DOI:
10.1039/C1MD00072A
(Concise Article)
Med. Chem. Commun., 2011,
2, 780-788
Received
11th March 2011
, Accepted 23rd May 2011
First published on 28th June 2011
Abstract
A series of COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundcarbazole–COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundpyrrolobenzodiazepine conjugates (4a–g and 5a–f) have been designed, and synthesized as anticancer agents. These compounds are prepared by linking the C8-position of DC-81 with a COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundcarbazole moiety through simple alkane spacers as well as COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundpiperazine side-armed alkane spacers in good yields. The DNA binding ability of these conjugates has been determined by thermal denaturation studies and also supported by molecular docking studies. These conjugates showed potent anticancer activity with GI50 ranging from 5.27–0.01 μM. The FACS analysis and BrdU assay of selected conjugates (4c, 4f, 5a and 5f) on MCF-7 cell lines disclosed the increased G1 cell cycle arrest and one of the conjugates 5f has exhibited significant anticancer activity. The analysis of the intrinsic factors involved in causing the G1 arrest in MCF-7 cell lines by 5f conjugate has been demonstrated on the proteins which play a vital role in G1 arrest followed by apoptosis (Cyclin D1, CDK4, c-Jun, JunB, CREB, p53, JNK1/2, procaspase-7, cleaved PARP, pRb, and COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundBAX). Thus, these PBD conjugates (in particular 5f) have promising potency for combating human carcinoma.
Introduction
Sequence selective DNA intercalators have enormous potential for the therapy of genetic diseases including some cancers and also as research tools for functional genomic studies.1–3 The antitumor antibiotic pyrrolo[2,1-c][1,4]-benzodiazepines (PBDs) is found in naturally occurring Streptomyces species, the well known members of this family are COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundanthramycin, sibiromycin, tomaymycin and DC-81 (1, Fig. 1).4,5 These compounds are known to exhibit their cytotoxic potency by covalent binding to the C2–NH2 group of a COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundguanine base through the electrophilic C11-position of the PBD in the minor groove of DNA.6 The PBDs are known interact to sequence-selectively with DNA and preferentially target 5′-Pu-G-Pu sequences.7,8 Moreover, PBDs perfectly fit into the minor groove of DNA by its natural right-handed twist due to the S-configuration at their C11a position. These unique features highlight the importance of PBDs in anticancer drug designing. Structure–activity relationship (SAR) studies have suggested that the linkage at C8-position of PBD with another DNA intercalator would greatly influence the biological activity. The main rationale for making these PBD conjugates is to produce a compound with two pharmacophoric heads with different DNA sequence specificities, which is likely to enhance the DNA binding ability with increased sequence selectivity and promising cytotoxicity.9,10
On the other hand, carbazoles belong to the unusual class of DNA binding agents. These molecules contain a planar chromophore, which is the characteristic of DNA intercalators.11 The COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundcarbazole scaffold found in many synthetic anticancer agents and a novel DNA interactive 3,6-disubstituted COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundcarbazole with interesting biological action via binding to A–T sequences in the minor groove of DNA and subsequent inhibition of DNA-directed enzyme helicases have been recently reported (Fig. 1).12–14 Moreover, some of the COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundcarbazole conjugates also act as antimitotic agents,15 cyclooxygenase-2 inhibitors16 and sigma (σ) receptor ligands.17
These findings encouraged us to synthesize a new class of COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundcarbazole linked PBD conjugate. Several research groups around the world have explored the chemical and biological aspects of such compounds.18–31 As a part of our ongoing exploration aimed towards the discovery of new PBD conjugates, we have investigated several potent PBD conjugates.32–46 In continuation of these efforts, in this article we have synthesized and evaluated a series of PBD–COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundcarbazole conjugates to unravel their anticancer potential. The significant DNA binding ability and the proficient cytotoxicity of these conjugates prompted us to further evaluate detailed biological effects. The FACS analysis data and BrdU assay on one of the conjugates 5f have shown profound activity towards cell cycle arrest at G1 phase among the tested conjugates. The conjugate 5f which exhibits promising anticancer activity among tested conjugates was chosen for in vivo efficacy studies and the conjugate showed good efficacy.
Results and discussion
Chemistry
The COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundcarbazole bromoalkyl spacers were prepared by n-alkylation of COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundcarbazole (6a) or COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound3,6-bis(4-methoxyphenyl)-9H-carbazole (6b)14 with dibromoalkanes by refluxing at 85 °C in COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundacetonitrile for 24 h using K2CO3 to afford the desired bromoalkoxy carbazoles in good yield (90–95%) (7a–d, Scheme 1).
The commercially available COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundcarbazole (6a) or prepared COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound3,6-bis(4-methoxyphenyl)-9H-carbazole (6b) was joined to the (2S)-N-{4-[n-bromoalkoxy]-5-methoxy-2-nitrobenzoyl}pyrrolidine-2-carboxaldehyde diethylthioacetal (8a–d),30 by using K2CO3 in COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundacetone at 80 °C to provide the corresponding nitrothioacetal intermediates (9a–g) in good yield (70–78%). Further, reduction of the nitro compounds by using SnCl2·2H2O in COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundmethanol followed by deprotection using HgCl2/COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundCaCO3 in COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundMeCN–COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundwater afforded the desired COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundcarbazole–PBD conjugates 4a–g as shown in Scheme 2.
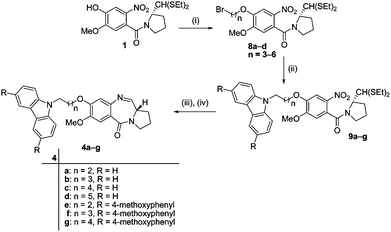 |
| | Scheme 2 Synthetic route of COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundcarbazole–PBD conjugates (4a–g). Reagents and conditions: (i) dibromoalkanes, K2CO3, COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundacetone, reflux, 12 h (70–78%); (ii) compounds 6a–b, K2CO3, COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundMeCN, reflux, 12 h (70–78%); (iii) SnCl2·2H2O, COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundMeOH, reflux, 6 h (85–90%); (iv) HgCl2, COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundCaCO3, COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundMeCN![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) COMPOUND LINKS COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundMeOH (4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1), 12 h (50–60%). 1), 12 h (50–60%). | |
The synthesis of C8-linked COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundcarbazole–PBD conjugates linked through COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundpiperazine (COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound5a–d) was carried out from the n-alkylation of (2S)-N-{4-[n-bromoalkoxy]-5-methoxy-2-nitrobenzoyl}pyrrolidine-2-carboxaldehyde diethylthioacetal (8a,b) with COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundN-Boc-piperazine in COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundacetone at 80 °C for 12 h to afford the coupled intermediate (10a,b) in good yield (90%). These intermediates were deprotected by using COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundTFA over a period overnight giving methoxy-(2S)-[N-{3-[4-piperazino]alkyloxy}-5-methoxy-2-nitrobenzoyl]pyrrolidine-2-carboxaldehyde diethylthioacetal (11a,b) (yield 90–95%). Compounds 11a,b on alkylation with COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundcarbazole precursors (7a–d) using K2CO3 in COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundacetone at 80 °C for 24 h provided the corresponding nitro thioacetals (12a–f) in good yield (70–75%). Further, reduction of nitro compounds by using SnCl2·2H2O in COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundmethanol at 85 °C for 5 h reflux followed by deprotection using HgCl2/COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundCaCO3 in COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundMeCN–COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundwater at room temperature for 12 h afforded the desired COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundcarbazole–PBD conjugates 5a–f (yield 50–55%) as shown in Scheme 3.
 |
| | Scheme 3 Synthetic route of COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundcarbazole–piperzinyl–PBD conjugates (5a–f). Reagents and conditions: (i) COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundN-Boc-piperazine, K2CO3, COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundacetone, reflux, 24 h (90–95%); (ii) COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundTFA, COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundDCM, 12 h (90–95%); (iii) Compounds 7a–d, K2CO3, COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundacetone, reflux, 24 h (80–85%); (iv) SnCl2·2H2O, COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundMeOH, reflux, 6 h (85–90%); (v) HgCl2, COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundCaCO3, COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundMeCN![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) COMPOUND LINKS COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundMeOH (4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1), 12 h (50–60%). 1), 12 h (50–60%). | |
Biological activity
Molecular modelling studies
The melting temperature studies indicated that these PBD conjugates possess enhanced DNA binding affinity, hence it was considered of interest to investigate them for their ability to interact with oligonucleotide duplexes (5′-CGCAGAATTCTGCG-3′) that contain potential target binding sites.47 The PBD subunit has a chiral center at the C11 position and a DNA-reactive imine moiety at the N10 and C11 positions. PBD monomers typically span ∼3 bp of DNA with an alkylating preference for the 5′-AGA sequence. These molecules are the covalent minor groove binders and can covalently bind to a COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundguanine base at its exocyclic 2-amino group in double-helical B-DNA (AGA). These molecules are accommodated within the minor groove covering the centrally located 5–6 bp. The molecular docking and minimization protocols were used to study the DNA–ligand interactions. Initial coordinates for the molecular minimization of the PBD hybrid were taken from the COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundGOLD docked ligand–DNA complexes. The covalent bond between the C11 positions of the ligand with exocyclic 2-amino group of a COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundguanine base at its double-helical B-DNA (AGA) was built manually using Sybyl 6.9.48 The free binding energies were then calculated by subtracting the minimization energies of DNA and ligand from the total energy of the covalently bonded complex (DNA + ligand). These energies of the ligands, DNA and covalent complexes of DNA were minimized using the Tripos force field with MMFF94 partial charges and Powell's conjugate gradient energy minimization method was used until a convergence criterion of 0.001 kcal mol−1 was reached. The COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundGOLD score and the free binding energy of the complexes are given in Table 2. The COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundGOLD scores and free binding energy of the systems have clearly shown that the 5f complex has highest COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundGOLD score with lowest free binding energy and is more stable than other ligands.
| S. No. |
Molecule |
COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundGOLD score |
Free binding energy/kcal mol−1 |
| 1 |
4a
|
62.43 |
−162.225 |
| 2 |
4b
|
62.43 |
−170.474 |
| 3 |
4c
|
72.07 |
−165.197 |
| 4 |
4d
|
72.46 |
−208.943 |
| 5 |
4e
|
69.45 |
−144.717 |
| 6 |
4f
|
66.96 |
−175.353 |
| 7 |
4g
|
73.95 |
−194.774 |
| 8 |
5a
|
73.64 |
−198.390 |
| 9 |
5b
|
70.84 |
−215.986 |
| 10 |
5c
|
68.80 |
−209.702 |
| 11 |
5d
|
76.02 |
−202.395 |
| 12 |
5e
|
66.15 |
−205.602 |
| 13 |
5f
|
76.04 |
−228.136 |
The DNA–ligand (5f) adduct depicted in Fig. 2 clearly reveals that the PBD conjugate tightly spans within the centrally located 6–8 base pairs of the minor groove region. The H-bonding between the COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundcytosine and modified COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundguanine was disrupted and is displaced along the helix axis. It is observed that there is no significant change in the modified COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundguanine, however A4 tries to approach G5 through non-bonded stacking interaction, which results in an opening toward the major groove and a simultaneous local expansion of the minor groove around the 7-membered ring of the ligand. As a consequence the significant helical distortions by the covalent binding of the PBD conjugate are mostly confined to the covalent binding site and at the junction of the linker binding region of DNA.
Anticancer activity.
The COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundcarbazole–PBD conjugates 4c and 5a have been evaluated for their anticancer activity against 60 cancer cell lines derived from nine different types of human cancer (lung, leukemia, colon, CNS, melanoma, ovarian, renal, prostate and breast cancer) at the National Cancer Institute (NCI), Bethesda, and the results are expressed as percentage of growth inhibition (GI50) determined relative to that of untreated control cells. These conjugates 4c and 5a exhibited a wide spectrum of activity against different cancer cell lines with the mean GI50 values of 1.97 and 0.10 μM respectively. Interestingly compound 5a that has a COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundpiperazine moiety in its alkane spacer has shown ten fold higher activity than 4c and the results are illustrated in Table 3.
Table 3
In vitro cytotoxicity of compounds 4c (NSC: 744990-G)b and 5a (NSC: 744992-I)b in human cancer cell linesa
| Cancer panel per cell line |
GI50/μM |
Cancer panel per cell line |
GI50/μM |
|
4c
|
5a
|
4c
|
5a
|
|
Data obtained from NCI's in vitro anticancer activity cells screen.
NCI numbering.
Mean values over 60 cell lines tested.
|
|
Leukemia
|
Ovarian
|
| CCRF-CEM |
1.97 |
0.02
|
IGROV1 |
2.08 |
<0.01
|
| HL-60(TB) |
1.72 |
0.03
|
OVCAR-3 |
1.88 |
0.069
|
| K-562 |
2.60 |
— |
OVCAR-4 |
3.09 |
0.10 |
| MOLT-4 |
2.15 |
0.02
|
OVCAR-5 |
2.42 |
0.03
|
| RPMI-8226 |
1.72 |
— |
OVCAR-8 |
2.55 |
— |
| SR |
1.54 |
0.03
|
SK-OV-3 |
4.57 |
0.29 |
| Non-small cell lung |
Renal
|
| A549/ATCC |
1.57 |
— |
786-0 |
1.73 |
0.19 |
| EKVX |
5.27 |
0.14 |
A498 |
1.74 |
0.17 |
| HOP-62 |
1.80 |
0.08
|
ACHN |
1.77 |
0.25 |
| HOP-92 |
1.31 |
0.27 |
CAKI-1 |
1.93 |
0.12 |
| NCI-H226 |
2.49 |
0.13 |
RXF 393 |
4.34 |
0.22 |
| NCI-H23 |
2.69 |
0.26 |
SN12C |
1.94 |
0.27 |
| NCI-H322M |
3.83 |
0.25 |
TK-10 |
2.25 |
0.12 |
| NCI-H460 |
1.96 |
0.25 |
UO-31 |
1.52 |
0.12 |
| NCI-H522 |
1.65 |
<0.01
|
|
|
|
|
Colon
|
Breast
|
| COLO 205 |
2.11 |
0.07
|
MCF7 |
2.33 |
0.04
|
| HCC-2998 |
1.71 |
0.13 |
MDA-MB- |
2.14 |
0.95 |
| HCT-116 |
1.65 |
0.06
|
HS 578T |
1.97 |
0.16 |
| HCT-15 |
1.67 |
0.28 |
MDA-MB-435 |
1.73 |
<0.01
|
| HT29 |
2.08 |
0.05
|
BT-549 |
2.03 |
0.13 |
| KM12 |
1.96 |
0.15 |
T-47D |
2.79 |
0.17 |
| SW-620 |
1.85 |
0.04
|
MDA-MB-468 |
2.26 |
0.11 |
|
CNS
|
Prostate
|
| SF-268 |
1.91 |
0.05
|
PC-3 |
2.36 |
0.03
|
| SF-539 |
1.85 |
0.12 |
DU-145 |
2.23 |
0.04
|
| SNB-19 |
2.05 |
0.21 |
|
|
|
| SNB-75 |
1.05 |
0.15 |
|
|
|
| U251 |
1.59 |
0.03
|
|
|
|
|
Melanoma
|
Melanoma
|
| LOX IMVI |
2.08 |
0.03
|
SK-MEL-28 |
1.80 |
0.06
|
| MALME-3M |
2.20 |
0.15 |
SK-MEL-5 |
1.73 |
0.23 |
| M14 |
2.10 |
0.15 |
UACC-257 |
2.29 |
0.15 |
| SK-MEL-2 |
1.67 |
0.19 |
UACC-62 |
1.86 |
0.15 |
|
Mean
|
1.97
|
0.1
|
|
|
|
The promising activity shown by these conjugates prompted us to evaluate the anticancer activity of the other conjugates (4a–g and 5a–f) in selected human cancer cell lines. The compounds that exhibit GI50 ≤10−5 μM have been considered as active on the respective cell lines. Table 4 illustrates that compounds 4a–g and 5a–f exhibited promising anticancer activity with GI50 values ranging from <0.1 to 2.21 μM (DC-81 and COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundadriamycin were used as positive controls). The conjugates 4a–g that contain simple alkane chain spacers exhibit substantial anticancer activity. However, the conjugates (4a, 4c, 4e and 4g) with odd number of alkane chain spacers (3,5) show enhanced activity compared to their counterparts that have an even number of alkane chain spacers (4b, 4d and 4f). Further, substitution in the COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundcarbazole ring also enhances the activity compared to the un-substituted moiety in these COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundcarbazole conjugates. It appears that the incorporation of a COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundpiperazine moiety in the linker further enhances the activity of these conjugates. Some of the potent molecules (4c, 4f, 5a and 5f) were taken up for the evaluation of the cell viability studies against MCF-7 cancer cell line in view of its resistance towards chemotherapy and radiation. MTT cytotoxicity assay was conducted both in a concentration dependent (2–16 μM) as well as time dependent manner (6–48 h) on conjugates 1, 6b, 1 + 6b, 4c, 4f, 5a and 5f. It has been observed that there is an increase in the cytotoxicity with regard to the combination of parent compounds (1 + 6b) in MCF-7 cell line at all concentrations studied than individual parent compounds (1 and 6b). However the effectiveness of the conjugates towards causing cytotoxicity is superior to the parent compounds, in particular conjugate 5f. There was a gradual increase in cytotoxicity during the 6–48 h study, the optimum standardized time to obtain effective cytotoxicity was 24 h at 4 μM concentration. From this study we have standardized the optimum time duration and concentration as 24 h and 4 μM respectively. Thus further downstream experiments have been carried out with this concentration and time period. The cell viability results of these compounds are shown in the ESI†, it is observed that the conjugates 5a and 5f have shown pronounced cytotoxicity. Similar observations were made from COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundATP dependent cell viability assays (data not shown).
Table 4 GI50 values (in μM) for compounds 4a–g and 5a–f in selected human cancer cell linesa
| Comp. |
HOP-62b |
A-549b |
KBc |
Guravc |
A-2780d |
PC-3e |
SiHaf |
Colo-205g |
Zr-75-1h |
MCF7h |
|
50% growth inhibition and the values are mean of three determinations.
Lung cancer.
Oral cancer.
Ovarian cancer.
Prostate cancer.
Cervix cancer.
Colon cancer.
Breast cancer.
Not tested.
|
|
4a
|
1.93 |
1.84 |
1.87 |
1.60 |
1.21 |
—i |
1.70 |
1.68 |
1.89 |
1.85 |
|
4b
|
1.93 |
1.92 |
1.87 |
1.60 |
1.57 |
— |
1.70 |
1.70 |
1.81 |
2.21 |
|
4c
|
1.80 |
1.42 |
1.80 |
1.61 |
1.32 |
2.30 |
1.68 |
1.64 |
1.76 |
1.64 |
|
4d
|
1.58 |
1.50 |
1.95 |
1.71 |
1.70 |
— |
1.58 |
1.62 |
2.14 |
1.65 |
|
4e
|
0.11 |
0.18 |
0.18 |
— |
0.15 |
0.16 |
0.17 |
0.17 |
— |
0.13 |
|
4f
|
0.14 |
0.19 |
0.19 |
— |
0.16 |
0.16 |
0.17 |
0.19 |
— |
0.15 |
|
4g
|
0.12 |
0.16 |
0.17 |
— |
0.14 |
0.15 |
0.17 |
0.18 |
— |
0.14 |
|
5a
|
0.10 |
0.14 |
0.13 |
0.12 |
0.12 |
0.14 |
0.14 |
0.16 |
0.13 |
0.08 |
|
5b
|
0.10 |
0.12 |
0.14 |
0.13 |
0.11 |
0.11 |
0.13 |
0.14 |
0.15 |
0.15 |
|
5c
|
0.11 |
0.14 |
0.16 |
0.15 |
0.14 |
0.14 |
0.16 |
0.17 |
0.16 |
0.15 |
|
5d
|
0.08 |
0.13 |
0.15 |
0.17 |
0.16 |
0.10 |
0.15 |
0.16 |
0.15 |
0.12 |
|
5e
|
0.10 |
0.13 |
0.11 |
0.12 |
0.01 |
0.12 |
0.12 |
0.08 |
0.13 |
0.04 |
|
5f
|
0.01 |
0.09 |
0.10 |
0.11 |
0.01 |
0.01 |
0.11 |
0.07 |
0.12 |
0.01 |
| ADR |
0.14 |
7.25 |
0.17 |
0.16 |
0.16 |
1.83 |
0.17 |
0.14 |
1.79 |
0.17 |
| DC-81 |
0.15 |
0.16 |
0.17 |
0.17 |
0.14 |
0.20 |
0.17 |
0.11 |
2.34 |
0.17 |
To investigate the mechanism underlying the anti-proliferative action, these COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundcarbazole–PBD conjugates (4c, 4f, 5a and 5f) are further analyzed for the cell-cycle distribution by treating MCF-7 cells at 4 μM concentration for 24 h by flow cytometry using DC-81 (1) and compound 6b as positive controls. The control cells treated with COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundDMSO (the solvent in which the compound is dissolved) showed 60.75%, 14.30% and 24.95% of G0/G1, S and G2/M phase respectively. Compounds 4c, 4f, 5a and 5f showed 71.69%, 67%, 86.60% and 88.34% of G0/G1 phase, whereas the positive controls DC-81 (1) and 6b showed 67.74% and 66.63% of G0/G1 phase respectively (Table 5). The increase in G0/G1 phase and decrease in G2/M phase indicate that G1 cell cycle arrest is caused by these compounds. Moreover the G0 cells, which indicate the apoptotic cell death are observed more in the case of 5f (24%) than 5a (16%) as seen in Fig. 3b. In view of the high cytotoxicity of 5f we were interested to investigate this conjugate at the molecular level.
Table 5 Cell cycle distribution of MCF 7 cell line using 1, 6b, 4c, 4f, 5a and 5f at 4 μM concentrations
| Compound |
Cell cycle distribution (%) |
| G0 |
G1 |
S |
G2/M |
G0/G1 |
| Control |
2.77 |
57.98 |
14.30 |
24.95 |
60.75 |
|
1
|
3.95 |
63.79 |
12.20 |
20.07 |
67.74 |
|
6b
|
4.39 |
62.24 |
11.84 |
21.53 |
66.63 |
|
4c
|
3.45 |
68.24 |
13.03 |
15.38 |
71.69 |
|
4f
|
4.40 |
62.6 |
11.12 |
21.88 |
67.00 |
|
5a
|
17.13 |
69.47 |
9.53 |
3.87 |
86.60 |
|
5f
|
25.31 |
63.03 |
5.44 |
6.20 |
88.34 |
 |
| | Fig. 3 Apoptotic percentage of MCF-7 cells treated with conjugates (1, 6b, 4c, 4f, 5a and 5f) at 4 μM concentration. | |
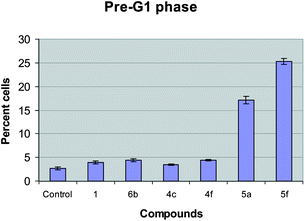
An increase in the percentage of cells in the pre-G1 phase was observed for the tested compounds (4c, 4f, 5a and 5f). Among the tested conjugates compounds 5a and 5f were found to be the most effective ones and the percentage of pre-G1 phase cells was found to be 2.77, 3.95, 4.39, 3.45, 4.4, 17.13 and 25.31 for control, DC-81 (1), 6b, 4c, 4f, 5a and 5f respectively (Fig. 3).
TUNEL assay
Conjugate 5f is highly cytotoxic and causes good G1 cell cycle arrest in MCF-7 cells. Thus it was considered of interest to investigate the apoptosis mediated by these conjugates employing a TUNEL assay wherein 1 and 6b are used as positive controls. Results of the tunnel assay reveal that the Apoalert DNA fragmentation is more prominent in the case of 5f than the controls as seen in Fig. 4.
 |
| | Fig. 4 TUNEL staining of MCF-7 cells after exposure to conjugates (1, 6b, and 5f) at 4 μM concentration after 24 h time period. Green colour stained cells represent TUNEL positive cells. Here control cells act as control for staining procedure. Lack of staining in control (untreated) cells represents that the cells are actively proliferating, without apoptotic cell death. | |
Read more about this on ChemSpider
Download mol file of compounduridine) cell proliferation assay was carried out. BrdU cell proliferation assay is based on the principle that BrdU is incorporated into newly synthesized DNA strands of actively proliferating cells. It is observed that MCF-7 cells treated with 4 μM of 5a and 5f for 24 h incorporated less BrdU than the untreated control cells. The levels of proliferation decreased to 35.19% and 28% in the case of 5a and 5f than the control cells 100% (Fig. 5). These results clearly indicate the decrease in the percentage of cells reaching a S-phase and indirectly supporting the G1 cell cycle arrest.
 |
| | Fig. 5 The BrdU cell proliferation assay: the MCF-7 cells were seeded in 96 well plate at a density of 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells and were grown for 24 h, BrdU was incubated for 5 h at RT followed by treatment with conjugates (1, 6b, 5a and 5f) for 24 h at a final concentration of 4 μM. The intensity of blue colour at 450 nm was measured and is proportional to the amount of BrdU incorporated in proliferating cells. Higher O.D. reading denotes the higher proliferation rate. 000 cells and were grown for 24 h, BrdU was incubated for 5 h at RT followed by treatment with conjugates (1, 6b, 5a and 5f) for 24 h at a final concentration of 4 μM. The intensity of blue colour at 450 nm was measured and is proportional to the amount of BrdU incorporated in proliferating cells. Higher O.D. reading denotes the higher proliferation rate. | |
Effect on Cyclin D1 and associated proteins
We have evaluated protein expression studies on MCF-7 cancer cell lines by treating with the starting materials as positive controls (1, 6b and 1 + 6b) and the effective compound obtained from the MTT studies (5f) for 24 h at 2 and 4 μM concentrations. The levels of Cyclin D1, whose activation is essential for rapid cell proliferation from G1 to S phase transition,49 were down regulated at both 2 and 4 μM, with 4 μM being more effective. Furthermore, we have also conducted the Western blot analysis on p53 (the key protein in tumor suppression)50 and Bax (the mitochondrial protein whose activation is alone sufficient to cause apoptotic event).51 It is observed that there is up-regulation of both p53 and Bax protein at 4 μM concentration rather than 2 μM for the apoptosis event by these conjugates. It is observed that there is an increase in apoptosis in the case of conjugate 5f which is more prominent than the effect by the starting materials (1 + 6b) as shown in Fig. 6a.
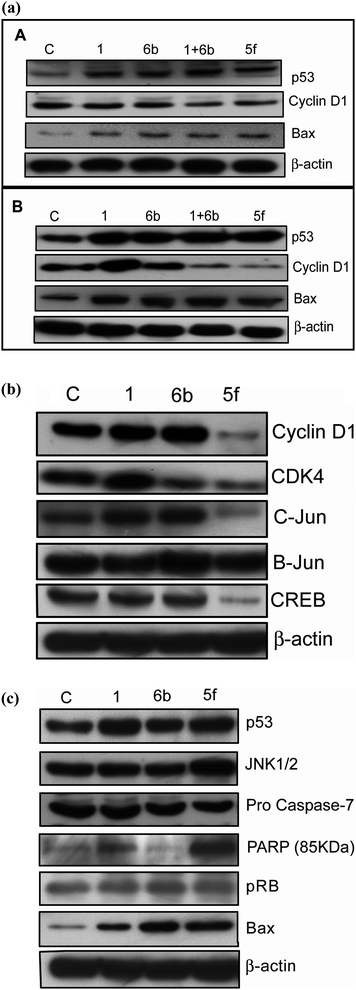 |
| | Fig. 6 (a) The effect of compounds on the expression of key proteins involved in cell cycle arrest and apoptosis. Treatments were conducted at 2 and 4 μM concentrations for 24 h time period in MCF-7 breast cancer cell line conjugates 1, 6b, 1 + 6b, and 5f. Here 1 and 6b are the starting materials used. 1 + 6b is the treatment wherein we have used both starting materials. Panel (A) shows the protein expression pattern of Cyclin D1, p53 and Bax proteins at 2 μM concentration. Panel (B) represents the protein expression pattern of Cyclin D1, p53 and Bax proteins at 4 μM concentration. (b) Effect of the compounds on the expression of Cyclin D1 and associated proteins (CDK-4, C-Jun, Jun B and CREB (c COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundAMP response element binding protein)). MCF-7 cells were treated with 4 μM concentration of conjugates (1, 6b, and 5f) for 24 h. The cell lysates were collected and observed for expression of Cyclin D1, CDK-4, C-Jun, JunB and CREB protein levels using specific antibodies. β-Actin was used as a loading control. C: control (untreated). (c) Effect of compounds on the expression level of tumor suppressor and apoptotic specific proteins. MCF-7 cells were treated with 4 μM concentration of conjugates (1, 6b, and 5f) for 24 h. The cell lysates have been collected and observed for the levels of proteins (p53, JNK1/2, procaspase-7, cleaved PARP, pRb and COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundBAX) using specific antibodies. β-Actin was used as the loading control. C: control (untreated). | |
In order to further understand the G1 cell arrest at the molecular level, we initiated our studies with G1 Cyclin called Cyclin D1. Overexpression of Cyclin D1 leads to a malignant form of human breast cancer. Moreover, about 50% of invasive breast-carcinoma have shown increase in the expression of Cyclin D1 and is accumulated during the late G1 phase.52,53 Based on these previous findings and the results of the present investigation, the FACS data (G1 arrest) suggest a possible involvement of Cyclin D1. To understand this, MCF-7 cells are treated with 4 μM concentration of DC-81 (1), 6b, and 5f for 24 h and then the lysates are subjected to Western blot analysis. It is observed that there is a drastic down regulation of Cyclin D1 protein in the case of 5f.
It is established that MCF-7 cell line is an estrogen responsive (ER) positive cell line, and during the process Cyclin D1 and the proteins get up regulated. Therefore the attention has been focused towards Cyclin D1 and its associated proteins such as c-Jun, Jun B and CREB that are known to play a vital role in activating its promoter.54–56 To investigate this aspect the compounds are treated for 24 h at 4 μM concentration and Western blot analysis is carried out. It is interesting to observe that c-Jun, Jun B and CREB protein levels are down regulated in the case of the conjugate 5f. These analyses unveil the possible involvement of Cyclin D1 protein in this event. Furthermore, Cyclin D-CDK4/CDK6 are important protein members that have a crucial role in the progression of cells through the G1 phase of the cell cycle.57 Therefore the CDK4 level was further examined for DC-81 (1), 6b and 5f at 4 μM concentration. It is observed that these levels are down regulated, by enforcing the G1 cell cycle arrest in the case of 5f treated MCF-7 cells (Fig. 6b).
Effect on tumor suppressors and apoptotic proteins
Based on the apoptotic inducing property of 5f it was considered of interest to investigate this compound at the molecular level in order to understand the type of proteins that are involved in this apoptotic event.
The p53 tumor suppressor is a potent transcription factor that is activated in response to various DNA-damaging agents leading to cell cycle arrest and/or apoptosis,58 we examine some of the important factors, including COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundBAX,59,60 poly(ADP-ribose)polymerase (PARP),61 JNK1/2,62,63 pRB, and procaspases-764,65 that are associated with p53 and related proteins, as these could play a vital role in regulating apoptotic events mediated by this COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundcarbazole–PBD conjugate (5f). Therefore MCF-7 cells are treated with 4 μM concentration of DC-81, 6b and 5f, and incubated for 24 h to probe the effect against these proteins by conducting western blot analysis. Interestingly p53, JNK1/2, COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundBAX and PARP protein levels were highly up-regulated in the case of 5f in comparison to controls (DC-81, 6b). Cleavage of Procaspase-7 and pRb is more in the case of 5f treated compound, indirectly depicting the release of caspase-7 (Fig. 6c). These findings indicate that there is a possibility of involvement of cell cycle regulatory proteins in this event, however the intricacies involved further need to be evaluated.
In vivo tumor xenograft studies of compound 5f
The preliminary in vitro anticancer activities revealed that compound 5f has shown significant anticancer activity among the series. These encouraging results provided an impetus to carry out in vivo efficacy studies using a xenograft model of human prostate cancer cells (PC-3) in male NOD-SCID mice. The dose determination studies demonstrated the maximally tolerated dose (COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundMTD) of compound 5f as <50 mg kg−1. Hence, compound 5f was administered at 20 mg kg−1 intravenous (I.V.), on days 1, 5 and 9 (q4d) (total drug administered was 20 mg kg−1). It was interesting to observe that at this dosage there was no toxicity in terms of weight loss or mortality of the experimental mice (Fig. 7a and b).
 |
| | Fig. 7 (a) NOD-SCID male mice treated with compound 5f. Survival data of compound 5f on PC-3 xenograft mice. (b) Tumor weight curves in NOD-SCID male mice treated with compound 5f. | |
Conclusion
In the present investigation, we have designed and synthesized a new class of COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundcarbazole–PBD conjugates. The DNA binding affinity of these conjugates has been evaluated by thermal denaturation studies and further confirmed by molecular modeling studies. The cytotoxicity results indicate that these conjugates have potency to deplete the growth of various human cancer cells and these can be considered as broad spectrum anticancer agents. However, simple alkane linked COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundcarbazole–PBD conjugates (4a–g) have exhibited comparably lower cytotoxicity (GI50 value in the ranges of 5.27–0.12 μM) than the COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundpiperazine incorporated PBD–COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundcarbazole conjugates (5a–f, GI50 value in the ranges of 0.28–0.01 μM). The cell cycle distribution studies of selected conjugates (4c, 4f, 5a, and 5f (4 μM)) on MCF-7 cell lines showed G1 cell cycle arrest with apoptotic inducing ability. Among the conjugate, 5f has shown remarkable cytotoxicity as shown by MTT assay, FACS analysis and BrdU assays. To understand the molecular mechanism behind the G1 cell cycle arrest and apoptotic inducing nature of 5f, Western blot analysis has been carried out against Cyclin D1, CDK-4, c-Jun, B-JUN, CREB, p53, JNK1/2, procaspase, pRb, PARP and COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundBAX molecules in MCF-7 cancer cells, and results of these studies were very promising, hence from these observations we can conclude that the COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundcarbazole–PBD conjugates have shown improved antiproliferative activity against a wide range of cancers, and have potential for further development.
Acknowledgements
The authors R.V.C.R.N.C.S, P.S, and A.M.R thank CSIR and the Department of Biotechnology, New Delhi (BT/PR/7037/Med/14/933/2006), for financial assistance.
References
- D. E. Thurston, Br. J. Cancer, 1999, 80, 65–85 CAS
 .
.
-
New Targets for Cancer Chemotherapy, S. Neidle, D. E. Thurston, D. J. Kerr and P. Workmann, CRC, London, 1994, p. 159 Search PubMed
 .
.
- T. Berge, N. S. Jenkins, R. B. Hopkirk, M. J. Waring and J. M. Edwardson, Nucleic Acids Res., 2002, 30, 2980–2986 CrossRef CAS
 .
.
- L. H. Hurley, J. Antibiot., 1977, 30, 349–370 CAS
 .
.
-
Molecular Aspects of Anticancer Drug–DNA Interactions 1, D. E. Thurston, S. Neidle and M. J. Waring, The Macmillan press Ltd., London, 1993, vol. 1, pp. 54–88 Search PubMed
 .
.
- L. H. Hurley and R. L. Petrusek, Nature, 1979, 282, 529–531 CrossRef CAS
 .
.
- L. H. Hurley, T. Reck, D. E. Thurston, D. R. Langley, K. G. Holden, R. P. Hertzberg, J. R. E. Hoover, G. Gallagher, Jr, L. F. Faucette, S. M. Mong and R. K. Johnson, Chem. Res. Toxicol., 1988, 1, 258–268 CrossRef CAS
 .
.
- F. L. Boyd, D. Stewart, W. A. Remers, M. D. Barkley and L. H. Hurley, Biochemistry, 1990, 29, 2387–2403 CrossRef CAS
 .
.
- D. E. Thurston and D. S. Bose, Chem. Rev., 1994, 94, 433–465 CrossRef CAS
 .
.
- A. Kamal, S. Azeeza, E. V. Bharathi, M. S. Malik and R. V. C. R. N. C. Shetti, Mini-Rev. Med. Chem., 2010, 10, 405–435 CrossRef CAS
 .
.
- A. T. Farial, D. Ding, A. P. Donald, C. Bailly, R. T. Richard and W. W. David, Biochemistry, 2000, 39, 12091–12101 CrossRef
 .
.
- S. Akinaga, K. Nomura, K. Gomi and M. Okabe, J. Antibiot., 1993, 46, 1767–1771 CAS
 .
.
- D. Nathalie, J. Ulrich, B. Brigtte, T. Christelle, L. Amele, C. Pierre, T. Farial, W. W. David, R. Sylvain, B. Chrisine, B. Christian and M. Jean-yves, Biochemistry, 2004, 43, 15169–15178 CrossRef
 .
.
- U. Jacquemard, S. Routier, A. Tatibouët, J. Kluza, W. Laine, C. Bal, C. Bailly and J.-Y. Mérour, Org. Biomol. Chem., 2004, 2, 1476–1483 CAS
 .
.
- L. Hu, Z.-R. Li, Y. Li, J. Qu, Y.-H. Ling, J.-D. Jiang and D. W. Boykin, J. Med. Chem., 2006, 49, 6273–6282 CrossRef CAS
 .
.
- R. Narlawar., B. I. P. Revuelta, C. Haass, H. Steiner, B. Schmidt and K. Baumann, J. Med. Chem., 2009, 49, 7588–7591 CrossRef
 .
.
- F. Savina, A. Carmen, A. C. Nicola, R. Mauro, I. Carmela, B. Francesco and P. Roberto, J. Med. Chem., 2007, 50, 4648–4655 CrossRef
 .
.
- B. Gerratana, Med. Res. Rev., 2010 DOI:10.1002/med.20212
 .
.
- K. M. Rahman, V. Mussa, M. Narayanaswamy, C. H. James, P. W. Howard and D. E. Thurston, Chem. Commun., 2009, 227–229 RSC
 .
.
- G. B. Jones, C. L. Davey, T. C. Jenkins, A. Kamal, G. G. Kneale, S. Neidle, G. D. Webster and D. E. Thurston, Anti-Cancer
Drug Des., 1990, 5, 249–264 CAS
 .
.
- G. Wells, M. Suggitt, M. Coffils, M. A. H. Baig, P. W. Howard, P. M. Loadman, J. A. Hartley, T. C. Jenkins and D. E. Thurston, Bioorg. Med. Chem. Lett., 2008, 18, 2147–2151 CrossRef CAS
 .
.
- K. M. Rahman, A. S. Thompson, C. H. James, M. Narayanaswamy and D. E. Thurston, J. Am. Chem. Soc., 2009, 131, 13756–13766 CrossRef CAS
 .
.
- D. Antonow, M. Kaliszczak, G.-D. Kang, M. Coffils, A. C. Tiberghien, N. Cooper, T. Barata, S. Heidelberger, C. H. James, M. Zloh, T. C. Jenkins, A. P. Reszka, S. Neidle, S. M. Guichard, D. I. Jodrell, J. A. Hartley, P. W. Howard and D. E. Thurston, J. Med. Chem., 2010, 53, 2927–2941 CrossRef CAS
 .
.
- M. Tercel, S. M. Stribbling, H. Sheppard, B. G. Siim, K. Wu, S. M. Pullen, K. J. Botting, W. R. Wilson and W. A. Denny, J. Med. Chem., 2003, 46, 2132–2151 CrossRef CAS
 .
.
- J.-J. Wang, Y.-K. Shen, W.-P. Hu, M.-C. Hsieh, F.-L. Lin, M.-K. Hsu and M.-H. Hsu, J. Med. Chem., 2006, 49, 1442–1449 CrossRef CAS
 .
.
- Z. V. Zhilina, A. J. Ziemba, J. O. Trent, M. W. Reed, V. Gorn, Q. Zhou, W. Duan, L. Hurley and S. W. Ebbinghaus, Bioconjugate Chem., 2004, 15, 1182–1192 CrossRef CAS
 .
.
- W.-P. Hu, J.-J. Ling, C.-L. Kao, Y.-C. Chen, C.-Y. Chen, F.-Y. Tsai, M.-J. Wu, L.-S. Chang and J.-J. Wang, Bioorg. Med. Chem., 2009, 17, 1172–1180 CrossRef CAS
 .
.
- B. Purnell, A. Sato, A. O'Kelley, C. Price, K. Summerville, S. Hudson, C. O'Hare, K. Kiakos, T. Asao, M. Lee and J. A. Hartley, Bioorg. Med. Chem. Lett., 2006, 16, 5677–5681 CrossRef CAS
 .
.
- A. C. Tiberghien, D. A. Evans, K. Kiakos, C. R. H. Martin, J. A. Hartley, D. E. Thurston and P. W. Howard, Bioorg. Med. Chem. Lett., 2008, 18, 2073–2077 CrossRef CAS
 .
.
- D. E. Thurston, V. S. Murty, D. R. Langley and G. B. Jones, Synthesis, 1990, 81–84 CrossRef CAS
 .
.
- R. Kumar and J. W. Lown, Org. Biomol. Chem., 2003, 1, 3327–3342 CAS
 .
.
- A. Kamal, G. Ramesh, N. Laxman, P. Ramulu, O. Srinivas, K. Neelima, A. K. Kondapi, V. B. Sreenu and H. Nagarajaram, J. Med. Chem., 2002, 45, 4679–4688 CrossRef CAS
 .
.
- M. Rettig, M. Weingarth, W. Langel, A. Kamal, P. P. Kumar and K. Weisz, Biochemistry, 2009, 48, 12223–12232 CrossRef CAS
 .
.
- A. Kamal, V. Tekumalla, A. Krishnan, M. Pal-Bhadra and U. Bhadra, ChemMedChem, 2008, 3, 794–802 CrossRef CAS
 .
.
- A. Kamal, R. Ramu, V. Tekumalla, G. B. R. Khanna, M. S. Barkume, A. S. Juvekar and S. M. Zingde, Bioorg. Med. Chem., 2008, 16, 7218–7224 CrossRef CAS
 .
.
- A. Kamal, G. Ramesh, P. Ramulu, O. Srinivas, T. Rehana and G. Sheelu, Bioorg. Med. Chem. Lett., 2003, 13, 3451–3454 CrossRef CAS
 .
.
- A. Kamal, N. Shankaraiah, S. Prabhakar, C. R. Reddy, N. Markandeya, K. L. Reddy and V. Devaiah, Bioorg. Med. Chem. Lett., 2008, 18, 2434–2439 CrossRef CAS
 .
.
- A. Kamal, G. Ramesh, O. Srinivas and P. Ramulu, Bioorg. Med. Chem. Lett., 2004, 14, 471–474 CrossRef CAS
 .
.
- A. Kamal, R. Ramu, G. B. K. Ramesh, A. K. Saxena, M. Shanmugavel and R. M. Pandita, Bioorg. Med. Chem. Lett., 2004, 14, 4907–4909 CrossRef CAS
 .
.
- A. Kamal, M.N.A. Khan, K. S. Reddy, S. K. Ahmed, M. S. Kumar, A. Juvekar, S. Sen and S. Zingde, Bioorg. Med. Chem. Lett., 2007, 17, 5345–5348 CrossRef CAS
 .
.
- A. Kamal, O. Srinivas, P. Ramulu, G. Ramesh and P. P. Kumar, Bioorg. Med. Chem. Lett., 2004, 14, 4107–4111 CrossRef CAS
 .
.
- A. Kamal, E. V. Bharathi, M. J. Ramaiah, J. S. Reddy, D. Dastagiri, A. Viswanath, F. Sultana, S. N. C. V. L. Pushpavalli, M. Pal-Bhadra, A. Juvekar, S. Sen and S. Zingde, Bioorg. Med. Chem. Lett., 2010, 20, 3310–3313 CrossRef CAS
 .
.
- A. Kamal, K. Sreekanth, P. P. Kumar, N. Shankaraiah, G. Balakishan, M. J. Ramaiah, S. N. C. V. L. Pushpavalli, P. Ray and M. Pal Bhadra, Eur. J. Med. Chem., 2010, 45, 2173–2181 CrossRef CAS
 .
.
- A. Kamal, J. S. Reddy, M. J. Ramaiah, D. Dastagiri, E. V. Bharathi, M. A. Azhar, F. Sultana, S. N. C. V. L. Pushpavalli, M. Pal-Bhadra, A. Juvekar, S. Sen and S. Zingde, Eur. J. Med. Chem., 2010, 45, 3924–3937 CrossRef CAS
 .
.
- A. Kamal, K. S. Reddy, M. N. A. Khan, R. V. C. R. N. C. Shetti, M. J. Ramaiah, S. N. C. V. L. Pushpavalli, C. Srinivas, M. Pal-Bhadra, M. Chourasia, G. N. Sastry, A. Juvekar, S. Zingde. and M. Barkume, Bioorg. Med. Chem., 2010, 18, 4747–4761 CrossRef CAS
 .
.
- A. Kamal, N. Shankaraiah, C. R. Reddy, S. Prabhakar, N. Markandeya, H. K. Srivastava and G. N. Sastry, Tetrahedron, 2010, 16, 5498–5506 CrossRef
 .
.
- M. Smellie, D. S. Bose, A. S. Thompson, T. C. Jenkins, J. A. Hartley and D. E. Thurston, Biochemistry, 2003, 42, 8232–8239 CrossRef CAS
 .
.
- SYBYL 6.9 Tripos Associates Inc 1699, St Hanley Road St Louis, MO, USA.
- J. P. Alao, Mol. Cancer, 2007, 6, 24 CrossRef
 .
.
- T. Ozaki and A. Nakagawara, J. Biomed. Biotechnol., 2011 DOI:10.1155/2011/603925
 .
.
- T. Kobayashi, H. Sawa, J. Morikawa, S. Ueno, N. Katayama, W. Zhang and H. Shiku, Int. J. Oncol., 2002, 20, 723–728 CAS
 .
.
- C. J. Sherr, Science, 1996, 274, 1672–1677 CrossRef CAS
 .
.
- M. Fu, C. Wang, Z. Li, T. Sakamaki and R. G. Pestell, Endocrinology, 2004, 145, 5439–5447 CrossRef CAS
 .
.
- C. J. Sherr and J. M. Robert, Genes Dev., 2004, 18, 2699–2711 CrossRef CAS
 .
.
- A. Sunters, P. A. Madureira, K. M. Pomeranz, M. Aubert, J. J. Brosens, S. J. Cook, M. T. Boudewijn, R. Burgerging, C. R. Coombes and E. W.-F. Lam, Cancer Res., 2006, 66, 212–220 CrossRef CAS
 .
.
- Y. T. Siu and D. Y. Jin, FEBS J., 2007, 274, 3224–3232 CrossRef CAS
 .
.
- R. G. Pestell, C. Albanese, A. T. Reutens, J. E. Segall, R. J. Lee and A. Arnold, Endocr. Rev., 1999, 20, 501–534 CrossRef CAS
 .
.
- C. L. Wei, Q. Wu, V. B. Vega, K. P. Chiu, P. Ng, T. Zhang, A. Shahab, H. C. Yang, Y. Fu, Z. Weng, J. Liu, X. D. Zhao, J. L. Chew, Y. L. Lee, V. A. Kuznetsov, W. K. Sung, L. D. Miller, B. Lim, E. T. Liu, Q. Yu, H. H. Ng and Y. Ruan, Cell, 2006, 4, 207–219 CrossRef
 .
.
- L. Zhang, J. Yu, B. H. Park, K. W. Kinzler and B. Vogelstein, Science, 2000, 290, 989–992 CrossRef CAS
 .
.
- J. E. Chipuk, T. Kuwana, L. Bouchier-Hayes, N. M. Droin, D. D. Newmeyer, M. Schuler and D. R. Green, Science, 2004, 303, 1010–1014 CrossRef CAS
 .
.
- N. A. Berger, Radiat. Res., 1985, 101, 4–15 CrossRef CAS
 .
.
- R. J. Davis, Cell, 2000, 103, 239–252 CrossRef CAS
 .
.
- C. Tournier, P. Hess, D. D. Yang, J. Xu, T. K. Turner, A. Nimnual, D. Bar-sagi, S. N. Jones, R. A. Flavell and R. J. Davis, Science, 2000, 288, 870–874 CrossRef CAS
 .
.
- C. L. Fattman, S. M. Delach, Q. P. Dou and D. E. Johnson, Oncogene, 2001, 20, 2918–2926 CrossRef CAS
 .
.
- R. U. Janicke, M. L. Sprengart, M. R. Wati and A. G. Porter, J. Biol. Chem., 1998, 17, 3876–3888 Search PubMed
 .
.
Footnote |
| † Electronic supplementary information (ESI) available: Spectral data of compounds 4a–g, 5a–f, 7a–d, 8a–d, 9a–g, 10a,b, 11a–b and 12a–f and experimental procedures for synthesis and biological evaluations. See DOI: 10.1039/c1md00072a |
|
| This journal is © The Royal Society of Chemistry 2011 |
Click here to see how this site uses Cookies. View our privacy policy here. 


![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) MeOH (4
MeOH (4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1), 12 h (50–60%).
1), 12 h (50–60%).
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) MeOH (4
MeOH (4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1), 12 h (50–60%).
1), 12 h (50–60%).![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5. Interestingly, all the PBD conjugates elevate the helix melting temperature of CT-DNA in the range of 3.0–9.6 °C and were also examined after 18 h incubation at 37 °C. Conjugates linked with simple alkane chain spacers have shown ΔTm values in 3.0–5.9 range, it is observed that the odd number alkane chain spacers containing conjugates have shown high ΔTm when compared to the even number of alkane chain spacer containing conjugates. Moreover, substituted carbazole–PBD conjugates have better binding affinity than their simple carbazole counterparts. Whereas, the conjugates with the piperazine moiety in the linker (5a–f) have shown the highest ΔTm of 9.6 °C at 0 h and increased upto 10.2 °C after 18 h incubation, however, the naturally occurring DC-81 exhibits a ΔTm of 0.7 °C after incubation under similar conditions (Table 1). This result indicates the effect on DNA binding affinity introducing the carbazole moieties to PBD through piperazine containing alkanes as well as simple alkane spacers at C8-position of the DC-81.
5. Interestingly, all the PBD conjugates elevate the helix melting temperature of CT-DNA in the range of 3.0–9.6 °C and were also examined after 18 h incubation at 37 °C. Conjugates linked with simple alkane chain spacers have shown ΔTm values in 3.0–5.9 range, it is observed that the odd number alkane chain spacers containing conjugates have shown high ΔTm when compared to the even number of alkane chain spacer containing conjugates. Moreover, substituted carbazole–PBD conjugates have better binding affinity than their simple carbazole counterparts. Whereas, the conjugates with the piperazine moiety in the linker (5a–f) have shown the highest ΔTm of 9.6 °C at 0 h and increased upto 10.2 °C after 18 h incubation, however, the naturally occurring DC-81 exhibits a ΔTm of 0.7 °C after incubation under similar conditions (Table 1). This result indicates the effect on DNA binding affinity introducing the carbazole moieties to PBD through piperazine containing alkanes as well as simple alkane spacers at C8-position of the DC-81.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) [DNA] molar ratiob
[DNA] molar ratiob![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 molar ratio of [PBD]/[DNA], where CT-DNA concentration = 100 μM and ligand concentration = 20 μM in aqueous sodium phosphate buffer [10 mM sodium phosphate + 1 mM EDTA, pH 7.00 ± 0.01].
c The ΔTm for PBD hybrids 4a–g and 5a–f at a [PBD]
5 molar ratio of [PBD]/[DNA], where CT-DNA concentration = 100 μM and ligand concentration = 20 μM in aqueous sodium phosphate buffer [10 mM sodium phosphate + 1 mM EDTA, pH 7.00 ± 0.01].
c The ΔTm for PBD hybrids 4a–g and 5a–f at a [PBD]![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) [DNA] molar ratio of 1
[DNA] molar ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 increased to a value of 2.0 °C, 2.1 °C, 2.3 °C, 2.0 °C, 2.3 °C and 2.1 °C after 18 h incubation respectively.
5 increased to a value of 2.0 °C, 2.1 °C, 2.3 °C, 2.0 °C, 2.3 °C and 2.1 °C after 18 h incubation respectively.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5
5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5
5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5
5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5
5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5
5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5
5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5
5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5
5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5
5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5
5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5
5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5
5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5
5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5
5



![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells and were grown for 24 h, BrdU was incubated for 5 h at RT followed by treatment with conjugates (1, 6b, 5a and 5f) for 24 h at a final concentration of 4 μM. The intensity of blue colour at 450 nm was measured and is proportional to the amount of BrdU incorporated in proliferating cells. Higher O.D. reading denotes the higher proliferation rate.
000 cells and were grown for 24 h, BrdU was incubated for 5 h at RT followed by treatment with conjugates (1, 6b, 5a and 5f) for 24 h at a final concentration of 4 μM. The intensity of blue colour at 450 nm was measured and is proportional to the amount of BrdU incorporated in proliferating cells. Higher O.D. reading denotes the higher proliferation rate.

.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
