combining low line]](https://www.rsc.org/images/entities/char_0056_0332.gif) ![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif) ![[E with combining low line]](https://www.rsc.org/images/entities/char_0045_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[E with combining low line]](https://www.rsc.org/images/entities/char_0045_0332.gif) ![[V with combining low line]](https://www.rsc.org/images/entities/char_0056_0332.gif) ![[P with combining low line]](https://www.rsc.org/images/entities/char_0050_0332.gif) ![[S with combining low line]](https://www.rsc.org/images/entities/char_0053_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[E with combining low line]](https://www.rsc.org/images/entities/char_0045_0332.gif) ![[E with combining low line]](https://www.rsc.org/images/entities/char_0045_0332.gif) ![[P with combining low line]](https://www.rsc.org/images/entities/char_0050_0332.gif) ![[S with combining low line]](https://www.rsc.org/images/entities/char_0053_0332.gif) ![[P with combining low line]](https://www.rsc.org/images/entities/char_0050_0332.gif) ![[V with combining low line]](https://www.rsc.org/images/entities/char_0056_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) ![[E with combining low line]](https://www.rsc.org/images/entities/char_0045_0332.gif) ![[V with combining low line]](https://www.rsc.org/images/entities/char_0056_0332.gif) ![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) ![[E with combining low line]](https://www.rsc.org/images/entities/char_0045_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[Q with combining low line]](https://www.rsc.org/images/entities/char_0051_0332.gif) ![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) ![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif) ![[D with combining low line]](https://www.rsc.org/images/entities/char_0044_0332.gif) ![[V with combining low line]](https://www.rsc.org/images/entities/char_0056_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[P with combining low line]](https://www.rsc.org/images/entities/char_0050_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) ![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[E with combining low line]](https://www.rsc.org/images/entities/char_0045_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif) ![[V with combining low line]](https://www.rsc.org/images/entities/char_0056_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[E with combining low line]](https://www.rsc.org/images/entities/char_0045_0332.gif) ![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[V with combining low line]](https://www.rsc.org/images/entities/char_0056_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[P with combining low line]](https://www.rsc.org/images/entities/char_0050_0332.gif) ![[E with combining low line]](https://www.rsc.org/images/entities/char_0045_0332.gif) ![[D with combining low line]](https://www.rsc.org/images/entities/char_0044_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[E with combining low line]](https://www.rsc.org/images/entities/char_0045_0332.gif) ![[E with combining low line]](https://www.rsc.org/images/entities/char_0045_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[E with combining low line]](https://www.rsc.org/images/entities/char_0045_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[P with combining low line]](https://www.rsc.org/images/entities/char_0050_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[E with combining low line]](https://www.rsc.org/images/entities/char_0045_0332.gif) ![[V with combining low line]](https://www.rsc.org/images/entities/char_0056_0332.gif) ![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[E with combining low line]](https://www.rsc.org/images/entities/char_0045_0332.gif) ![[D with combining low line]](https://www.rsc.org/images/entities/char_0044_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif) ![[P with combining low line]](https://www.rsc.org/images/entities/char_0050_0332.gif) ![[I with combining low line]](https://www.rsc.org/images/entities/char_0049_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[E with combining low line]](https://www.rsc.org/images/entities/char_0045_0332.gif) ![[E with combining low line]](https://www.rsc.org/images/entities/char_0045_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[P with combining low line]](https://www.rsc.org/images/entities/char_0050_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[E with combining low line]](https://www.rsc.org/images/entities/char_0045_0332.gif) ![[E with combining low line]](https://www.rsc.org/images/entities/char_0045_0332.gif) ![[V with combining low line]](https://www.rsc.org/images/entities/char_0056_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[E with combining low line]](https://www.rsc.org/images/entities/char_0045_0332.gif) ![[V with combining low line]](https://www.rsc.org/images/entities/char_0056_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[E with combining low line]](https://www.rsc.org/images/entities/char_0045_0332.gif) ![[I with combining low line]](https://www.rsc.org/images/entities/char_0049_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[E with combining low line]](https://www.rsc.org/images/entities/char_0045_0332.gif) ![[E with combining low line]](https://www.rsc.org/images/entities/char_0045_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[E with combining low line]](https://www.rsc.org/images/entities/char_0045_0332.gif) ![[P with combining low line]](https://www.rsc.org/images/entities/char_0050_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[E with combining low line]](https://www.rsc.org/images/entities/char_0045_0332.gif) ![[V with combining low line]](https://www.rsc.org/images/entities/char_0056_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[E with combining low line]](https://www.rsc.org/images/entities/char_0045_0332.gif) ![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif) ![[P with combining low line]](https://www.rsc.org/images/entities/char_0050_0332.gif) c,d c,d |
10/16 | MAP1B (582–705) | 1.0 × 10−19 |
| 2 | NS![[P with combining low line]](https://www.rsc.org/images/entities/char_0050_0332.gif) ![[E with combining low line]](https://www.rsc.org/images/entities/char_0045_0332.gif) ![[D with combining low line]](https://www.rsc.org/images/entities/char_0044_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[E with combining low line]](https://www.rsc.org/images/entities/char_0045_0332.gif) ![[E with combining low line]](https://www.rsc.org/images/entities/char_0045_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) SQR SQR![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) WLKRRTKHLSRRRKNQKRKRd WLKRRTKHLSRRRKNQKRKRd |
2/16 | MAP1B (646–653) | 2.0 × 102 |
| 3 | NS![[P with combining low line]](https://www.rsc.org/images/entities/char_0050_0332.gif) ![[E with combining low line]](https://www.rsc.org/images/entities/char_0045_0332.gif) ![[D with combining low line]](https://www.rsc.org/images/entities/char_0044_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[E with combining low line]](https://www.rsc.org/images/entities/char_0045_0332.gif) ![[E with combining low line]](https://www.rsc.org/images/entities/char_0045_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[E with combining low line]](https://www.rsc.org/images/entities/char_0045_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[P with combining low line]](https://www.rsc.org/images/entities/char_0050_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[E with combining low line]](https://www.rsc.org/images/entities/char_0045_0332.gif) ![[V with combining low line]](https://www.rsc.org/images/entities/char_0056_0332.gif) ![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[E with combining low line]](https://www.rsc.org/images/entities/char_0045_0332.gif) ![[D with combining low line]](https://www.rsc.org/images/entities/char_0044_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif) ![[P with combining low line]](https://www.rsc.org/images/entities/char_0050_0332.gif) ![[I with combining low line]](https://www.rsc.org/images/entities/char_0049_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[E with combining low line]](https://www.rsc.org/images/entities/char_0045_0332.gif) ![[E with combining low line]](https://www.rsc.org/images/entities/char_0045_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[P with combining low line]](https://www.rsc.org/images/entities/char_0050_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[E with combining low line]](https://www.rsc.org/images/entities/char_0045_0332.gif) ![[E with combining low line]](https://www.rsc.org/images/entities/char_0045_0332.gif) ![[V with combining low line]](https://www.rsc.org/images/entities/char_0056_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[E with combining low line]](https://www.rsc.org/images/entities/char_0045_0332.gif) ![[V with combining low line]](https://www.rsc.org/images/entities/char_0056_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[E with combining low line]](https://www.rsc.org/images/entities/char_0045_0332.gif) ![[I with combining low line]](https://www.rsc.org/images/entities/char_0049_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[E with combining low line]](https://www.rsc.org/images/entities/char_0045_0332.gif) ![[E with combining low line]](https://www.rsc.org/images/entities/char_0045_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[E with combining low line]](https://www.rsc.org/images/entities/char_0045_0332.gif) ![[P with combining low line]](https://www.rsc.org/images/entities/char_0050_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[E with combining low line]](https://www.rsc.org/images/entities/char_0045_0332.gif) ![[V with combining low line]](https://www.rsc.org/images/entities/char_0056_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[E with combining low line]](https://www.rsc.org/images/entities/char_0045_0332.gif) ![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif) ![[P with combining low line]](https://www.rsc.org/images/entities/char_0050_0332.gif) ![[P with combining low line]](https://www.rsc.org/images/entities/char_0050_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[E with combining low line]](https://www.rsc.org/images/entities/char_0045_0332.gif) ![[V with combining low line]](https://www.rsc.org/images/entities/char_0056_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[E with combining low line]](https://www.rsc.org/images/entities/char_0045_0332.gif) ![[V with combining low line]](https://www.rsc.org/images/entities/char_0056_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[E with combining low line]](https://www.rsc.org/images/entities/char_0045_0332.gif) ![[E with combining low line]](https://www.rsc.org/images/entities/char_0045_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[E with combining low line]](https://www.rsc.org/images/entities/char_0045_0332.gif) ![[V with combining low line]](https://www.rsc.org/images/entities/char_0056_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[E with combining low line]](https://www.rsc.org/images/entities/char_0045_0332.gif) ![[E with combining low line]](https://www.rsc.org/images/entities/char_0045_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[E with combining low line]](https://www.rsc.org/images/entities/char_0045_0332.gif) ![[P with combining low line]](https://www.rsc.org/images/entities/char_0050_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[E with combining low line]](https://www.rsc.org/images/entities/char_0045_0332.gif) ![[I with combining low line]](https://www.rsc.org/images/entities/char_0049_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[L with combining low line]](https://www.rsc.org/images/entities/char_004c_0332.gif) ![[P with combining low line]](https://www.rsc.org/images/entities/char_0050_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[D with combining low line]](https://www.rsc.org/images/entities/char_0044_0332.gif) ![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[S with combining low line]](https://www.rsc.org/images/entities/char_0053_0332.gif) ![[S with combining low line]](https://www.rsc.org/images/entities/char_0053_0332.gif) ![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif) ![[P with combining low line]](https://www.rsc.org/images/entities/char_0050_0332.gif) ![[L with combining low line]](https://www.rsc.org/images/entities/char_004c_0332.gif) ![[S with combining low line]](https://www.rsc.org/images/entities/char_0053_0332.gif) ![[E with combining low line]](https://www.rsc.org/images/entities/char_0045_0332.gif) ![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[P with combining low line]](https://www.rsc.org/images/entities/char_0050_0332.gif) ![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) ![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) ![[L with combining low line]](https://www.rsc.org/images/entities/char_004c_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[P with combining low line]](https://www.rsc.org/images/entities/char_0050_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[V with combining low line]](https://www.rsc.org/images/entities/char_0056_0332.gif) ![[P with combining low line]](https://www.rsc.org/images/entities/char_0050_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[E with combining low line]](https://www.rsc.org/images/entities/char_0045_0332.gif) ![[E with combining low line]](https://www.rsc.org/images/entities/char_0045_0332.gif) ![[S with combining low line]](https://www.rsc.org/images/entities/char_0053_0332.gif) ![[V with combining low line]](https://www.rsc.org/images/entities/char_0056_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[D with combining low line]](https://www.rsc.org/images/entities/char_0044_0332.gif) ![[S with combining low line]](https://www.rsc.org/images/entities/char_0053_0332.gif) ![[V with combining low line]](https://www.rsc.org/images/entities/char_0056_0332.gif) ![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) ![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[P with combining low line]](https://www.rsc.org/images/entities/char_0050_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) LAAALEd LAAALEd |
1/16 | MAP1B (646–777) | 1.6 × 10−28 |
| 4 | NSKKKRQQ![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) ![[R with combining low line]](https://www.rsc.org/images/entities/char_0052_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif) ![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) ![[S with combining low line]](https://www.rsc.org/images/entities/char_0053_0332.gif) ![[V with combining low line]](https://www.rsc.org/images/entities/char_0056_0332.gif) ![[L with combining low line]](https://www.rsc.org/images/entities/char_004c_0332.gif) ![[S with combining low line]](https://www.rsc.org/images/entities/char_0053_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[D with combining low line]](https://www.rsc.org/images/entities/char_0044_0332.gif) ![[D with combining low line]](https://www.rsc.org/images/entities/char_0044_0332.gif) ![[V with combining low line]](https://www.rsc.org/images/entities/char_0056_0332.gif) ![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) ![[P with combining low line]](https://www.rsc.org/images/entities/char_0050_0332.gif) ![[E with combining low line]](https://www.rsc.org/images/entities/char_0045_0332.gif) ![[S with combining low line]](https://www.rsc.org/images/entities/char_0053_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[D with combining low line]](https://www.rsc.org/images/entities/char_0044_0332.gif) ![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif) ![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif) ![[V with combining low line]](https://www.rsc.org/images/entities/char_0056_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[P with combining low line]](https://www.rsc.org/images/entities/char_0050_0332.gif) ![[E with combining low line]](https://www.rsc.org/images/entities/char_0045_0332.gif) ![[S with combining low line]](https://www.rsc.org/images/entities/char_0053_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[E with combining low line]](https://www.rsc.org/images/entities/char_0045_0332.gif) ![[Q with combining low line]](https://www.rsc.org/images/entities/char_0051_0332.gif) ![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif) ![[P with combining low line]](https://www.rsc.org/images/entities/char_0050_0332.gif) ![[E with combining low line]](https://www.rsc.org/images/entities/char_0045_0332.gif) ![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[R with combining low line]](https://www.rsc.org/images/entities/char_0052_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[R with combining low line]](https://www.rsc.org/images/entities/char_0052_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) ![[Q with combining low line]](https://www.rsc.org/images/entities/char_0051_0332.gif) ![[V with combining low line]](https://www.rsc.org/images/entities/char_0056_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) ![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif) ![[N with combining low line]](https://www.rsc.org/images/entities/char_004e_0332.gif) ![[E with combining low line]](https://www.rsc.org/images/entities/char_0045_0332.gif) ![[S with combining low line]](https://www.rsc.org/images/entities/char_0053_0332.gif) ![[E with combining low line]](https://www.rsc.org/images/entities/char_0045_0332.gif) ![[D with combining low line]](https://www.rsc.org/images/entities/char_0044_0332.gif) ![[E with combining low line]](https://www.rsc.org/images/entities/char_0045_0332.gif) ![[I with combining low line]](https://www.rsc.org/images/entities/char_0049_0332.gif) ![[P with combining low line]](https://www.rsc.org/images/entities/char_0050_0332.gif) ![[Q with combining low line]](https://www.rsc.org/images/entities/char_0051_0332.gif) ![[L with combining low line]](https://www.rsc.org/images/entities/char_004c_0332.gif) ![[V with combining low line]](https://www.rsc.org/images/entities/char_0056_0332.gif) ![[P with combining low line]](https://www.rsc.org/images/entities/char_0050_0332.gif) ![[I with combining low line]](https://www.rsc.org/images/entities/char_0049_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif) ![[P with combining low line]](https://www.rsc.org/images/entities/char_0050_0332.gif) ![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) ![[N with combining low line]](https://www.rsc.org/images/entities/char_004e_0332.gif) ![[E with combining low line]](https://www.rsc.org/images/entities/char_0045_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[V with combining low line]](https://www.rsc.org/images/entities/char_0056_0332.gif) ![[E with combining low line]](https://www.rsc.org/images/entities/char_0045_0332.gif) ![[I with combining low line]](https://www.rsc.org/images/entities/char_0049_0332.gif) ![[Q with combining low line]](https://www.rsc.org/images/entities/char_0051_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) ![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) ![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[S with combining low line]](https://www.rsc.org/images/entities/char_0053_0332.gif) ![[P with combining low line]](https://www.rsc.org/images/entities/char_0050_0332.gif) ![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[S with combining low line]](https://www.rsc.org/images/entities/char_0053_0332.gif) ![[P with combining low line]](https://www.rsc.org/images/entities/char_0050_0332.gif) ![[N with combining low line]](https://www.rsc.org/images/entities/char_004e_0332.gif) ![[P with combining low line]](https://www.rsc.org/images/entities/char_0050_0332.gif) ![[S with combining low line]](https://www.rsc.org/images/entities/char_0053_0332.gif) ![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif) ![[P with combining low line]](https://www.rsc.org/images/entities/char_0050_0332.gif) ![[R with combining low line]](https://www.rsc.org/images/entities/char_0052_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[R with combining low line]](https://www.rsc.org/images/entities/char_0052_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) ![[L with combining low line]](https://www.rsc.org/images/entities/char_004c_0332.gif) ![[P with combining low line]](https://www.rsc.org/images/entities/char_0050_0332.gif) ![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) ![[S with combining low line]](https://www.rsc.org/images/entities/char_0053_0332.gif) ![[E with combining low line]](https://www.rsc.org/images/entities/char_0045_0332.gif) ![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif) ![[P with combining low line]](https://www.rsc.org/images/entities/char_0050_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) ![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) ![[E with combining low line]](https://www.rsc.org/images/entities/char_0045_0332.gif) ![[S with combining low line]](https://www.rsc.org/images/entities/char_0053_0332.gif) ![[E with combining low line]](https://www.rsc.org/images/entities/char_0045_0332.gif) ![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif) ![[P with combining low line]](https://www.rsc.org/images/entities/char_0050_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[S with combining low line]](https://www.rsc.org/images/entities/char_0053_0332.gif) ![[P with combining low line]](https://www.rsc.org/images/entities/char_0050_0332.gif) ![[E with combining low line]](https://www.rsc.org/images/entities/char_0045_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) ![[P with combining low line]](https://www.rsc.org/images/entities/char_0050_0332.gif) SLRPHSSNd SLRPHSSNd |
3/16 | Ribosomal L1 domain-containing protein (309–432) | 3.3 × 10−39 |
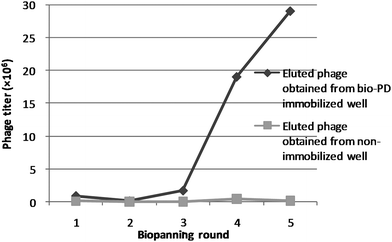 | ||
| Fig. 2 Enrichment of phage particles binding to bio-PD. A phage library displaying human liver cancer cell line cDNA was screened by affinity selection using bio-PD. The solution of the phage library was incubated in wells with or without bio-PD for 1 h. After washing the wells with 0.05% (v/v) Tween-20 in TBS (TBS-T), the phage particles eluted with 1% (w/v) SDS in TBS (TBS-S) were counted and subsequently amplified by infecting Escherichia coli BLT5615. The amplified phage particles were then subjected to the next round of selection. | ||
PD 113,271 binds to the microtubule-binding domain of MAP1B
It was expected that phage clone #1, expressing a 646–777 amino acid (a.a) region of MAP1B, and/or phage clone #4, expressing a 309–432 a.a. region of RSL1D1, would bind to PD 113,271 (Table 1). Since clones #1–4 had highly basic regions including arginine, histidine, and lysine, it is possible that the phosphate monoester in PD 113,271 non-specifically bound to these clones. Therefore, we confirmed the affinity for immobilized bio-PD and biotin-labeled phosphate monoester (bio-P; Fig. 1) using phage clones #1 and #4. Phage clone #1 showed a significant affinity for the immobilized bio-PD (Fig. 3A, clone #1) but showed no interaction with bio-P. Phage clone #4 showed moderate affinity for non-ligand wells compared with those for bio-PD and bio-P (Fig. 3A, clone #4), suggesting that clone #4 was more strongly bound to the wells than PD 113,271. These results demonstrated that phage clone #1 that expressed MAP1B had a highly specific affinity for PD 113,271.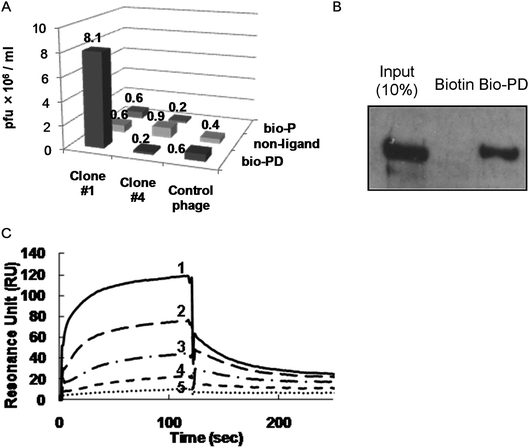 | ||
| Fig. 3 PD 113,271 binding to the microtubule binding domain of MAP1B. (A) Single phage clone binding assay. Phage titers of single phage clones (clone #1, clone #4 or control phage expressing the short peptide sequence Asp-Ser) bound to streptavidin-coated wells with or without bio-PD or bio-P was measured. (B) Pull-down assay with biotin or bio-PD in cell lysate. Biotin- or bio-PD-immobilized beads were treated with PC12 cell lysates. Proteins bound to biotin or bio-PD were visualized by western blotting analysis using the anti-MAP1B antibody. (C) SPR analysis of the binding between PD 113,271 and MAP1B-HBR (partial recombinant MAP1B; 589–790 a.a.). Analytes were injected over flow cells on the immobilized PD 113,271 using bio-PD. The background resulting from an injection of running buffer was subtracted from the data before plotting. Response units (RU) were generated by subtraction of the background signal generated simultaneously on the control flow cell. Binding of various concentrations of MAP1B-HBR (1, 1 μM; 2, 0.5 μM; 3, 0.25 μM; 4, 0.13 μM; 5, 0.063 μM) to the immobilized PD 113,271 was analyzed. Kinetic studies were performed using the BIA evaluation software. | ||
Next, we examined whether bio-PD bound to native MAP1B. We performed a pull-down assay using bio-PD-immobilized beads in rat PC12 cell lysate. Western blotting using the anti-MAP1B antibody indicated that PD 113,271 bound to native MAP1B in the cell lysate (Fig. 3B). Using surface plasmon resonance (SPR) (Fig. 3C), the equilibrium dissociation constant for immobilized bio-PD and the recombinant highly basic region of MAP1B (MAP1B-HBR, 589–790 a.a; Fig. 4) was determined to be 0.21 μM. These results indicated that PD 113,271 bound to MAP1B through a highly basic region that is also known to bind to microtubules.39
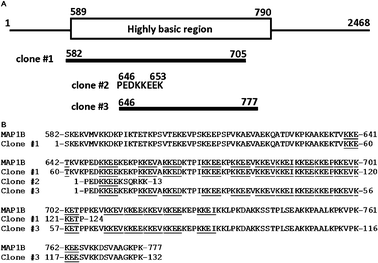 | ||
| Fig. 4 (A) Schematic diagram of human MAP1B and clones #1–3. (B) MAP1B and clones #1–3 were aligned. The KKE(E/V/D/I/P) motifs in each sequence are underlined. | ||
PD 113,271 dissociates MAP1B-HBR from tubulin and induces an abnormal distribution of MAP1B in cells
Yamauchi et al. reported that acidic phospholipids such as phosphatidic acid dissociated MAP1B from tubulin.40 Since PD 113,271 is an acidic phospholipid, we subsequently surveyed the interaction between MAP1B-HBR and tubulin in the absence and presence of PD 113,271. SPR analysis of tubulin and MAP1B-HBR with or without PD 113,271 demonstrated that binding of PD 113,271 to MAP1B-HBR dissociated MAP1B-HBR from tubulin (Fig. 5A).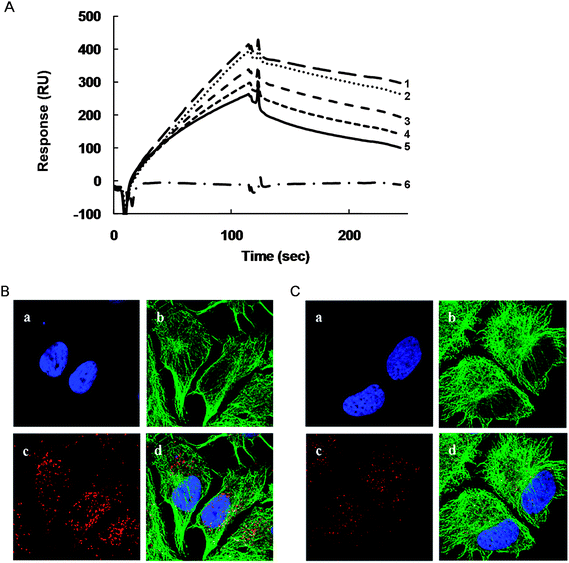 | ||
| Fig. 5 PD 113,271 dissociates MAP1B-HBR from tubulin and induces abnormal distribution of MAP1B in cells. (A) SPR analysis of the binding between PD 113,271, tubulin, and MAP1B-HBR (partial recombinant MAP1B; 589–790 a.a.). Analytes were injected over flow cells on the immobilized tubulin. The background resulting from an injection of running buffer was subtracted from the data before plotting. Response units (RU) were generated by subtraction of the background signal generated simultaneously on the control flow cell. Binding of MAP1B-HBR (1 μM) to immobilized tubulin under various concentrations of PD 113,271 (1, 0 μM; 2, 1 μM; 3, 5 μM; 4, 10 μM; 5, 20 μM), and binding of PD 113,271 (6, 20 μM) to tubulin were analyzed. (B and C) MCF7 cells were immunostained using the anti-tubulin and anti-MAP1B antibodies. (a; DAPI, b; anti-tubulin Alexa 488, c; anti-MAP1B Alexa 568, d; merge) with PD 113,271 (1 μM) (B) or without PD 113,271 (C). | ||
To characterize the relationship between PD 113,271-induced growth arrest and the inhibitory effect of PD 113,271 on the binding of MAP1B-HBR to tubulin, we observed a PD 113,271-induced morphological change using immunofluorescent microscopy. In this technique, we used fixed MCF7 cells with antibodies against MAP1B and tubulin and demonstrated that, in MCF7 cells, MAP1B was localized throughout the cytoplasm and co-localized with tubulin in several areas (Fig. 5C). On the other hand, when cells were treated with PD 113,271 (1 μM) majority of MAP1B was localized around the perinuclear region. Moreover, MAP1B was shown to increase in size in the presence of PD 113,271 compared with untreated cells (Fig. 5B).
Discussion
In the present study, we isolated a phage clone expressing a highly basic region of microtubule-associated protein (MAP1B-HBR) that had an affinity for PD 113,271. MAP1B promotes microtubule assembly and stabilizes microtubules by binding to them.39 This function is regulated by the degree of phosphorylation, which is controlled by the activities of protein kinases and phosphatases such as PP2A and 2B.41,42 The inhibition of PP2A increases the phosphorylated MAP1B and decreases MAP1B microtubule-binding activity.42 While MAP1B lacks the microtubule-binding motifs common to other MAPs such as MAP2 and tau, it has a unique, highly basic region comprising multiple and repeated copies of a short motif, KKE(E/V/D/I/P) (Fig. 4B, underlined) that is known to bind with tubulin.39MAP1B-HBR contains 18 KKE(E/V/D/I/P) motifs and has a calculated isoelectric point of 10.1 (Fig. 4B). Since a crystal structure of MAP1B has not been determined, we performed a modeling analysis of the short motifs of MAP1B, such as KKE(E/V/D/I/P), predicted from molecular dynamic calculations. Model structures of KKEE and KKED indicated that lysine and glutamic acid or aspartic acid side chains interacted intramolecularly and constructed a bi-ring structure (data not shown). Model structures of KKE(V/I/P) indicated that a lysine side chain and a glutamic acid side chain interact intramolecularly, and another lysine side chain can interact with an acidic residue (data not shown). Consideration of the report that MAP1B may interact with a phospholipid bilayer through microtubule-binding domain39 suggests that compounds containing a phosphate monoester can non-specifically bind to MAP1B. However, a binding analysis of MAP1B-HBR and bio-P revealed that bio-P showed no interaction with MAP1B (Fig. 3A). We calculated the binding energies of the modeled KKEV peptide with bio-PD and bio-P to be −1321 and −903 kcal mol−1, respectively. The binding model of KKEV with bio-PD showed a stronger binding energy because the hydrophobic side chain of PD 113,271 interacted with that of valine (data not shown). MAPs can bind through their microtubule wall and stabilize microtubules,39 and the interaction of MAPs with microtubules is regulated by phosphorylation. Inhibition of PP2A causes MAPs to dissociate from the microtubule wall leading to reduced microtubule stability.41,42 PD 113,271 belongs to the fostriecin family and is known to be a PP2Ac inhibitor, and its parent compound, fostriecin, inhibits PP2A (IC50 = 3.2 nM). PD 113,271 may inhibit both PP2A and the interactions between MAP1B and microtubules and induce a morphological abnormality in the distribution of MAP1B.
Conclusion
In the present study, we used phage display biopanning to isolate a binding protein of PD 113,271 from a phage library displaying human liver cancer cell line cDNA. We subsequently identified that PD 113,271 bound to the highly basic region of MAP1B, which is known to bind to tubulin. Direct interaction of PD 113,271 and MAP1B was confirmed using phage clone binding and pull-down assays. Moreover, PD 113,271 inhibited the binding of MAP1B to tubulin. Furthermore, we showed that PD 113,271 induced a morphological abnormality in the distribution of MAP1B. This study opens up new opportunities for the development of new tools for biological studies and cancer drugs.Experimental section
Construction of a T7 phage library from the liver cancer cell line MH14
T7Select10-3 OrientExpress cDNA Cloning System, Random Primer, and T7Select10-3 Cloning Kit were purchased from Novagen. Construction of the phage library was performed according to the manufacturer's instructions. In brief, an aliquot (117.5 μg) of total RNA, extracted from human liver cancer MH14 cells, was used to construct a cDNA library. Total RNA was treated with an mRNA purification kit (GE Healthcare) to produce poly(A) + RNA, and 4 μg was used to synthesize first-strand DNA by using a HindIII random primer, MMLV-RT, and methylation dNTP. Second-strand cDNA was synthesized with Escherichia coli DNA polymerase I and RNase H. The double-stranded cDNA was then treated with T4 DNA polymerase to generate flush ends and ligated with an EcoRI/HindIII linker. After linker ligation, the cDNA was sequentially digested with HindIII and EcoRI. The small cDNA fragments were then removed using a Mini Column Fraction Kit. The cDNA was then inserted into the EcoRI/HindIII site of the T7 phage 10-3b vector and packaged. The titer of the primary recombinants was 4.8 × 105 PFU mL−1.T7 phage biopanning procedure and DNA sequence analysis
Streptavidin-coated enzyme-linked immunosorbent assay (ELISA) wells (Nalge Nunc International, Wiesbaden, Germany) were treated with biotin-labeled fostriecin (10 μM in 50 mM tris-buffered saline (TBS), 200 μL, 2.0 nmol) or TBS (200 μL) at room temperature for 30 min, and the wells were washed with 200 μL of TBS 3 times to produce PD 113,271-immobilized or non-immobilized wells, respectively. Next, the T7 phage library (1.0 × 1010 PFU mL−1, 200 μL) was added to each well. After incubation of the wells with gentle rotation for 1 h, the wells were washed with 200 μL of 0.1% Tween-20 (v/v) in TBS wash buffer (TBS-T) 10 times, and bound phages were eluted from the wells with 200 μL of elution buffer (1% SDS (w/v) in TBS) to give eluates of the phages. The number of phages in each eluate was counted. The phages in the eluate (100 μL) from the PD 113,271-immobilized wells were amplified by infecting the E. coli strain BLT5615 grown in 10 mL of LB medium containing 1 mM isopropyl-β-D-thiogalactoside (IPTG). Amplified phages were applied to subsequent rounds of selection. After 5 rounds of selection, the phages eluted from the PD 113,271-immobilized wells in the fifth round were used to infect BLT5615 grown on Luria-Bertani (LB) plates containing 50 μg mL−1 carbenicillin. Sixteen plaques were randomly picked up from the plate. Each DNA inserted into a phage was amplified by polymerase chain reaction (PCR) using T7 SelectUP and T7 SelectDown primers (T7 Select Cloning kit, Novagen). Free primers and dNTPs in the PCR products were removed using ExoSAP-IT (usb) and sequenced using a PRISM BigDye Terminator Cycle Sequencing Ready Reaction kit (version 3.1) (PE-ABI, Foster City, CA) and an ABI3100 sequencer (Applied Biosystems, Foster City, CA). A homology search with the BLASTP algorithm (BLAST; http://www.ncbi.nlm.nih.gov/blast/Blast.cgi) was used to determine the peptide sequences expressed on the T7 phage particles.Single phage clone binding assay
Streptavidin-coated ELISA plates were treated with or without bio-PD or bio-P for 1 h at room temperature and were washed with 200 μL of TBS. Next, 200 μL of a single phage clone displaying clone #1, clone #4, or the peptide sequence Asp-Ser (control phage), containing 0.01% SDS, was added to each well. After incubation for 1 h with gentle rotation, the plates were washed with TBS-T 10 times, and bound phages were eluted with 200 μL of 1% SDS in TBS (TBS-S). The phage titer of eluted phages was measured.Protein expression and purification
DNA encoding the highly basic region of MAP1B (589–790 a.a.) was amplified from the cDNA library of MH14 cells using the forward primer containing the NdeI site (5′-CATATGAGCAAAGAAAAGGTAATGGT-3′) and the reverse primer containing the SacI site (5′-GAGCTCTCAGCCAATGGCTGCTATTCCA-3′). The DNA fragment was inserted into the NdeI and SacI sites of the pET28a expression vector (Novagen). E. coli BL21(DE3) was transformed with the resulting plasmid and grown in 200 mL of LB medium containing 30 μg mL−1 kanamycin at 37 °C. When the absorbance at 600 nm reached 0.5, gene expression was induced with 1 mM IPTG for 4 h. The cells were then harvested by centrifugation at 5000 × g for 10 min and disrupted by sonication in 15 mL of ice-cold column binding buffer (8.1 mM Na2HPO4, 500 mM NaCl, 2.68 mM KCl, 1.47 mM KH2PO4, 20 mM imidazole). Cell debris was removed by centrifugation at 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g for 10 min, and the resulting crude extract was filtered with a 0.45 μm filter (Millipore). Next, the soluble protein fraction was loaded onto a 1 mL HisTrap HP column (GE Healthcare) of the FPLC system (AKTA 1 explorer, GE Healthcare) with a flow rate of 0.5 mL min−1. Bound proteins were washed with binding buffer and eluted using a gradient method with the elution buffer (8.1 mM Na2HPO4, 500 mM NaCl, 2.68 mM KCl, 1.47 mM KH2PO4, 500 mM imidazole). The purity of the eluted fraction was confirmed by 15% SDS (v/v) polyacrylamide gel electrophoresis. The concentration of the purified MAP1B-HBR was determined using the Bio-Rad Protein Assay (BioRad).
000 × g for 10 min, and the resulting crude extract was filtered with a 0.45 μm filter (Millipore). Next, the soluble protein fraction was loaded onto a 1 mL HisTrap HP column (GE Healthcare) of the FPLC system (AKTA 1 explorer, GE Healthcare) with a flow rate of 0.5 mL min−1. Bound proteins were washed with binding buffer and eluted using a gradient method with the elution buffer (8.1 mM Na2HPO4, 500 mM NaCl, 2.68 mM KCl, 1.47 mM KH2PO4, 500 mM imidazole). The purity of the eluted fraction was confirmed by 15% SDS (v/v) polyacrylamide gel electrophoresis. The concentration of the purified MAP1B-HBR was determined using the Bio-Rad Protein Assay (BioRad).
Surface plasmon resonance (SPR) analysis
SPR analysis was performed on a BIAcore 3000 system (GE Healthcare). Bio-PD 113,271 was immobilized on a SA sensor chip (GE Healthcare) according to the manufacturer's instructions. Bio-P was immobilized on a control flow cell. Binding analysis of the interaction between PD 113,271 and MAP1B-HBR (1, 0.5, 0.25, 0.13, 0.63 μM) was performed in running buffer (PBS) at 25 °C. Association and dissociation were each measured for 120 s at a flow rate of 20 μL min−1. Kinetic parameters of the interaction between PD 113,271 and MAP1B-HBR were obtained by global fitting of the sensorgrams to a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 Langmuir binding model using the BIAevaluation 4.1 software (GE Healthcare).
1 Langmuir binding model using the BIAevaluation 4.1 software (GE Healthcare).
Cell culture and pull-down assay
Rat PC12 cells were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% (v/v) fetal bovine serum and 5% horse serum in a humidified atmosphere containing 5% CO2. A bio-PD or biotin was immobilized on streptavidin agarose beads (Sigma) for 1 h at 4 °C. The beads were washed with PBS containing 1% Tween-20 (PBS-T) 3 times to remove unbound PD 113,271 or biotin. Simultaneously, PC12 cells were harvested and lysed using the lysis buffer (50 mM Tris–HCl (pH 7.5), 150 mM NaCl, 1 mM EDTA, 2.5 mM EGTA, 0.5% NP-40, 0.1 mM PMSF) at 4 °C for 1 h, and the resulting lysate was centrifuged to remove precipitants. The supernatant of the lysate was added to the PD 113,271- or biotin-immobilized beads, and the beads were incubated at 4 °C for 2 h with gentle rotation. After centrifugation, the supernatant was removed, and the beads were washed with PBS-T. These beads were boiled in SDS-sample buffer (75 mM Tris–HCl (pH 6.8), 2.5% SDS, 10% glycerol) at 100 °C for 5 min, and the bound proteins were separated on 5% (v/v) SDS polyacrylamide gel electrophoresis. After electrophoresis, MAP1B was detected with western blotting analysis using a mouse IgG monoclonal antibody for anti-MAP1B (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500; Chemicon International) as well as sheep anti-mouse Ig HRP-linked whole Ab (1
500; Chemicon International) as well as sheep anti-mouse Ig HRP-linked whole Ab (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3000; GE Healthcare) as the secondary antibody. Immunoreacted signals were visualized using Chemi-Lumi One (Nacalai Tesque).
3000; GE Healthcare) as the secondary antibody. Immunoreacted signals were visualized using Chemi-Lumi One (Nacalai Tesque).
SPR analysis of the binding between PD 113,271, tubulin, and MAP1B-HBR
The surface of a CM5 sensor chip (GE Healthcare) was activated using an activation solution (0.2 M N-ethyl-N′-(3-dimethylaminopropyl) carbodiimide, 50 mM N-hydroxysuccinimide) for 7 min. Tubulin (0.2 mg mL−1), stabilized with 20 nM paclitaxel and 2 mM GTP, was diluted in the ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 with 10 mM sodium acetate buffer at pH 4.0 and then injected. The surface of the sensor chip was then blocked by injecting 1 M ethanolamine at pH 8.5 for 7 min. All immobilizing methods were performed at a flow rate of 10 μL min−1. MAP1B-HBR (1 μM in PBS) was prepared, and various concentrations of PD 113,271 (20, 10, 5, 1, 0 μM) were added to each vial. The preparations were then introduced at a flow rate of 20 μL min−1. The response unit of each curve was analyzed using the BIAevaluation 4.1 software.
10 with 10 mM sodium acetate buffer at pH 4.0 and then injected. The surface of the sensor chip was then blocked by injecting 1 M ethanolamine at pH 8.5 for 7 min. All immobilizing methods were performed at a flow rate of 10 μL min−1. MAP1B-HBR (1 μM in PBS) was prepared, and various concentrations of PD 113,271 (20, 10, 5, 1, 0 μM) were added to each vial. The preparations were then introduced at a flow rate of 20 μL min−1. The response unit of each curve was analyzed using the BIAevaluation 4.1 software.
Cell culture and immunofluorescence procedure
Human breast adenocarcinoma MCF7 cells were grown in DMEM supplemented with 10% (v/v) fetal bovine serum in a humidified atmosphere containing 5% CO2 in the presence of antibiotics (1% penicillin–streptomycin mixed solution, Nacalai Tesque). Cells (5 × 104) were plated on a glass bottom dish (35 mm × 10 mm; Corning), and PD 113,271 (1 μM) was added. After incubation for 24 h, the cells were washed with PBS and fixed with 4% paraformaldehyde for 20 min at room temperature. Washing with PBS was repeated, and the cell membranes were penetrated with 0.1% Triton-X in PBS for 20 min at room temperature; 1% BSA in PBS was used for blocking. The mouse anti-MAP1B IgG (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100) and anti-β tubulin (Sigma; 1
100) and anti-β tubulin (Sigma; 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500) antibodies were used. Antibodies against MAP1B and tubulin were labeled with Alexa568 and Alexa488, respectively, using Zenon Mouse IgG1 Labeling Kits (Invitrogen). After washing with PBS-T, the nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI). Cells were examined using a confocal Zeiss LSM 5 Pascal Exciter in multitrack configuration. Confocal image stacks were processed using the LSM Image Browser software (Carl Zeiss).
500) antibodies were used. Antibodies against MAP1B and tubulin were labeled with Alexa568 and Alexa488, respectively, using Zenon Mouse IgG1 Labeling Kits (Invitrogen). After washing with PBS-T, the nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI). Cells were examined using a confocal Zeiss LSM 5 Pascal Exciter in multitrack configuration. Confocal image stacks were processed using the LSM Image Browser software (Carl Zeiss).
References
- T. B. Tunac, B. D. Graham and W. E. Dobson, J. Antibiot., 1983, 36, 1595–1600 CAS.
- S. S. Stampwala, R. H. Bunge, T. R. Hurley, N. E. Willmer, A. J. Brankiewicz, C. E. Steinman, E. A. Smitka and J. C. French, J. Antibiot., 1983, 36, 1601–1605 CAS.
- G. C. Hokanson and J. C. French, J. Org. Chem., 1985, 50, 462 CrossRef CAS.
- S. W. Mamber, W. G. Okasinski, C. D. Pinter and J. B. Tunac, J. Antibiot., 1986, 39, 1467–1472 CAS.
- S. S. Stampwala, J. C. French, J. B. Tunac, T. R. Hurley and R. H. Bunge, U.S. pat., 447, 544, 1986.
- S. S. Stampwala, J. C. French, J. B. Tunac, T. R. Hurley and R. H. Bunge, EP Appl. 87021 A2, 1983.
- D. S. Lewy, C.-M. Gauss, D. R. Soenen and D. L. Boger, Curr. Med. Chem., 2002, 9, 2005–2032 CAS.
- R. C. Jackson, D. W. Fry, T. J. Boritzki, B. J. Roberts, K. E. Hook and W. R. Leopold, Adv. Enzyme Regul., 1985, 23, 193–215 CrossRef CAS.
- W. Scheithauer, D. D. von Hoff, G. M. Clark, J. L. Shillis and E. F. Elslager, Eur. J. Cancer Clin. Oncol., 1986, 22, 921–926 CrossRef CAS.
- T. J. Boritzki, T. S. Wolfard, J. A. Besserer, R. C. Jackson and D. W. Fry, Biochem. Pharmacol., 1988, 37, 4063–6068 CrossRef CAS.
- R. S. de Jong, E. G. E. de Vries and N. H. Mulder, Anti-Cancer Drugs, 1997, 8, 413–418 CAS.
- M. Amemiya, T. Someno, R. Sawa, H. Naganawa, M. Ishizuka and T. Takeuchi, J. Antibiot., 1994, 47, 541–544 CAS.
- M. Amemiya, M. Ueno, M. Osono, T. Masuda, N. Kinoshita, C. Nishida, M. Hamada, M. Ishizuka and T. Takeuchi, J. Antibiot., 1994, 47, 536–540 CAS.
- S. Fushimi, S. Nishikawa, A. Shimazu and H. Seto, J. Antibiot., 1989, 42, 1019–1025 CAS.
- S. Fushimi, K. Furihata and H. Seto, J. Antibiot., 1989, 42, 1026–1036 CAS.
- M. Roberge, C. Tudan, S. M. F. Hung, K. W. Harder, F. R. Jirik and H. Anderson, Cancer Res., 1994, 54, 6115–6121 CAS.
- X. W. Guo, J. P. Th'ng, R. A. Swank, H. J. Anderson, C. Tudan, E. M. Bradbury and M. Roberge, EMBO J., 1995, 14, 976–985 CAS.
- M. Kawada, M. Amemiya, M. Ishizuka and T. Takeuchi, Biochim. Biophys. Acta, Mol. Cell Res., 1999, 1452, 209–217 CrossRef CAS.
- T. Usui, G. Marriott, M. Inagaki, G. Swarup and H. Osada, J. Biochem., 1999, 125, 960–965 CAS.
- D. W. Fry, J. A. Besserer and T. J. Boritzki, Cancer Res., 1984, 44, 3366–3370 CAS.
- J. H. Connor, T. Kleeman, S. Barik, R. E. Honkanen and S. J. Shenolikar, J. Biol. Chem., 1999, 274, 22366–22372 CrossRef CAS.
- L. Bialy and H. Waldmann, Angew. Chem., Int. Ed., 2002, 41, 1748–1751 CrossRef CAS.
- S. B. Buck, C. Hardouin, S. Ichikawa, D. R. Soenen, C. M. Gauss, I. Hwang, M. R. Swingle, K. M. Bonness, R. E. Honkanen and D. L. Boger, J. Am. Chem. Soc., 2003, 125, 15694–15695 CrossRef CAS.
- L. Bialy and H. Waldmann, Chem.–Eur. J., 2004, 10, 2759–2780 CrossRef CAS.
- T. Teruya, S. Simizu, N. Kanoh and H. Osada, FEBS Lett., 2005, 579, 2463–2468 CrossRef CAS.
- K. Maki, R. Motoki, K. Fujii, M. Kanai, T. Kobayashi, S. Tamura and M. Shibasaki, J. Am. Chem. Soc., 2005, 126, 17111–17117 CrossRef.
- B. G. Lawhorn, S. B. Boga, S. E. Wolkenberg, D. A. Colby, C.-M. Gauss, M. R. Swingle, L. Amable, R. E. Honkanen and D. L. Boger, J. Am. Chem. Soc., 2006, 128, 16720–16732 CrossRef.
- M. R. Swingle, L. Amable, B. G. Lawhorn, S. B. Buck, C. P. Burke, P. Ratti, K. L. Fischer, D. L. Boger and R. E. Honkanen, J. Pharmacol. Exp. Ther., 2009, 331, 45–53 CrossRef CAS.
- T. Takeuchi, N. Takahashi, K. Ishi, T. Kusayanagi, K. Kuramochi and F. Sugawara, Bioorg. Med. Chem., 2009, 17, 8113–8122 CrossRef CAS.
- S. Simizu, T. Teruya, T. Nogawa, H. Aono, M. Ueki, M. Uramoto, Y. Kobayashi and H. Osada, Biochem. Biophys. Res. Commun., 2009, 383, 406–410 CrossRef CAS.
- D. T. Ho and M. Roberge, Carcinogenesis, 1996, 17, 967–972 CrossRef CAS.
- A. H. Walsh, A. Cheng and R. E. Honkanen, FEBS Lett., 1997, 416, 230–234 CrossRef CAS.
- C. J. Hastie and P. T. Cohen, FEBS Lett., 1998, 431, 357–361 CrossRef CAS.
- A. Cheng, R. Balczon, Z. Zuo, J. S. Koons, A. H. Walsh and R. E. Honkanen, Cancer Res., 1998, 58, 3611–3619 CAS.
- R. E. Honkanen, Handb. Exp. Pharmacol., 2005, 167, 295–317 Search PubMed.
- G. P. Smith, Science, 1985, 228, 1315–1317 CrossRef CAS.
- G. P. Smith and V. A. Petrenko, Chem. Rev., 1997, 97, 391–410 CrossRef CAS.
- P. P. Sche, K. M. McKenzie, J. D. White and D. J. Austin, Chem. Biol., 1999, 6, 707–716 CrossRef CAS.
- M. Noble, S. A. Lewis and N. J. Cowan, J. Cell Biol., 1989, 109, 3367–3376 CrossRef CAS.
- E. Yamauchi, K. Titani and H. Taniguchi, J. Biol. Chem., 1997, 272, 22948–22953 CrossRef CAS.
- L. Ulloa, V. Dombrádi, J. Díaz-Nido, K. Szücs, P. Gergely, P. Friedrich and J. Avila, FEBS Lett., 1993, 330, 85–89 CrossRef CAS.
- C.-X. Gong, J. Wegiel, T. Lidsky, L. Zuck, J. Avila, H. M. Wisniewski, I. Grundke-Iqbal and K. Iqbal, Brain Res., 2000, 853, 299–309 CrossRef CAS.
Footnote |
| † These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2011 |
