Discovery of novel selective Sigma-1 ligands as cognitive enhancers†
Anne
Valade
*a,
Sophie Binet
Cross
b,
Claire
Brown
b,
Eric
Detrait
a,
Doina
Ene
b,
Michel
Gillard
a,
Michel
Guyaux
a,
Yves
Lamberty
a,
Martin
Maguire
b,
Nivedita
Namdev
b,
Laurent
Provins
a,
Eric
Schwartz
b and
Céline
Vermeiren
a
aUCB Pharma, UCB NewMedicines, Chemin du Foriest, B-1420, Braine-L'Alleud, Belgium. E-mail: anne.valade@ucb.com
bFormer UCB Research Inc., Cambridge, USA
First published on 10th June 2011
Abstract
We disclose the identification and SAR of phenylcyclopropylcarboxamide compounds as novel, selective Sigma-1 ligands presenting excellent drug-like properties, high solubility and metabolic stability. A selected representative, compound 14, demonstrated in vivo very good brain exposure and antiamnesic effect in a mouse model of recognition memory.
Introduction
The Sigma-1 receptor is an endoplasmic reticular protein, widely distributed centrally and peripherally.1 With the discovery of N-allylnormetazocine (SKF-10,047) in 1976 by Martin,2 this receptor was initially mis-characterised as a new type of opioid receptor. The cloning of the guinea pig Sigma-1 binding site in 19963 revealed that this protein is unique and does not share any homology with other mammalian proteins.In 2007, Hayashi and Su described the Sigma-1 protein as a chaperone protein modulating a variety of key intracellular targets leading to regulation of intracellular calcium, increased neuronal excitability, or neuroprotection.4 Various drugs, including antipsychotics (morphine analogs), neuroleptics (haloperidol) and neuroactive steroids (progesterone, DHEA), bind to the Sigma-1 receptor.1Ligands that modulate the Sigma-1 receptor are proposed to display effects in several therapeutic areas such as stroke, neurodegenerative disorders, schizophrenia, depression, pain, amnesia and Alzheimer's disease.5
Since the 1970s, pharmaceutical companies have put tremendous efforts in the identification of potent Sigma-1 ligands. Several non-selective preclinical and fewer clinical candidates were discovered, demonstrating some improvement of cognitive impairment induced by scopolamine, age or β-amyloid peptides in rodents, such as (+)-SKF-10,0476 or SA-45037 (Fig. 1). Despite promising preclinical data, no clinical proof of concept has been reported to date.
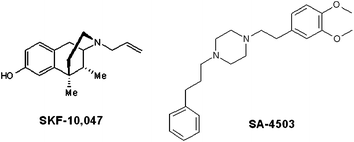 | ||
| Fig. 1 Synthetic Sigma-1 receptor ligands. | ||
A common pharmacophoric feature of the Sigma-1 receptor ligands is an alkylamine moiety, which can explain how challenging it can be to design selective Sigma-1 ligands.8 Here we disclose the identification of a novel series of selective and drug-like phenylcyclopropylcarboxamide Sigma-1 ligands.
Results and discussion
In the framework of one of our projects targeting cough,9 compound (±)-1 was identified as an interesting new hit binding Sigma-1 receptor, following medium throughput screening and scaffold hopping (Table 1). Despite moderate in vitro binding potency towards the Sigma-1 receptor (pKi = 6.7 using human CHO cell membranes),10 compound (±)-1 was already a very good starting point, with a lipophilic ligand efficiency (LLE)11 above 5. However, similarly to most of the Sigma-1 ligands, it exhibited low selectivity, especially versuscalcium channels, serotoninergic and adrenergic receptors. The separation of the trans-enantiomers of compound (±)-1 led to compounds (+)-1 and (−)-1,12 which both displayed the same level of Sigma-1 affinity (pKi = 6.8) as the racemic mixture. When profiled, pure isomer (+)-1 was found to be poorly selective versus a large number of targets and particularly versus Sigma-2 receptor (pKi = 6.6), whereas enantiomer (−)-1 seemed more selective from the internal off-targets profiling. Despite low lipophilicity, compound (+)-1 was poorly stable metabolically (intrinsic clearances in rat liver microsomes: Clint = 147 μL min−1 mg protein−1). Moreover, these compounds are strongly basic (pKa > 10), but were considered appropriate starting points to initiate a lead generation program on this chemical series with the aim of improving Sigma-1 binding affinity, selectivity and metabolic stability.| Compound |
|
R3 | σ1 pKia | σ2 pKib | Off-targets profiling | log D [ACD_log D]c | LLE [ cLLE ] d | pKa[ACD_pKa]e | Clint (rat)f |
|---|---|---|---|---|---|---|---|---|---|
| a Binding affinities (pKi) were assessed by displacement of [3H](+)pentazocine (in CHO cell membranes expressing human σ1 receptors).10Kd[3H](+) pentazocine = 37 ± 21 nM (n = 7); Bmax [3H](+)pentazocine = 28 ± 15 pmol mg−1 of proteins (n = 7). Assay concentration = 1–2 nM (29–35 Ci mmol−1). pKihaloperidol = 8.3 ± 0.2 nM (n = 126). Results are mean of 2–3 experiments. Each pKi value is given with its confidence interval (CI) at 95%, which was calculated based on the variance and SEM values. b Binding affinities (pKi) were assessed (by CEREP) by displacement of [3H]1,3-di-O-tolylguanidine (+300 nM (+)pentazocine) (in rat cerebral cortex). Kd[3H]1,3-di-O-tolylguanidine = 32 nM. Assay concentration = 5 nM. IC50haloperidol = 120 nM (at 10 μM). c log D values were measured at pH 7.4.15 When the log D values were not available, log D predictions at pH 7.4 were calculated from ACD/Labs software [ACD_log D].16 d LLE 11 = lipophilic ligand efficiency = pKi − log D. When the log D values were not available, cLLE was calculated using the following expression: cLLE = pKi − ACD_log D. e pKa values were experimentally measured.17 When the pKa values were not available, pKa predictions at pH 7.4 were calculated from ACD/Labs software [ACD_pKa].16 f Clint = intrinsic clearances in rat liver microsomes (μL min−1 mg protein−1). g Internal off-targets profiling (about 20 targets): target named when at least 50% displacement observed at 10 μM. Target in bold when displacement at 10 μM is higher then 90%. h Extended off-targets profiling (CEREP, http://www.cerep.fr/, about 50 targets): target named when at least 50% displacement observed at 10 μM. Target in bold when displacement at 10 μM is higher then 90%. Opiate stands for the opioid (non-selective) assay from CEREP ([3H]naloxone in rat cerebral cortex). Muscarinic stands for the muscarinic (non-selective) assay from CEREP ([3H]QNB in rat cerebral cortex). | |||||||||
| (±)-1 |
|
H | 6.7 ± 0.38 | nt | Ca 2+ , 5HT2a, α1g | 1.3 | 5.4 | 10.1 | nt |
| (+)-1 |

|
H | 6.8 ± 0.38 | 6.6 | 5HT2a , Na+, α1, α2c, α2b, Ca2+h | 1.1 | 5.7 | 10.4 | 147 |
| (−)-1 |

|
H | 6.8 ± 0.27 | nt | Na+g | 1.1 | 5.7 | 10.4 | nt |
| 2 |
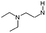
|
H | 6.9 ± 0.38 | nt | Opiate, 5HT2ah | 0.6 | 6.3 | 8.9 | nt |
| 3 |
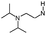
|
H | 7.2 ± 0.38 | nt | 5HT2a g | 1.0 | 6.2 | 9.7 | >300 |
| 4 |

|
H | 7.2 ± 0.27 | nt | 5HT2a g | 0.9 | 6.3 | 9.2 | nt |
| 5 |
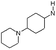
|
H | 5.9 ± 0.38 | nt | nt | 0.9 | 5.0 | 9.6 | nt |
| 6 |
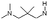
|
H | 6.4 ± 0.38 | nt | nt | 0.8 | 5.6 | 8.9 | nt |
| 7 |

|
H | 7.8 ± 0.27 | 5.9 | D4, 5HT2a, 5HT6, 5HT7, kappa, mu, NE transporter, DA transporterh | 2.8 | 5 | [7.4] | 122 |
| 8 |
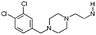
|
H | 8.0 ± 0.27 | 6.2 | 5HT2a , H1, Na+g | >4 | <4 | [7.4] | 200 |
| 9 |
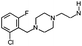
|
H | 7.6 ± 0.22 | nt | nt | 3.7 | 3.9 | [7.4] | 283 |
| 10 |

|
H | 6.2 ± 0.38 | nt | 5HT2a g | 2.3 | 3.9 | [6.2] | 81 |
| 11 |
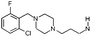
|
H | 7.6 ± 0.38 | nt | 5HT2a , Ca2+, H1, α1, α2cg | 3.5 | 4.1 | [7.8] | 305 |
| 12 |

|
H | 5.9 ± 0.38 | nt | nt | 3.8 | 2.1 | <2.5 | nt |
| 13 |

|
H | 6.7 ± 0.38 | nt | Cleang | 0.7 | 6 | 9.3 | 25 |
| 14 |

|
H | 7.1 ± 0.22 | 50% inhib. at 10 μM | H3, Ca2+h | 1.1 | 6.0 | 9.3 | 32 |
| 15 |
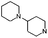
|
F | 7.0 ± 0.27 | 61% inhib. at 10 μM | Cleanh | [1.4] | [5.6] | [9.3] | nt |
| 16 |

|
H | 7.1 ± 0.38 | nt | Na+, muscarinic, 5HT1ah | [0.5] | [6.6] | [10.6] | 17 |
| (+)-SKF-10,047 | — | 6.8 ± 0.38 | 29% inhib. at 10 μM | kappa , mu, AChEh | [2.1] | [4.7] | [10.1] | 313 | |
| SA-4503 | — | 8.5 ± 0.38 | 99% inhib. at 10 μM | NE transporter , α1, α2c, 5HT2a, 5HT transporter, D2, D3, D4, DA transporter, H3h | [2.7] | [5.8] | [8.2] | nt |
All the phenylcyclopropylcarboxamide analogues were synthesised as shown in Scheme 1.13,14 Treatment of commercially available trans-2-phenylcyclopropane carboxylic acid with 1 equiv. of carbonyldiimidazole in tetrahydrofuran and subsequent addition of 1 equiv. of commercially available primary or secondary amines gave access to the desired amides in moderate to very high yields (26–95%).
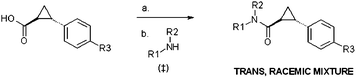 | ||
| Scheme 1 Preparation of phenylcyclopropylcarboxamide analogs. Reagents and conditions: (a) carbonyldiimidazole (1 equiv.), THF, reflux, 1 h 30 min; (b) R1R2NH (1 equiv.), room temperature, 8 h, 26–95%. ‡ Key acids and amines used were commercially available (from Sigma Aldrich). | ||
Structure–activity relationship around other secondary amides, linked by an ethyl or n-propyl chain to different alkylamine moieties, was explored (compounds 2–11, Table 1). Since there were no differences in terms of binding affinities between enantiomers (+)-1 and (−)-1, we decided to pursue our investigation in the racemate series. Replacement of the cyclohexylamine moiety of compound (±)-1 by cyclic and non-cyclic tertiary amines gave rise to small molecules (compounds 2–4), displaying good affinities (pKi = 6.9–7.2) and a better selectivity profile (mainly touching 5HT2a) than compounds (±)-1 and (+)-1.
Unfortunately, despite low lipophilicity (LLE = 6.2), compound 3 was still highly metabolised on rat microsomes (Clint > 300 μL min−1 mg protein−1). Rigidification of the n-propyl linker (compounds 5 and 6) seemed detrimental to the Sigma-1 affinity. The introduction of substituted benzylpiperazines instead of the alkylamine induced a high increase of Sigma-1 affinities (compounds 7–9 and 11, pKi from 7.6 to 8). Unfortunately these compounds appeared to be poorly selective and characterised by higher molecular weight, high lipophilicity (LLE < 5) and high metabolic instabilities.
Interestingly, with the exception of compounds 10 and 12, which highlight the need for a basic moiety, the replacement of the secondary amide by rigidified cyclic tertiary amides (compounds 13–16) gave access to ligands exhibiting:
-good affinity (compounds 14versus5),
-improved selectivity (compounds 13versus4),
-lower lipophilicity (LLE > 5.6),
-acceptable metabolic stability (Clint ≤ 32 μL min−1 mg protein−1; compounds 13, 14 and 16).
The combination of a cyclic amine directly linked to a piperidine led to our best ligand, compound 14, which was quite potent, highly selective, poorly lipophilic (LLE = 6) and metabolically stable. A para-fluoro phenyl substitution was also tolerated: no affinity loss was observed with compound 15 and high overall selectivity was also achieved. It is interesting to note that compounds 14 and 15 fit quite well to Glennon's pharmacophore (distance between the amine site and the primary hydrophobic region ∼8 to 10 Å, with amine moiety embedded in a cyclic structure).8
With the aim to compare our ligands with Sigma-1 reference compounds, (+)-SKF-10,047 and SA-4503 were included in the study. They both displayed binding affinities similar to those reported (Table 1).18 These two ligands, the initial hit (compound (+)-1), piperazine7 and compounds 14 and 15, were tested against a large number of targets (extended off-targets profiling of about 50 targets). (+)-SKF-10,047 displayed the same level of affinity as our initial hit compound (+)-1, while binding κ opioid receptor with more than 90% inhibition at 10 μM (Fig. 2). As observed for compound 7, SA-4503, sharing the same piperazine pattern, exhibited much higher affinity towards the Sigma-1 receptor (pKi = 8.5), but an extremely poor overall selectivity profile (four targets are hit with at least 90% inhibition at 10 μM). Satisfyingly, secondary amides14 and 15 demonstrated a fully selective profile, while maintaining good Sigma-1 potency.
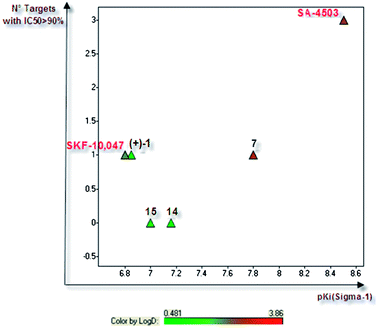 | ||
| Fig. 2 Off-targets profile: number of targets with at least 90% displacement observed at 10 μM (extended off-targets profiling of about 50 targets) vs. Sigma-1 affinity (pKi measured using human CHO cell membranes).10 | ||
In order to assess the drug-like properties of our best phenylcyclopropylcarboxamide analogs, we decided to use two prospective design tools: the lipophilic ligand efficiency (LLE)11 and the recent Central Nervous System Multiparameter Optimisation (CNS MPO) algorithm proposed by Wager et al.19 (Fig. 3). The CNS MPO function is the sum of desirability scores of six physicochemical parameters: calculated lipophilicity (clog P), distribution coefficient (log D), molecular weight, polar surface area (TPSA), number of hydrogen bond donors (HBDs) and basicity. This calculation enables a balanced, multi-parameter assessment of drug-like properties.
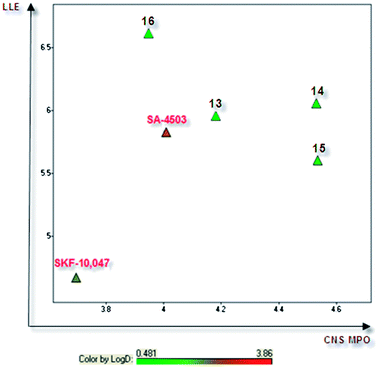 | ||
| Fig. 3 Lipophilic ligand efficiency (LLE) vs. CNS MPO scores (calculated following the functions reported in ref. 19; 74% of marketed CNS drugs displayed a CNS MPO score above 4). | ||
While the reference compounds (+)-SKF-10,047 and SA-4503 were both poorly scored using the CNS MPO algorithm (≤4), our best compounds (14 and 15) displayed CNS MPO desirability scores of 4.5, together with very high LLE values (>5.5). The CNS MPO global and individual scores of compound 14, (+)-SKF-10,047 and SA-4503 are detailed in Table 2. Firstly, it is noteworthy that these three Sigma-1 ligands are systematically characterised by very low polar surface areas. This observation seems to be quite frequent within Sigma-1 ligands.
| Compound 14 | (+)-SKF-10,047 | SA-4503 | ||||
|---|---|---|---|---|---|---|
| Value | T0a | Value | T0a | Value | T0a | |
| a T0 = individual transformed scores were calculated following the functions reported in ref. 19. b clog P values were calculated from ACD/Labs software.16 c log D values were measured at pH 7.4.15 When the log D values were not available, log D predictions at pH 7.4 were calculated from ACD/Labs software.16 d Topological Polar Surface Area (TPSA) values were calculated from ACD/Labs software.16 e Number of hydrogen bond donors (HBD). f pKa values were experimentally measured.17 When the pKa values were not available, pKa predictions were calculated from ACD/Labs software.16 g CNS MPO scores were calculated following the functions reported in ref. 19. | ||||||
| clog Pb | 2.6 | 1 | 3.5 | 0.75 | 4.5 | 0.25 |
| log D [ACD_log D]c | 1.1 | 1 | [2.1] | 0.94 | [2.7] | 0.66 |
| TPSA d | 23.6 | 0.18 | 23.5 | 0.17 | 24.9 | 0.24 |
| M w | 312.4 | 1 | 257.3 | 1 | 368.5 | 0.94 |
| HBD e | 0 | 1 | 1 | 0.83 | 0 | 1 |
| pKa[ACD_pKa]f | 9.3 | 0.35 | [10.1] | 0 | [8.2] | 0.91 |
| CNS MPO scoresg | 4.5 | 3.7 | 4.0 | |||
Compound 14 displayed optimal scores for 4 out of 6 properties and clearly demonstrates improved drug-like properties (CNS MPO score of 4.5) in comparison with reference compounds (+)-SKF-10,047 and SA-4503 (respective CNS MPO scores of 3.7 and 4). The CNS MPO scoring of compound 14 was negatively impacted by its high basicity (pKa = 9.3). Nevertheless, we observed that while a basic part is necessary to maintain Sigma-1 potency (see compounds 10 and 12), high basicity is not mandatory (see piperazine compounds 7–9 and 11). If pKa were to become an issue later in the lead optimisation process (e.g. for toxicity or DMPK reasons), there should still be room for optimisation.
Overall, compound 14 represented the best analog for further drug candidate profiling. It exhibited very good solubility (1.1 mg mL−1 at pH 7.4). Some in vitro ADME endpoints as well as brain to plasma ratios were evaluated for compound 14 and reference compound (+)-SKF-10,047 (Table 3). Both compounds showed low to moderate intrinsic clearances on mice liver microsomes, good cell permeability profiles in a Caco-2 assay and, despite high basicity, high to very high brain to plasma ratios in mice (B/P ratio of 13.5 for compound 14 and B/P ratio of 5.6 for (+)-SKF-10,047, after intraperitoneal administration of a 1 mg kg−1 dose), supporting their use in in vivo models of cognition.
| Compound 14 | (+)-SKF-10,047 | |
|---|---|---|
| a Papps (permeability coefficient determined using Caco-2 cell permeability assay): apical-to-basolateral flux following incubation of the given compound at 20 μM with Caco-2 monolayers for up to 3 h. b Mean brain-to-plasma ratios at 40 min after single intraperitoneal dosing to male C57BL6/J mice at 1 mg kg−1 (n = 3 per group). | ||
| Microsomal stability | ||
| Human Clint/μL min−1 mg prot.−1 | 56 | nt |
| Rat Clint/μL min−1 mg prot.−1 | 32 | 313 |
| Mouse Clint/μL min−1 mg prot.−1 | 50 | 17 |
| Permeability | ||
| Caco-2: Papps/nm s−1a | 166 | 202 |
| Brain penetration | ||
| Brain/plasma ratiob | 13.5 | 5.64 |
Consequently, compound 14 and (+)-SKF-10,047 were evaluated in the mouse two-trial object recognition assay (Fig. 4).20 This test is based on the observation that rodents, when confronted several times with various objects, may distinguish familiar from novel objects. Compound 14 and (+)-SKF-10,047, administered intraperitoneally, displayed the capacity to counteract scopolamine-induced memory deficit in this model (Fig. 4). Indeed, contrary to scopolamine only treated mice, mice treated with scopolamine (0.3 mg kg−1, i.p. 30 min prior acquisition trial) and compound 14 (1–23 mg kg−1, i.p. 40 min prior acquisition trial) or (+)-SKF-10,047 (0.1–1 mg kg−1, i.p. 40 min prior acquisition trial) showed a significant increase of the exploration difference between the familiar and the novel object during the retention trial.
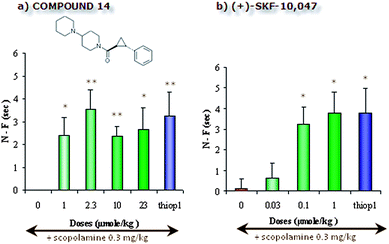 | ||
| Fig. 4 Anti-amnesic effect of compound 14 (a) and reference compound (+)-SKF-10,047 (b) versusthioperamide in scopolamine-induced amnesia using the two-trial object recognition test20 in male C57Black 6J mice. N–F = Time (in seconds) spent in active exploration of the Novel (N) object—time (in seconds) spent in active exploration of the Familiar (F) object. Compound 14 was administered i.p. at 1–23 mg kg−1, as indicated; (+)-SKF-10,047 was administered i.p. at 0.03–1 mg kg−1, as indicated; thioperamide, used as a positive control, was administered i.p. at 0.5 mg kg−1; scopolamine was administered i.p. at 0.3 mg kg−1, as indicated; vehicle = 1% MC/DMSO; statistical analysis: sample size = 9–11; one way ANOVA followed by Dunnett's post-hoc test: * = p < 0.05; ** = p < 0.01 compared to scopolamine treated group. | ||
Conclusion
We have discovered a novel class of potent Sigma-1 ligands among which some key leads demonstrated a higher level of selectivity (vs. Sigma-2, GPCR's, ion channels) than the tested reference compounds (+)-SKF-10,047 and SA-4503. These compounds present excellent drug-like properties (low molecular weight, low lipophilicity, very high lipophilic ligand efficiency, CNS MPO scores > 4.5) including high solubility and metabolic stability. A selected representative, compound 14, demonstrated very good brain exposure. Efficacy of compound 14 in a mouse model of recognition memory is comparable to that of (+)-SKF-10,047, therefore supporting the therapeutic potential of this chemotype for the symptomatic treatment of cognitive disorders.Acknowledgements
We thank Mrs A. Descamps, Mrs G. Longfils, Dr B. Mathieu for physchem measurements; Mr A. Fauconnier, Mr J. Claessens for analytical assistance; Mr. E. Hanon for statistical calculations; Mr P. Collart for in vitro pharmacokinetic measurements; Mrs V. Bertaina-Anglade, Mrs E. Enjuanes and Dr C. Drieu la Rochelle (Biotrial) are greatly acknowledged for the testing of compounds in the two-trial object recognition model.Notes and references
- T. Hayashi and T.-P. Su, Expert Opin. Ther. Targets, 2008, 12, 45 Search PubMed.
- W. R. Martin, C. G. Eades, J. A. Thompson, R. E. Huppler and P. E. Gilbert, J. Pharmacol. Exp. Ther., 1976, 197, 517 Search PubMed.
- M. Hanner, F. F. Moebius, A. Flandorfer, H.-G. Knaus, J. Striessnig, E. Kempner and H. Glossmann, Proc. Natl. Acad. Sci. U. S. A., 1996, 93, 8072 CrossRef CAS.
- T. Hayashi and T.-P. Su, Cell, 2007, 131, 596 CrossRef CAS.
- T. Maurice and T.-P. Su, Pharmacol. Ther., 2009, 124, 195 Search PubMed.
- K. Matsuno, T. Senda, T. Kobayashi and S. Mita, Brain Res., 1995, 690, 200 Search PubMed.
- T. Maurice and A. Privat, Eur. J. Pharmacol., 1997, 328, 9 Search PubMed.
- R. A. Glennon, Mini-Rev. Med. Chem., 2005, 5, 927 CrossRef CAS.
- C. I. Brown, M. Fezoui, W. M. Selig, E. Schwartz and J. L. Ellis, Br. J. Pharmacol., 2004, 141, 233 Search PubMed.
- Affinity of the test compounds for human Sigma-1 receptor was measured by competition with [3H]pentazocine. This binding assay was performed essentially as described by M. Hanner et al. (1996)21 with few modifications. Briefly, membranes Chinese Hamster Ovary (CHO) cells expressing human recombinant Sigma-1 receptors were incubated at 25 °C for 180 min in 0.2 mL of a 50 mM Tris–HCl buffer (pH 7.4) containing 2 mM MgCl2, 1.5 nM [3H]pentazocine and increasing concentrations of the test compounds. The nonspecific binding was defined as the residual binding observed in the presence of 10 μM haloperidol. Membrane-bound and free radioligand were separated by rapid filtration through glass fiber filters presoaked in 0.1% polyethylenimine. Samples and filters were rinsed by at least 6 mL of ice-cold 50 mM Tris–HCl buffer (pH 7.4). The entire filtration procedure did not exceed 10 seconds per sample. Radioactivity trapped onto the filters was counted by liquid scintillation in a β-counter.
- P. D. Leason and B. Springthorpe, Nat. Rev. Drug Discovery, 2007, 6, 881 CrossRef CAS.
- Trans-enantiomers of compound (±)-1 were separated by preparative chiral high pressure liquid chromatography on a CHIRALPAK AD column (Daicel, 10 × 500 mm) with EtOH 10%/benzine 90%/diethylamine as eluent. Compound (+)-1 was isolated with an enantiomeric excess of 100% and an optical rotation value of +208.5° at 22 °C. Compound (−)-1 was isolated with an enantiomeric excess of 100% and an optical rotation value of −203.4° at 22 °C. Since there were no differences in terms of binding affinities between enantiomers (+)-1 and (−)-1, we did not push further investigation towards absolute configuration assignment.
- J. Ellis, D. Ene, E. Schwartz, N. Namdev, C. Brown, S. Binet-Cross and D. Meyers, PCT Int. Appl., WO2006/027252. All physical and spectroscopic details, including salt forms, melting points and mass spectroscopic data, are reported for all the described compounds in this patent.
- Example: synthesis of compound 14 (trans-1′-[(2-phenylcyclopropyl)carbonyl]-1,4′-bipiperidine): commercially available trans-2-phenylcyclopropanecarboxylic acid (10.37 g, 64.0 mmol) was dissolved in anhydrous tetrahydrofuran (250 mL) and carbonyldiimidazole (10.37 g, 64.0 mmol) was added to it. The reaction mixture was allowed to reflux for 3 h followed by addition of commercially available 4-piperidinopiperidine (10.78 g, 64.0 mmol) and the reaction mixture was stirred at room temperature for 48 h. The solvent was removed in vacuo, dissolved in dichloromethane and washed with water. The crude residue obtained after removal of solvent was purified by flash chromatography using methanol/ethylacetate as eluent to provide 18.7 g of a white solid, which was then recrystallised from hexanes. 6 g (30%) were kept as free base and the rest was dissolved in ether. A 2 N solution of HCl in ether was added to form a salt, which was filtered, washed with ether and dried to afford 8.2 g (37%) of the HCl salt (1/1 HCl); white solid; mp 86 °C; MS [M + H]: 313; 1H NMR (CDCl3, δ ppm): 7.36–7.06 (m, 5H), 4.67 (m, 1H), 4.16 (m, 1H), 3.04 (m, 1H), 2.58 (m, 1H), 2.48 (m, 6H), 1.98 (m, 1H), 1.85 (m, 2H), 1.75–1.37 (m, 9H), 1.26 (m, 1H).
- log D measurements (octanol/H2O): test compound (1 mg) was dissolved in octanol (2 mL) saturated with H2O (PBS buffer solution at a given pH) or H2O (2 mL) saturated with octanol then filtrated. This solution (0.5 mL) was added to 0.5 mL of the other phase and sonicated. The samples were then centrifuged for 1 h. Quantification of each phase was performed by HPLC. The distribution coefficient is the logarithm of the ratio between HPLC trace surface areas from the octanol phase over those of the aqueous phase at a given pH.
- ACD_log D (log D predictions at pH 7.4), ACD_pKa (pKa predictions) and TPSA were calculated from ACD/Labs software, version 12.01, Advanced Chemistry Development, Inc., Toronto.
- pKa measurements: all titrations were performed using GlpKa™ (Sirius, Forest Row, UK) in aqueous solution or methanol–water mixture with 0.15 M KCl at 25 °C using standardised 0.5 M HCl titrant.
- J. R. Lever, J. L. Gustafson, R. Xu, R. L. Allmon and S. Z. Lever, Synapse, 2006, 59, 350 Search PubMed.
- T. T. Wager, X. Hou, P. R. Verhoest and A. Villalobos, ACS Chem. Neurosci., 2010, 1, 435 Search PubMed.
- V. Bertaina-Anglade, E. Enjuanes, D. Morillon and C. Drieu la Rochelle, J. Pharmacol. Toxicol. Methods, 2006, 54, 99 Search PubMed.
- M. Hanner, F. F. Moebius, A. Flandorfer, H. G. Knaus, J. Striessnig, E. Kempner and H. Glossmann, Proc. Natl. Acad. Sci. U. S. A., 1996, 15, 8072 Search PubMed.
Footnote |
| † In memory of Dr Patrice Talaga. |
| This journal is © The Royal Society of Chemistry 2011 |