DOI:
10.1039/C0MD00266F
(Concise Article)
Med. Chem. Commun., 2011,
2, 579-584
Received
19th December 2010
, Accepted 8th March 2011
First published on 24th March 2011
Introduction
The development of self-cleaving chemical nucleases is regarded as the paradigm of redox active metal-based chemotherapeutics. DNA targeting agents capable of inducing single stranded or double stranded scission have found clinical application within cancer chemotherapy.1 Other applications within this class of compound include, the probing of DNA-specific structures, mapping of protein and DNA interactions, gene regulation and signal transduction.2,3 Thus, explorations toward the discovery and development of natural or synthetic chemical nucleases are major topics of interest. Redox-active, transition-metal-based chemical nucleases are particularly important due to their potential to catalytically support the one-electron oxidation/reduction reactions necessary to drive activation of C–H deoxyribose bonds.
In the presence of COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundCu2+, the oxidative formation of π radical cations within marine-based products, tambjamine E,4 prodigiosin5 and pyrimol,6 have recently been shown to mediate self-cleaving DNA damage, i.e. scission which does not require the presence of added oxidant or reductant. These COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundCu2+ compounds have also demonstrated significant in vitro chemotherapeutic potential against leukaemia and ovarian cancer cells, some of which were resistant to COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundcisplatin.5,6
The discovery of the first synthetic chemical nuclease, [Cu(phen)2]2+ (COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundphen = COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound1,10-phenanthroline, Fig. 1), has sparked intensive effort toward the development of new bis(phen) agents with enhanced DNA cleaving ability.7 The DNA cleaving limitations of [Cu(phen)2]2+ include, (i) a high dissociation constant of the second coordinated COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundphen ligand8 and (ii) the need for exogenous reductant to generate the active COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundCu+ species, [Cu(phen)2]+. The dissociation problem was solved by Meunier, Pitie et al.9–11 through the advent of clip-COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundphen, whereby two COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundphen ligands are connected at the 2′ or 3′ position by a COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundserinol bridge. Recently, we have reported the first self-cleaving bis-phen system, [Cu(phen)2(phthalate)] (COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundphthalate = o-, m-, COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundp-phthalate, Fig. 1), capable of inducing single-stranded DNA scission in the absence of exogenous reductant or oxidant.12
While mononuclear [Cu(phen)2(phthalate)] complexes displayed excellent chemotherapeutic potential against colon (HT29), breast (MC-F7) and prostate (DU145) cancer lines, their COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundwater solubility is poor. Of the COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundphthalate group of COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundCu2+ complexes the cationic, dinuclear species [Cu2(phen)4(μ2-p-phthalate)]2+ proved to be the most active DNA-binding, self-cleaving chemotherapeutic agent. To that end, in the current study, we have investigated the application of COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundwater soluble COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundCu2+ and COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundMn2+ dinuclear, cationic, bis-phen octanedioate (oda) (Fig. 1) systems as nuclease mimetics and determined their ability to induce cancer cell death through the redox-generation of reactive oxygen species (ROS). Arguments for the possible formation of a π carboxyl radical within the COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundCu2+ system, which cleaves DNA by self-activation, are also presented.
[Cu2(μ2-oda)(phen)4](ClO4)2 (1) (Fig. 2), {[Mn2(μ2-oda)(phen)4(oda)2]2−[Mn2(μ2-oda)(phen)4(H2O)2]2+} (2) (Fig. 3), were prepared according to the literature methods.13,14 The coordination environment about each of the COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundCu2+ ions in 1 is approximately square-pyramidal with the metals being linked via a bridging oda2− ligand. In the double complex salt, 2, the environment about each COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundMn2+ is octahedral with both metals in the cation and anion being bridged oda2− in a similar fashion to 1. Whereas each of the COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundMn2+ centres of the anionic subunit in 2 contain a unidentate oda2− ligand coordinated in the apical position, the cationic unit contains bound COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundwater at these equivalent positions.
![Molecular structure of the [Cu2(μ2-oda)(phen)4]2+ cation in complex 1.](/image/article/2011/MD/c0md00266f/c0md00266f-f2.gif) |
| | Fig. 2 Molecular structure of the [Cu2(μ2-oda)(phen)4]2+ cation in complex 1. | |
![Molecular structures of the dimeric cation and anion subunits in the MnII double salt complex {[Mn2(μ2-oda)(phen)4(oda)2]2−[Mn2(μ2-oda)(phen)4(H2O)2]2+} (2).](/image/article/2011/MD/c0md00266f/c0md00266f-f3.gif) |
| | Fig. 3 Molecular structures of the dimeric cation and anion subunits in the MnII double salt complex {[Mn2(μ2-oda)(phen)4(oda)2]2−[Mn2(μ2-oda)(phen)4(H2O)2]2+} (2). | |
Read more about this on ChemSpider
Download mol file of compoundethidium bromide displacement and fluorescence quenching experiments with calf thymus DNA (CT-DNAp) were conducted (Table 1).† Complexes 1 and 2 were found to have high apparent DNA binding constants (Kapp), and low fluorescence quenching values (Q), indicating that both systems have a high affinity for binding to DNA. High Q values (>20 μM) are generally found for “classical” DNA intercalants, whereas lower values between 2–15 μM are typically obtained for minor groove ligands or hybrid molecules.15,16
Table 1 Apparent DNA binding constants (Kapp) and fluorescence quenching (Q) values for 1 and 2.2 Assay conditions; C50/Kapp: final volume 2 mL, 1.2 μM EtBr, 1 μM CT-DNAp, 10 mM TES, 0.1 mM Na2EDTA, pH 7.0; Q: final volume 2 mL, 2.0 μM EtBr, 20 μM CT-DNAp, 2 mM NaOAc buffer, 9.3 mM COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundNaCl, 0.1 mM Na2EDTA, pH 5.0
DNA cleavage reactions
The relaxation of supercoiled pUC18 DNA (SC, Form I) into open circular (OC, Form II) and linear (LC, Form III) conformations was used to quantify the relative cleavage efficiency of 1 and 2.† To investigate the DNA self-cleaving ability of complexes 1 and 2, SC DNA was exposed to both complexes over a concentration range of 1–50 μM for 20 h in the absence of added H2O2 or reductant (Fig. 4(a)). Only complex 1 exhibits concentration-dependant self-cleaving of SC DNA (Form I) to OC (Form II), with complete depletion of the parent SC band (I → II) at 20 μM (lane 5 (a)). In the presence of added reductant (ascorbate) (Fig. 4(b)) both complexes exhibit enhanced DNA scission during a shorter time-frame (2 h) and at a lower concentration of added complex. While both 1 and 2 induced complete relaxation to OC Form II (lanes 4 and 8), only the COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundCu2+ complex, 1, was found to induce efficient double stranded scission (I → III) at a concentration of 20 μM, which was evident by the absence of a band in lane 5. In order to identify what role molecular O2 and the metal ions (COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundCu2+/COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundMn2+) play within DNA cleavage, experiments were conducted under anaerobic conditions in an atmosphere saturated with COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundargon and, separately, in the presence of 100 mM Na2EDTA, under aerobic conditions (Fig. 4 (c) and (d)). The COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundMn2+ system was examined at a concentration of 20 μM over a 2 h period in the presence of ascorbate, while the COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundCu2+ system was tested over a longer time period (20 h) and without added reductant. The COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundMn2+ complex, 2, did not cleave DNA in the absence of O2 and presence of COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundEDTA, however, complex 1 was found to induce some DNA damage in the presence of excess metal chelator (COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundEDTA) and slight relaxation in the absence of O2.
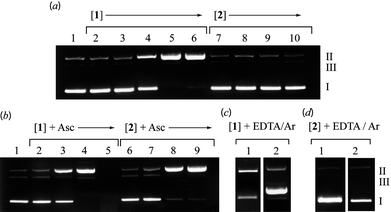 |
| | Fig. 4 Relaxation of pUC18 by 1 and 2.† Cleavage was carried out at 37 °C then analyzed by agarose gel electrophoresis (a) 20 h incubation in the absence of added oxidant or reductant. Lane 1: DNA alone; lanes 2–6: 1, 5, 10, 20, 50 μM 1; lanes 7–10: 5, 10, 20, 50 μM 2. (b) 2 h incubation in the presence of added ascorbate (at twice the complex concentration). Lane 1: DNA alone; lanes 2–5: 1, 5, 10, 20 μM 1; lanes 6–9: 1, 5, 10, 20 μM 2. (c) 20 h incubation of 20 μM 1 in the absence of added oxidant or reductant. Lane 1: 1 + 100 mM Na2EDTA; lane 2: sat. Ar atmosphere. (d) 2 h incubation of 20 μM 2 with added ascorbate (at twice the complex concentration). Lane 1: 2 + 100 mM Na2EDTA; lane 2: sat. Ar atmosphere. | |
Read more about this on ChemSpider
Download mol file of compoundsuperoxide radical (O2˙−) and COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundhydrogen peroxide (H2O2) are imperative for cleaving the phosphodiester backbone in DNA,17–19 we have examined the interaction of complexes 1 and 2 with both these species. COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundSuperoxide was generated enzymatically by the COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundxanthine/COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundxanthine-oxidase system and quantified photometrically by the detector molecule, nitro-blue-COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundtetrazolium (NBT). Both 1 and 2 show potent SOD (superoxide dismutase) mimetic activity, with the COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundMn2+ complex being an exceptional catalyst (1 U SOD = 24.6 nM) (Fig. 5 and Table 2). The catalase (CAT) mimetic activity of complexes 1 and 2 was determined by measuring the volume of evolved O2 from disproportionate H2O2 (30% v/v solution). Only the COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundMn2+ complex (2) was capable of decomposing H2O2 and its activity can also be described as exceptional in this regard (6 × 103 H2O2 molecules disproportionated in 5 min). Overall, both 1 and 2 interacted with COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundsuperoxide to produce COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundhydrogen peroxide (equation I), the COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundMn2+ complex being the most active. Furthermore, it was only the COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundMn2+ complex (2) which appeared capable of disproportionating the peroxide molecules (equation II) resulting from the COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundsuperoxide conversion.| | | 2O2˙− + 2H+ → 2H2O2 + O2 (SOD) | (I) |
| | | 2H2O2 → 2H2O + O2 (CAT) | (II) |
Table 2 Concentrations of complexes 1 and 2 equivalent to effect 1 U of bovine erythrocyte SOD activity (50% Inhibition) and the catalase mimetic potential of 1 and 2 examined as a function of H2O2 disproportionation
| Complex |
Concentration equivalent to 1 U bovine SOD (μM) |
Number of H2O2 molecules disproportionated by one molecule of complex in the first 5 min14 |
|
1
|
1.300 |
0 |
|
2
|
0.024 |
6 × 103 |
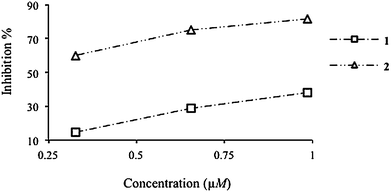 |
| | Fig. 5 SOD activity profiles for complexes 1 and 2. | |
Antitumour activity
The cytotoxicity of 1 and 2, the free COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundphen ligand and the clinical antitumour agent, COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundcisplatin, were measured at 24 and 96 h intervals in triplicate using a standard MTT assay against three progressive colorectal human-derived tumour cell lines {HT29, SW480 (Dukes' B) and SW620 (Dukes' C)} along with a non-cancerous normal human keratinocyte line (HaCaT).† The average cytotoxicity data and the associated SD obtained were then used to calculate LD50 values (±95% CI) for each cell line and time point (Table 3). Both complexes displayed remarkable cytotoxicity against all three cancerous lines. LD50 activities in the low micromolar range were found for both complexes after 24 h exposure and, significantly, these activities reached the nano- and picomolar level after 96 h. As the colorectal tumour lines progress (HT29 → SW480 → SW620) the LD50 value for 1, over a 96 h period, fell from pico- to low micromolar concentrations, while the activity of 2 remained consistently in the low- to mid-nanomolar region. It is notable that while COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundcisplatin displays significant low-micromolar cytotoxicity against all tumour lines after 96 h, the activity of 1 and 2, in the same time period is much superior (by a factor between approximately 1 × 101 – 5 × 103). Over a 96 h time period, the LD50 values of COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundcisplatin and COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundphen against the normal non-cancerous HaCaT cell line are approximately equivalent to the values obtained against cancerous lines HT29, SW480 and SW620. However, significant differences in the HaCaT cell line, were found for complexes 1 and 2, particularly when compared to the HT29 cell line. Compound 2 was found to be 9.25 times less cytotoxic toward HaCaT cells when compared to HT29, while, compound 1 was 700 times less cytotoxic when compared to the HT29 cell line. One possible way to explain the enhancement of toxicity towards cancer cells in this study may relate to the activation of the oncogene, p53, a vital tumour-suppressor gene that functions by inducing apoptosis and preventing gene amplification and which is found mutated in many forms of human cancer including HT29, SW480 and SW620 carcinomas.20 It has already been demonstrated that COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundphen and its coordinated metal adducts enhance the in vitro activity of p53 and can trigger apoptosis in p53 negative cell lines,21 however, it is not clear at present if this oncogene plays any significant role in the toxicity profiles of these COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundphen reagents.
In order to elucidate the relationship between cytotoxicity and reactive oxygen species (ROS) generation, complexes 1 and 2, along with metal-free COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundphen and COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundcisplatin, were exposed to HT29 colorectal cancer cells which had been pre-treated with the intracellular ROS indicator 2′,7′-dichlorofluorescin diacetate (DCFH-DA).† In the presence of endogenously generated ROS, DCFH-DA is oxidised to release the fluorophore 2′,7′-dichlorofluorescin (DFC). Results are shown as increases in cellular ROS levels after drug exposure and are expressed in comparison to the ROS level of unexposed controls (-Ctrl). A positive control of 0.5 μM COMPOUND LINKS
Read more about this on ChemSpider
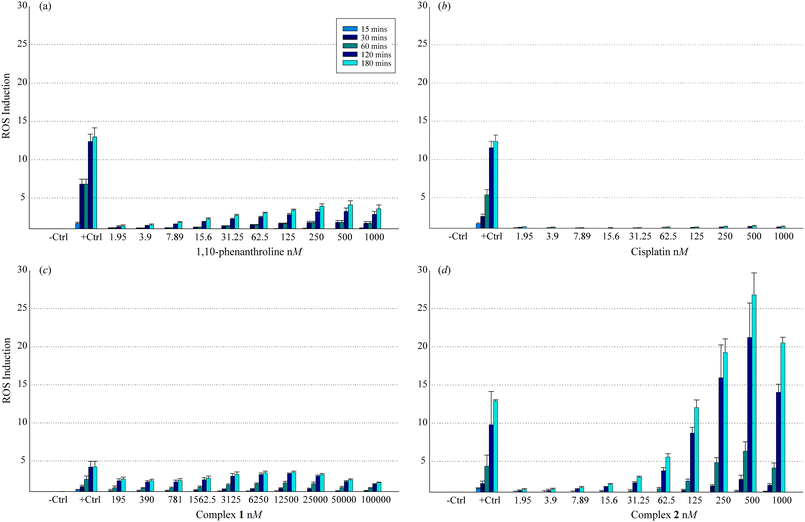
Download mol file of compoundhydrogen peroxide (+Ctrl) was utilised as it is considered a potent generator of ROS. Results were recorded at 15, 30, 60, 120 and 180 min intervals and are shown in Fig. 6. The COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundMn2+ complex 2 was found to be an exceptional ROS generator with greatest activity, relative to H2O2 (+Ctrl), across the concentration range 1000–250 nM and registering approximate equal activity to 0.5 μM COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundhydrogen peroxide at 125 nM. It is worth commenting that 2 was almost seven times more active than the next most effective ROS generator, COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundphen. The activity of the COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundCu2+ complex, (1), even when assessed across a much higher concentration range (100,000–195 nM) was considerably lower than that of metal-free COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundphen and complex 2. The clinical agent, COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundcisplatin, was the least active of all. This is not surprising considering that it is non-catalytic. Furthermore, it is known that COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundcisplatin only becomes cytotoxic upon hydrolysis to [Pt(NH3)2(OH)2], which generally occurs 48–96 h post intravenous administration.22
In vivo drug tolerance
Larvae of the insect Galleria mellonella (the greater wax moth) were employed to assess the in vivo cytotoxic tolerance of complexes 1 and 2, metal-free COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundphen and COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundcisplatin. Larvae of G. mellonella have been widely used as a convenient and inexpensive in vivo screening model to assess the therapeutic potential of novel antimicrobial drugs.23,24 They have yielded results that are considered comparable to those obtained using mammalian models.25 The innate defences of insects, including G. mellonella, like those of mammals consist of structural and passive barriers as well as humoral and cellular responses within the haemolymph (analogous to the blood of mammals).26 Indeed cellular responses within the haemolymph are often activated by signal transduction systems comparable to mice.27
Testing was carried out in triplicate using ten healthy G. mellonella larvae in the 6th developmental stage. Compounds were tested across the concentration range 5000−100 μg mL−1 (333–13 mg kg−1 average body weight) with sterile test solutions being administered via hypodermic injection. Larvae were incubated at 30 °C for 72 h with survival being monitored at 24 h intervals and significance being determined using the log rank (Mantel-Cox) method. Death was assessed by the lack of movement in response to stimulus together with discolouration. Results are presented (Table 4) as the mean % kill (± standard deviation) resulting from exposure to the tested compound. Larvae exposed to high concentrations of the compounds (5000 and 2000 μg mL−1) showed poor tolerance. However, at the lower concentration ranges (1000–200 μg mL−1) significant differences were observed. Larvae had the highest tolerance to complexes 1 and 2 (50 and 40% kill at 67 mg kg−1, respectively, and 0% kill at 33 mg kg−1). COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundCisplatin was the least well tolerated of the test compounds, with high toxicity (60% kill) being observed at 500 μg mL−1 (33 mg kg−1). This value for COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundcisplatin correlates well with the known LD50 value for this drug (32.7 mg kg−1 body weight) in the mouse model (oral exposure) but appears somewhat higher than the reported mouse intravenous LD50 value (11 mg kg−1 body weight).28 These differences could arise in mammals due to the dose-limiting toxicity of COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundcisplatin toward renal tubular damage.28
Proposed DNA self-cleaving mechanism
One possible mechanism by which the COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundCu2+ complex, 1, self-activates the phosphodiester backbone in DNA is via the formation of a π carboxylate radical which concomitantly leads to the formation of the reduced COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundCu+ species, [Cu(phen)2]+ (Scheme 1). Aliphatic carboxylate radicals are known to react by COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundhydrogen abstraction in close competition with decarboxylation.29,30 Generation of a π carboxylate radical within complex 1 would depend on the strength of the HOMO d orbital overlap on COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundCu2+ with the oxygen carboxylate. From previous X-ray crystallographic studies it is known that the Cu–O bond length in 1 is 1.974 Å and that it is significantly shorter than the equivalent Mn–O bond in 2 (2.147 Å).13,14 However, some degree of caution must be exercised when invoking this radical theory based on metal-COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundoxygen (carboxylate) bond lengths since the Cu–O bond length may alter when 1 binds to DNA.| | | Cu2+ → Cu+ (+0.15 eV) | (III) |
| | | Mn2+ → Mn0 (−1.18 eV) | (IV) |
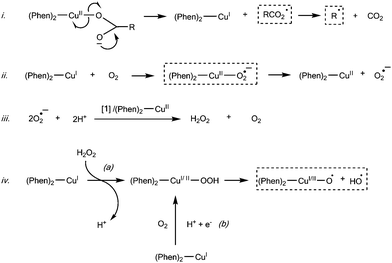 |
| | Scheme 1 Proposed mechanism of generating C–H bond activators (---). | |
Considering the standard reduction potentials for COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundCu2+ (III) and Mn2+ (IV) could explain why the COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundCu2+ complex, 1, self-activates SC DNA while the COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundMn2+ complex, 2, does not. Carboxylate anions could not be considered potent reducing agents and so metal ions that may show this effect are ions, like COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundCu2+, that have very low oxidation potentials between the Mn and Mn−1 state.
The proposed mechanism proceeds by the following steps: (i) after the intercalation of 1 to DNA (discussed in relation to Table 1), homolytic cleavage at the Cu–O bond generates the π carboxyl radical (---) and the reduced COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundCu+d10 complex [Cu(phen)2]+. The resultant carboxyl π radical (π-RCO2˙) undergoes rapid conversion to a σ radical (σ-RCO2˙) which decarboxylates to generate CO2 and R˙. (ii) [Cu(phen)2]+ reacts, as previously reported,31 with O2 to generate the COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundsuperoxide radical through an intermediate (phen)2-Cu2+-O2˙− which then decomposes to [Cu(phen)2]2+ and O2˙−. (iii) Either complex 1 or [Cu(phen)2]2+ subsequently react with O2˙− to generate H2O2 (as discussed in relation to Fig. 5 and Table 2). (iv) Since 1 does not disproportionate COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundhydrogen peroxide a subsequent reaction of [Cu(phen)2]+ with H2O2 in (a) or O2 in (b) can generate known metal-oxo and COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundhydroxyl radical bond activators ((phen)2-Cu+/2+-O2˙ and HO˙).31 Since some nuclease activity was detected for 1 in the presence of the powerful metal chelator COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundEDTA (Fig. 4(c)) it appears feasible that the carboxyl radical, and/or its breakdown product (R˙), is capable of abstracting H· from the phosphodiester backbone of DNA. In the absence of COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundoxygen, the self-cleaving nuclease potential of 1 is diminished (Fig. 4 (c)), and since, in the proposed mechanism, steps (ii)–(iv) are O2-dependent, it stands to reason that the nuclease potential of this COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundcopper complex would be less in the absence of these aerobically-generated oxo- and hydroxo- bond activators. Efforts are currently underway in our laboratory to establish direct experimental evidence for this proposed mechanism.
Conclusions
In summary, dinuclear, COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundwater-soluble [Cu2(μ2-oda)(phen)4](ClO4)2 (1) and {[Mn2(μ2-oda)(phen)4(oda)2]2−[Mn2(μ2-oda)(phen)4(H2O)2]2+} (2), in comparison to other mononuclear metal-COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundphen adducts, represent a significant advancement in the area of DNA-targeted chemotherapeutics. These COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundCu2+ and Mn2+ bis(phen) octanedioate complexes have powerful and unprecedented cytotoxicity, encouraging cytoselectivity and in vivo drug tolerance. Both complexes are avid binders of DNA, and the COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundCu2+ complex has the capacity to self-cleave DNA, possibly through the generation of a π carboxyl radical. The COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundMn2+ complex is an exceptional redox catalyst that generates remarkable levels of intracellular ROS within HT29 colorectal cancer cells compared to COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundhydrogen peroxide.
Acknowledgements
This paper is dedicated to the memory of Prof. Philip S. Skell (1918–2010), member, United States National Academy of Sciences. DDM would like to thank PSS for “teaching me that the best scientists have a broad base of knowledge in their field and open and curious minds in their exploration of nature's secrets”.
The authors wish to acknowledge financial support from the Dublin Institute of Technology Capacity Building Scheme for Strategic Research progamme (CaBS). This work has been carried out (in part) within the structures of the Focas Research Institute, DIT, funded under The Irish National Development Plan with assistance from the European Regional Development Fund.
Notes and references
- J. Stubbe and J. W. Kozarich, Chem. Rev., 1987, 87, 1107–1136 CrossRef CAS.
- F. Mancin, P. Scrimin, P. Tecilla and U. Tonellato, Chem. Commun., 2005, 2540–2548 RSC.
- J. A. Cowan, Curr. Opin. Chem. Biol., 2001, 5, 634–642 CrossRef.
- S. Borah, M. S. Melvin, N. Lindquist and R. A. Manderville, J. Am. Chem. Soc., 1998, 120, 4557–4562 CrossRef CAS.
- M. S. Melvin, J. T. Tomlinson, G. R. Saluta, G. L. Kucera, N. Lindquist and R. A. Manderville, J. Am. Chem. Soc., 2000, 122, 6333–6334 CrossRef CAS.
- P. U. Maheswari, S. Roy, H. den Dulk, S. Barends, G. van Wezel, B. Kozlevcar, P. Gamez and J. Reedijk, J. Am. Chem. Soc., 2006, 128, 710–711 CrossRef CAS.
- D. S. Sigman, D. R. Graham, V. D'Aurora and A. M. Stern, J. Biol. Chem., 1979, 254, 12269–12272 CAS.
- B. R. James and R. J. P. Williams, J. Chem. Soc., 1961, 2007–2019 RSC.
- M. Pitie, B. Donnadieu and B. Meunier, Inorg. Chem., 1998, 37, 3486–3489 CrossRef CAS.
- M. Pitie and B. Meunier, Bioconjugate Chem., 1998, 9, 604–611 CrossRef CAS.
- M. Pitié, B. Sudres and B. Meunier, Chem. Commun., 1998, 2597–2598 RSC.
- A. Kellett, M. O'Connor, M. McCann, M. McNamara, P. Lynch, G. Rosair, V. McKee, B. Creaven, M. Walsh, S. McClean, A. Foltyn, D. O'Shea, O. Howe and M. Devereux, Dalton Trans., 2011, 40, 1024–1027 RSC.
- M. Devereux, M. McCann, J. F. Cronin, G. Ferguson and V. McKee, Polyhedron, 1999, 18, 2141–2148 Search PubMed.
- M. T. Casey, M. McCann, M. Devereux, M. Curran, C. Cardin, M. Convery, V. Quillet and C. Harding, J. Chem. Soc., Chem. Commun., 1994, 2643–2645 RSC.
- A. W. McConnaughie and T. C. Jenkins, J. Med. Chem., 1995, 38, 3488–3501 CrossRef.
- C. Bailly, N. Pommery, R. Houssin and J. P. Henichart, J. Pharm. Sci., 1989, 78, 910–917 CrossRef CAS.
- L. E. Marshall, D. R. Graham, K. A. Reich and D. S. Sigman, Biochemistry, 1981, 20, 244–250 CrossRef CAS.
- T. B. Thederahn, M. D. Kuwabara, T. A. Larsen and D. S. Sigman, J. Am. Chem. Soc., 1989, 111, 4941–4946 CrossRef CAS.
- B. C. Bales, T. Kodama, Y. N. Weledji, M. Pitie, B. Meunier and M. M. Greenberg, Nucleic Acids Res., 2005, 33, 5371–5379 CrossRef CAS.
- N. R. Rodrigues, A. Rowan, M. E. Smith, I. B. Kerr, W. F. Bodmer, J. V. Gannon and D. P. Lane, Proc. Natl. Acad. Sci. U. S. A., 1990, 87, 7555–7559 CrossRef CAS.
- Y. Sun, J. Bian, Y. Wang and C. Jacobs, Oncogene, 1997, 14, 385–393 CrossRef CAS.
-
N. Farrell and S. Spinelli, Uses of Inorganic Chemistry in Medicine, Royal Society of Chemistry, 1999 Search PubMed.
- M. Brennan, D. Y. Thomas, M. Whiteway and K. Kavanagh, FEMS Immunol. Med. Microbiol., 2002, 34, 153–157 Search PubMed.
- R. Rowan, C. Moran, M. McCann and K. Kavanagh, BioMetals, 2009, 22, 461–467 Search PubMed.
- K. Kavanagh and E. P. Reeves, FEMS Microbiol. Rev., 2004, 28, 101–112 Search PubMed.
-
G. B. Dunphy and G. S. Thurston, Insect Immunity, CRC Press, Boca Raton, FL., 1990 Search PubMed.
- A. M. Fallon and D. Sun, Insect Biochem. Mol. Biol., 2001, 31, 263–278 Search PubMed.
- B. Rosenberg, Naturwissenschaften, 1973, 60, 399–406 Search PubMed.
- P. S. Skell and D. D. May, J. Am. Chem. Soc., 1981, 103, 967–968 Search PubMed.
- P. S. Skell and D. D. May, J. Am. Chem. Soc., 1983, 105, 3999–4008 Search PubMed.
- M. Pitie and G. Pratviel, Chem. Rev., 2010, 110, 1018–1059 CrossRef CAS.
Footnotes |
| † Electronic supplementary information (ESI) available: Experimental procedures and biological evaluation studies. See DOI: 10.1039/c0md00266f |
| ‡ Current address: School of Chemical Sciences, Dublin City University, Glasnevin, Dublin 9, Ireland. |
|
| This journal is © The Royal Society of Chemistry 2011 |
Click here to see how this site uses Cookies. View our privacy policy here. 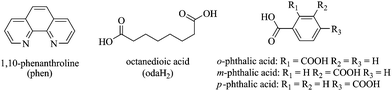
![Molecular structure of the [Cu2(μ2-oda)(phen)4]2+ cation in complex 1.](/image/article/2011/MD/c0md00266f/c0md00266f-f2.gif)
![Molecular structures of the dimeric cation and anion subunits in the MnII double salt complex {[Mn2(μ2-oda)(phen)4(oda)2]2−[Mn2(μ2-oda)(phen)4(H2O)2]2+} (2).](/image/article/2011/MD/c0md00266f/c0md00266f-f3.gif)