Erythromycin B: conformational analysis and antibacterial activity†
Paul
Tyson
a,
Abdolreza
Hassanzadeh‡
a,
Mohd. Nizam
Mordi§
a,
David G.
Allison
a,
Viter
Marquez¶
b and
Jill
Barber
*a
aSchool of Pharmacy and Pharmaceutical Sciences, University of Manchester, Stopford Building, Oxford Road, Manchester, UK M13 9PT. E-mail: jill.barber@manchester.ac.uk; Fax: +44-161-275-2396; Tel: +44-161-275-2369
bMax-Planck-Institut-für Molekulare Genetik, Ihnestr. 73, 14195, Berlin, Germany
First published on 3rd March 2011
Abstract
Erythromycin B, an acid-stable co-metabolite of the important antibiotic erythromycin A, differs from erythromycin A only in the absence of a hydroxyl group at C12, yet it has never been licensed for clinical use. We describe an NMR-based analysis of the conformation of erythromycin B, both free in aqueous solution and when weakly bound to bacterial ribosomes and show that it is conformationally similar to erythromycin A. The antibacterial activity of erythromycin B is shown to be similar to that of erythromycin A, but after acid-treatment, resembling exposure to the stomach, erythromycin B substantially retains antibacterial activity, whereas erythromycin A does not.
Introduction
Erythromycin B (1) is a biosynthetic co-metabolite of the clinically important macrolide antibiotic erythromycin A (2).1 The drug was first isolated from fermentation broths of Saccharopolyspora erythraea in 1954,2 and its structure was proposed in 1957.3 Like erythromycin A and its derivatives, the drug is basic, relatively insoluble in water but soluble in organic solvents.In common with other macrolide antibiotics, erythromycin B shows a broad range of activity against Gram-positive bacteria and limited activity against Gram-negative bacteria. It exerts its antibacterial activity by binding to the 50S subunit of the bacterial ribosome, thereby inhibiting microbial protein synthesis.4,5 Against a range of both Gram-negative and Gram-positive bacteria erythromycin B is reported to have comparable6 or slightly lower7 potency than erythromycin A.
Erythromycin A is more abundant and therefore easier to isolate than erythromycin B. Presumably because of the success of erythromycin A, erythromycin B has been largely neglected by the pharmaceutical industry. Recently, however, we have highlighted a number of advantages of erythromycin B over erythromycin A. Firstly, it is much more stable to acid, because it is unable to cyclize in a 12,9-direction; erythromycin A undergoes facile formation of the spiro-6,9:12,9-cyclized anhydroerythromycin A, which has little or no antibacterial activity.7 The 2′-esters of erythromycin B are also much more stable to acid than those of erythromycin A. This finding is potentially important in paediatric medicine, in which 2′-esters serve as pro-drugs. Finally, we have been able to derivatize erythromycin B to give pro-pro-drugs, for example erythromycin B enol ether ethyl succinate (3), which are almost insoluble in the medicine bottle and therefore unable to hydrolyse to yield the vile-tasting erythromycin free base. Compound 3 is activated in two stages, as shown in Scheme 1, to yield erythromycin B.8
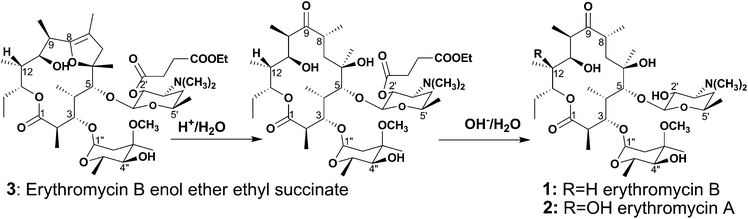 | ||
| Scheme 1 Activation of erythromycin B enol ether ethyl succinate to erythromycin B. | ||
These findings suggest that erythromycin B has potential as an independent entity in paediatric medicine, and prompted us to return to the fundamental chemistry of this drug, in particular to determine its conformation in free aqueous solution and when weakly bound to bacterial ribosomes.
It has previously been postulated that erythromycin A and some of its derivatives bind to the ribosome of Escherichia coli in a two stage process. A strong binding interaction has been detected by equilibrium dialysis and related techniques,9–11 and more recently characterized in detail by X-ray crystallography.12 A weak interaction has been characterized as a fast exchange process by NMR spectroscopy.13,14 Unlike the strong interaction, the weak interaction appears to be magnesium independent.15 It has also been shown that for erythromycin A, the E. coliribosome tautomerically recognizes and preferentially binds to the 9-ketone form of the drug.14
We now report the full assignments of the 1H and 13C NMR spectra of erythromycin B in organic and aqueous solutions. These assignments have allowed us to determine the conformational mix of the molecule in free aqueous solution and when bound (weakly) to the bacterial ribosome.
Results
Assignments of the 1H and 13C NMR spectra in aqueous solution
One dimensional 1H, DQF-COSY and tnTOCSY spectra were obtained using a 4 mM solution of erythromycin B in phosphate-buffered D2O. A saturated solution (∼20 mM) was used to obtain gradient HMBC and HMQC spectra.The one-dimensional 1H spectrum (Fig. 1 Supplementary Data†) showed immediately that, in aqueous solution, erythromycin B exists overwhelmingly as a single isomer. Both erythromycins A and C (a biosynthetic precursor of erythromycin A in which C8′′ bears OH instead of OCH3) form mixtures of 9-ketone and 12,9-hemiacetal in aqueous solution, and show characteristic doubling of each signal in the 1H NMR spectrum. The lack of the 12-OH prevents this in erythromycin B; the formation of the 6,9-hemiacetal is perfectly possible, but the presence of a single set of signals in the 1H NMR spectrum indicate that it is unfavoured.
The full assignments of the 1H and 13C spectra of erythromycin B and a description of the assignment process are given in the Supplementary Data.†
The free solution conformation of erythromycin B: The ROESY spectrum
Crystal structures of macrolide antibiotics have revealed a tendency for the molecules to adopt one of two distinct conformations, and several solution studies have led to similar conclusions. Conformational analyses of erythromycin A 9-ketone in CDCl316 and aqueous solutions17 suggest that, in both cases, the drug adopts predominantly the folded-out conformation, which is characterized by the close approach of H4–H11, H5–H318, H8–H318 and H315–H316. The folded-in conformation has been identified from the crystal structure of dirithromycin18 and is characterized by the close approach of H3–H8, H3–H11, H4–H318 and H316–H317. More recently, a third, intermediate conformation has been identified from analysis of the solution structure of erythromycin A 9-ketone. This conformation retains many of the same characteristics as the folded-out conformer, but is distinct owing to the close approach of H8–H11 so that the orientation of H8 is endo rather than exo to the macrolide ring, hence we term this the 8-endo-folded-out erythromycin 9-ketone.17Many previous studies of the conformations of macrolide antibiotics have made use of the 1H nuclear Overhauser effect (NOE) NMR experiment as the main means of obtaining conformational data.19–21 However, this method can be problematic when studying molecules of an intermediate size in aqueous solutions. At operating frequencies of 500–600 MHz, NOEs for erythromycins are close to zero and either slightly positive or negative. These effects make even semi-quantitative analysis of NOESY spectra unreliable when interpreting cross-peaks in terms of internuclear distances. We therefore chose to use the rotating frame Overhauser effect experiment,22 in which all the Overhauser signals are positive. When the spin-lock field and mixing time are adjusted accurately, artefacts are minimized and good data are obtained.
The ROESY spectrum of erythromycin B in phosphate buffered D2O (mixing time 250 ms) is shown in Fig. 1A. The cross-peaks were designated very small, small, medium or large and the corresponding connectivity table is shown in Table 2 (top) of the Supplementary Data.† Known internuclear distances (e.g. H2“proR-H2”proS, H4–H317) were used to predict internuclear distances for very small (3.5 ± 1.5 Å), small (2.5 ± 1.0 Å), medium (2.3 ± 0.5 Å) and large (2.0 ± 0.4 Å) cross-peaks. Medium H4–H11, H5–H318 and H8–H318 cross-peaks suggest that the folded-out conformation is highly represented in the solution, and H3–H8, H3–H11 and H316–H317 signals (indicative of the folded-in conformation) are absent. A small H8–H11 cross-peak suggests that the 8-endo-confimation might be represented. Curiously, no H315–H316 cross-peak was seen, although this had been found in folded-out clarithromycin and erythromycin A.19
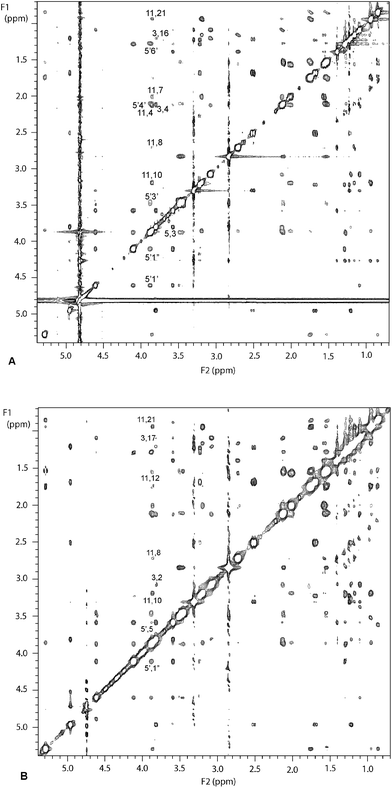 | ||
| Fig. 1 500 MHz NMR spectra of 4 mM erythromycin B in aqueous solution; A: ROESY spectrumB: TRNOESY spectrum in the presence of 0.48 μM E. coliribosomes. | ||
The experiment was repeated with a mixing time of 500 ms, with essentially identical results.
The NMR data therefore strongly suggest that erythromycin B exists exclusively or almost exclusively in the folded-out conformation in aqueous solution. Conformational flexibility leads to small amounts of the 8-endo-folded-out conformer, but we see no direct evidence for the presence of the folded-in conformer.
Unconstrained and constrained molecular modelling
An initial erythromycin B structure was constructed using Macromodel version 8.0 and erythromycin A23 as the starting point. The resulting structure was then submitted to an unconstrained Monte Carlo conformational search in water, using the AMBER force field, which is recommended for calculations in water. The global minimum was a conventional folded-out conformer with the sugars in the up-up orientation and represented a very close fit to the ROESY spectrum.Conformational searching is, at its best, satisfyingly rigorous, with all conformational space being explored. Macrolides are, however, rather large for this type of search and a 10,000 step calculation does not necessarily achieve an endpoint of a global minimum found 20 times. This calculation found no structure in which H8 approached H11, despite the presence of an H8–H11 crosspeak in the ROESY spectrum.
We therefore constrained the H8–H11 distance to 2.5 ± 0.5 Å and recalculated. The global minimum obtained actually had slightly lower energy than the folded-out global minimum (144 vs. 149 kJ mol−1), but was clearly a folded-in structure. This structure has an H3–H8 distance of 2.3 Å and an H4–H18 distance of 2.2 Å, but neither pair of hydrogens gives rise to a crosspeak in the ROESY spectrum.
A third calculation involved constraining both H8–H11 and H4–H11 to 2.5 ± 0.5 Å. This yielded the expected 8-endo-folded-out conformer with an energy of 158 kJmol−1, 9 kJ mol−1 above that of the folded-out conformer and consistent with the presence of this conformer at low abundance in aqueous solution.
The three global minima are shown in Fig. 2 and the full details of these structures are given in the Supplementary Data.†
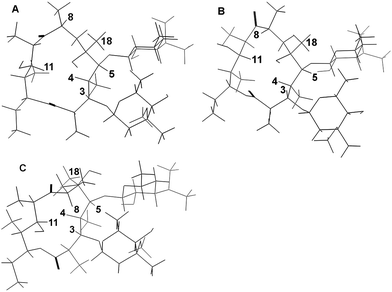 | ||
| Fig. 2 Results of conformational searches on erythromycin B A: folded-out conformer B 8-endo-folded-out conformer C folded-in conformer. | ||
The conformation of erythromycin B bound to bacterial ribosomes
Erythromycin A and some of its derivatives take part in a weak interaction with bacterial ribosomes, which may be detected by NMR. We now wished to determine whether erythromycin B could also take part in such an interaction and, if so, to determine the conformation of the bound drug.The conformation of the ligand bound in a fast exchange process to bacterial ribosomes may be determined using the transferred NOESY (TRNOESY) experiment. In this experiment the ligand is present in a large excess over ribosomes, and equilibrium exists between free and bound ligand. There is a very rapid build-up of NOEs in the bound state and this NOE information is transferred to the free state when the ligand molecule dissociates from the ribosome. Because NOE build-up in the free state is very slow, or even absent, it is relatively easy to choose a mixing time such that all the observable information pertains to the bound ligand. The resulting NOESY spectrum has chemical shifts due to the free drug, but NOE information due to the bound drug. The method is prone to spin-diffusion artefacts but these can be almost eliminated by the use of fully deuteriated ribosomes and an appropriate mixing time.
The transferred NOESY spectrum of erythromycin B (4 mM) in the presence of E. coli deuteriated ribosomes (0.48 μM – sufficient to cause an approximate doubling of the low frequency H315 linewidth) is shown in Fig. 1B. The mixing time was optimized empirically at 75 ms. Table 2 (bottom) of the Supplementary Data shows the connectivity table derived from these data. Signals were designated very small, small, medium or large on the basis of the size of the cross-peak, the abundance of information making a semi-quantitative analysis possible. Known internuclear distances were again used to predict the internuclear distances for very small (3.5 ± 1.5 Å), small (2.5 ± 1.0 Å) medium (2.3 ± 0.5 Å) and large (2.0 ± 0.4 Å) cross peaks.
More cross peaks were seen in the TRNOESY than in the ROESY spectrum of the free drug. This reflects the relative sensitivities of the two techniques. The maximum TRNOE has a relative intensity of −1, whereas the ROE has +½. The NOE build-up is very much faster in the TRNOESY than in the ROESY experiment, because of the very large size of the drug-ribosome complex. Most of the extra peaks in the TRNOESY spectrum were consistent with the structure of the free drug, with internuclear distances of 3–4 Å.
In the TRNOESY spectrum the H11–H8 signal is stronger than in the ROESY spectrum and signals corresponding to H3–H8 and H4–H18 are still absent, suggesting strongly that the 8-endo-folded-out conformer is present at low abundance and the folded-in conformer undetectable.
Three control experiments were carried out in order to control for non-specific binding to large molecules in the solution. Erythromycin B (4 mM) was incubated in the presence of 0.96 μM ribosomes and treated with Rnase-1-AS for four hours at 30 °C. The resulting TRNOESY spectrum (not shown) is essentially blank, containing only the cross-peaks seen in the NOESY spectrum of erythromycin B with the same mixing time.
To control for non-specific binding we have generally prepared ribosomal “core” particles from 50S ribosomal subunits. The subunits are treated with 2 M lithium chloride solution, as previously described,24 to remove 12 outer proteins, including L15.25Protein L15 has been shown, remarkably, to bind to erythromycin A in isolation, in the absence of other ribosomal components.26 It is also essential for the binding of erythromycin to reconstituted ribosomes.26 Fig. 3 in the Supplementary Data shows the NOESY spectrum obtained when erythromycin B (4 mM) was incubated with 0.96 μM LiCl treated 50S cores (a doubling of the molar amount of ribosomes used above, to compensate for the reduction in molecular weight). This spectrum again shows very few cross-peaks, illustrating clearly that the binding to whole ribosomes (or to 50S subunits13) that we observe in the TRNOESY experiment is specific, and not the result of random interactions with large macromolecular complexes.
The preparation of ribosomal core particles is a demanding and time-consuming procedure, requiring density gradient ultracentrifugation. We sought to simplify our control studies by treating whole ribosomes with lithium chloride. Surprisingly, this was unsuccessful. Whereas a single treatment with 1.3 M or 2 M LiCl was sufficient to prevent 50S subunits from binding in the NMR experiment, we typically required four treatments to prevent binding to 70S particles. Excellent TRNOESY spectra were obtained when 70S ribosomes were treated with lithium chloride only once or twice. 30S ribosomal subunits do not bind to macrolide antibiotics in the NMR experiment, whilst 50S subunits do.13 It seems therefore that the 30S subunits protect the 50S subunits from loss of the proteins involved in weak binding.
Finally, we used 30S ribosomal subunits in place of whole ribosomes. Macrolide antibiotics are known to bind to the 50S subunit of the ribosome, and binding to the 30S subunit was undetectable.
The antibacterial activity of erythromycin B
The NMR results, stability data obtained by us and others7,27 and our previous pro-drug results,8 suggest strongly that erythromycin B has potential has an independent therapeutic entity. Because of the uncertainty over erythromycinin vitro antibacterial activity,6,7 we revisited this, and included a protocol of acid exposure to mimic the action of the stomach. Minimum inhibitory concentrations of erythromycins A and B and clarithromycin were measured (using a serial dilution method) against four important opportunistic pathogens, namely the Gram-positive organisms Staphylococcus aureus ATCC 601065 and Streptococcus pyogenes, and Haemophilus influenzae ATCC 12699 and Burkholderia cepacia (Gram-negative). S. pyogenes and B. cepacia were lab isolates. The measurements were repeated using drug that had been pre-treated with 0.1 M hydrochloric acid for 40 min at 37 °C, then neutralized to pH 6.8 with sodium hydroxide solution. The results are shown in Table 1 None of the macrolides was effective against B. cepacia, an important pathogen in cystic fibrosis with known resistance to many types of antimicrobial agent (data not shown). Clarithromycin was slightly more effective than the two erythromycins in either the untreated or acid-treated form. Untreated erythromycin B showed comparable activity to erythromycin A. However, following acid treatment, erythromycin B retained a large proportion of its activity, the greatest loss being 50% of activity against S. aureus (1 doubling dilution), whereas erythromycin A showed a significant loss of activiity (ca. 87%) against both S. aureus and S. pyogenes and 50% against H. influenzae.| Minimum Inhibitory Concentrations (μg mL−1) | ||||||
|---|---|---|---|---|---|---|
| S. aureus | S. pyogenes | H. influenzae | ||||
| U | P | U | P | U | P | |
| Ery A | 0.9 | >8 | 0.2 | 1 | 2.2 | >4 |
| Ery B | 0.7 | 1.4 | 0.2 | 0.3 | 2.3 | 3.6 |
| Clari | 0.6 | 0.9 | 0.2 | 0.3 | 0.6 | 1.8 |
Discussion
It is clear from these data that the conformation adopted by erythromycin B when weakly bound to the bacterial ribosome is essentially the same as that adopted when the drug is free in aqueous solution. The folded-out conformation with “up-up” sugars described by Everett and Tyler is preferred in aqueous solution and by the bound drug. Both in the free and the bound state, the drug retains flexibility in the C6–C9 region, leading to small amounts of 8-endo-folded-out conformer, but not to the folded-in conformer. X-ray crystallographic studies by Steitz and co-workers12 have shown that even in the crystal erythromycin A retains this folded-out conformation.The nature of the NMR-detected binding remains unclear. Recent kinetic analysis suggests that erythromycin (erythromycin A in their study) binds to bacterial ribosomes in a two-stage process28 suggesting that the first stage of this binding is detected by NMR. This suggestion does, however, require that a single ribosome can bind two molecules of erythromycin simultaneously, one detectable by crystallography, the other by NMR. We are inclined to the view that the transferred NOESY experiment detects binding weakened by partial denaturation of the ribosome. This suggestion is (to a degree) supported by the observation that the weak binding cannot readily be eliminated by LiCl treatment of whole ribosomes.
Erythromycin B remains the Cinderella of the macrolide family, unlicenced for human or veterinary use. Yet, unlike erythromycin A, it is acid-stable. Unlike clarithromycin, it can be readily esterified at the 2′-position to give pro-drugs with acceptable solubility for paediatric use. We have described some simple chemistry that allows the preparation of taste-free pro-pro-drugs of erythromycin B.8 No other macrolide available at this time supports this chemistry. We and others have shown that erythromycin B has similar in vitro antibacterial properties to erythromycin A and clarithromycin.6,7 In the present paper we have shown that erythromycin B is conformationally similar to erythromycin A both in aqueous solution and when bound to bacterial ribosomes, and that the drug retains antibacterial activity after acid treatment mimicking the stomach.
Experimental
Erythromycin A, erythromycin B and clarithromycin in their free-base form were gifts from Dr Warren Mann (Abbot Laboratories, UK), and were used without further purification. 1H NMR spectra were acquired using a Varian Unity 500 spectrometer. 13C NMR spectra were acquired using a Jeol EX 270 spectrometer operating at 67.8 MHz. Spectra were referenced to TMS (tetramethylsilane) or TSP (3-(trimethylsilyl)-2,2,3,3,-tetradeuterio-propionic acid sodium salt) at δ 0.0.The Supplementary Data describes the protocols for the assignment of signals
ROESY spectra of erythromycin B in phosphate buffered D2O
Solutions of 8–10 mM erythromycin B in 50 mM phosphate buffered D20, apparent pH 7.4, were routinely used to acquire ROESY spectra. Mixing times of 250 ms and 150 ms were used with data matrices of 2048 x1024 complex points and the spectra were processed using a Gaussian window function. Essentially no differences in the observed signals were observed with the variations in mixing time and temperature.Molecular modelling of NMR spectral data
Conformational analysis of the free solution structure of erythromycin B in phosphate buffered D2O was performed using Macromodel version 8 software.29 The crystal structure for [9-(0–2,5,-dioxahexyl) oxime] erythromycin A hydrate23 was obtained from the Cambridge Crystallographic database and this was used to derive the starting structure for erythromycin B manually, the oxime giving a folded-out conformation. This structure was minimized using the Truncated Newtonian Conjugate Gradient method (TNCG) in order to obtain local minima.Monte Carlo searches using AMBER and MM2 force fields were then carried out to obtain global minimum structures, with water solvation. The cross peaks from the ROESY spectra were designated as very small (3.5 ± 1.5 Å), small (2.5 ± 1.0 Å), medium (2.3 ± 0.5 Å) or large (2.0 ± 0.4 Å), the magnitude of the constraints being determined empirically by referring to known interatomic distances within the molecule. The distance constraints from each ROESY and TRNOESY spectra were then compared with both global minimum erythromycin B conformers. Where the global minima did not satisfy all the NMR data, attention were turned to the higher energy structures and if no structure satisfying all the NMR data was found a constrained local minimization (or a restrained Monte Carlo search where appropriate) was performed to obtain conformers satisfying constraints which were not satisfied with the global minima.
TRNOESY spectra of erythromycin B bound to bacterial ribosomes in phosphate buffered D2O
Solutions of 4 mM erythromycin B in 50 mM sodium phosphate buffered D2O, apparent pH 7.4, were used throughout. All spectra were run at 30 °C. Mixing times were optimized empirically at 75 or 50 ms. Spectra were acquired with data matrices of 2048 x1024 complex points and were processed with Gaussian window functions.Control experiments
The control TRNOESY experiments were conducted with the same experimental parameters as for the 75 ms TRNOESY experiment, and were processed in the same manner.Preparation of whole 70S ribosomes, 30S ribosomal subunits and 50S subunit core particles
Whole 70S ribosomes were prepared as previously described25 from cultures of E.coliMRE 600, and were stored at −20 °C for no longer than 2 weeks in 50 mM phosphate buffered D2O. Fully deuteriated ribosomes were prepared from cultures of E. coliMRE 600D30 grown on a minimal medium based upon deuteriated sodium succinate and D2O.The 50S ribosomal subunits were prepared as described,24 and were used to prepare ribosomal core particles by treatment with 2 M LiCl solution.24 The core particles were stored in phosphate buffered D2O at −80 °C for no longer than 2 weeks, and were defrosted no more than once.
RNAase treatment of ribosomal solutions
A solution of 4 mM erythromycin B in 50 mM deuteriated phosphate buffer was prepared and 750 μl was transferred to a 5 mm NMR tube. To this, 32.6 μl of a 32 μM solution of ribosomes, pre-treated with 2 M lithium chloride,24 was added. RNase I-AS was added to a final concentration of 1 mg ml−1 and the solution incubated at 37 °C for 4 h prior to transferred NOESY experiments.Broth microdilution minimum inhibitory concentrations
These were estimated according to the National Committee for Clinical Laboratory Standards guidelines (2000).31 The protocol is described in full in the Supplementary Data.†Conclusions
Erythromycin B adopts a folded-out conformation both in aqueous solution and when weakly bound to bacterial ribosomes. Like erythromycin A, it retains some flexibility in the C7–C9 part of the molecule leading to a low abundance of the 8-endo-folded-out conformer. We speculate, based on the data presented here, that the weak NMR-detected binding of macrolide drugs to ribosomes arises from partial denaturation of the macrolide binding site on the ribosome, rather than from a preliminary binding. We present further evidence that erythromycin B has potential as an independent therapeutic compound; in particular, it retains antibacterial activity after treatment with acid, mimicking the stomach.Acknowledgements
We thank Dr Warren Mann (Abbott Laboratories, UK) for gifts of erythromycins A and B and clarithromycin. M.N.M. and A.H. were recipients of a Universiti Sains Malaysia fellowship and a Ministry of Health and Medical Education of Iran fellowship respectively.Notes and references
- J. M. McGuire, R. L. Bunch, R. C. Anderson, H. E. Boaz, E. H. Flynn, H. M. Powell and J. W. Smith, Antibiot. Chemother., 1952, 2, 281 Search PubMed.
- C. W. Pettinga, W. M. Stark and F. R. Van Abeele, J. Am. Chem. Soc., 1954, 76, 569 CrossRef CAS.
- P. F. Wiley, M. V. Sigal Jr., O. Weaver, R. Monahan and K. J. Gerzon, Erythromycin XI. Structure of erythromycin B, J. Am. Chem. Soc., 1957, 79, 6070 CrossRef CAS.
- J. C.-H. Mao and R. G. Wiegand, Biochem. Biophys. Acta, 1968, 157, 404 CAS.
- J. M. Wilhelm and J. W. Corcoran, Biochemistry, 1967, 6, 2578 CrossRef CAS.
- J. M. Wilhelm, N. L. Oleinick and J. W. Corcoran, Antmicrobi. Agents. Chemother., 1967, 10, 236 Search PubMed.
- I. O. Kibwage, J. Hoogmartens, E. Roets, H. Vanderhaeghe, L. Verbist, M. Dubost, C. Pascal, P. Petitjean and G. Levol, Antimicrob. Agent Chemother., 1985, 28, 630 CAS.
- P. K. Bhadra, G. A. Morris and J. Barber, J. Med. Chem., 2005, 48, 3878 CrossRef CAS.
- R. Fernandez-Munoz and D. Vasquez, J. Antibiot., 1973, 26, 107 CAS.
- S. Pestka, Antimicrob. Agents Chemother., 1974, 6, 474 CAS.
- R. C. Goldman, S. W. Fesik and C. C. Doran, Antimicrob. Agents Chemother., 1990, 34, 426 CAS.
- D. Tu, G. Blaha, P. B. Moore and T. A. Steitz, Cell, 2005, 121, 257 CrossRef CAS.
- J. Barber, J. I. Gyi and D. A. Pye, J. Chem. Soc., Chem. Commun., 1991, 1249 RSC.
- D. A. Pye, J. I. Gyi and J. Barber, J. Chem. Soc., Chem. Commun., 1990, 1143 RSC.
- G. Bertho, J. Gharbi-Benarous, M. Delaforge and J. P. Girault, Bioorg. Med. Chem., 1998, 6, 209 CrossRef CAS.
- J. R. Everett and J. W. Tyler, J. Chem. Soc., Perkin Trans. 2, 1987, 1659 RSC.
- A. Awan, R. J. Brennan, A. C. Regan and J. Barber, J. Chem. Soc., Perkin Trans. 2, 2000, 1645 RSC.
- G. A. Stephenson, J. G. Stowell, P. H. Toma, D. E. Dorman, J. R. Greene and S. R. Byrn, J. Am. Chem. Soc., 1994, 116, 5766 CrossRef CAS.
- J. R. Everett and J. W. Tyler, J. Chem. Soc., Perkin Trans. 2, 1988, 325–327 RSC.
- J. R. Everett, I. K. Hatton, E. Hunt and J. W. Tyler, J. Chem. Soc., Perkin Trans. 2, 1989, 1719 RSC.
- W. E. Steinmetz, R. Bersch, J. Townson and D. Pesiri, J. Med. Chem., 1992, 35, 4842 CrossRef CAS.
- A. Bax and D. G. Davis, J. Magn. Reson, 1985, 63, 207 CAS.
- B. Bachet, C. Brassy and J. Mornon, Acta. Crystallog, 1988, C44, 112 CAS.
- K. H. Nierhaus, Reconstitution of ribosomes. In Ribosomes and protein synthesis: a practical approach; Spedding, G., Ed.; Oxford University Press, Oxford, 1990, pp 165–168 Search PubMed.
- H. E. Homann and K. H. Nierhaus, Eur. J. Biochem., 1971, 20, 249 CAS.
- H. Teraoka and K. H. Nierhaus, J. Mol. Biol., 1978, 126, 185 CAS.
- M. N. Mordi, M. D. Pelta, V. Boote, G. A. Morris and J. Barber, J. Med. Chem., 2000, 43, 467 CrossRef CAS.
- A. D. Petropoulos, E. C. Kouvela, G. P. Dinos and Dimitrios L. Kalpaxis, J. Biol. Chem., 2008, 283, 4756 CAS.
- F. Mohamadi, N. G. J. Richards, W. C. Guida, R. Liskamp, M. Lipton, C. Caufield, G. Chang, T. Hendrickson and W. C. Still, J. Comput. Chem., 1990, 11, 440 CrossRef CAS.
- O. Paliy, D. Bloor, D. Brockwell, P. Gilbert and J. Barber, J. Appl. Microbiol., 2003, 94, 580 CrossRef CAS.
- National Committee for Clinical Laboratory Standards. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically – 5th Edition. Approved Standard M7-A5, 2000, NCCLS, Wayne, PA, USA Search PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: The full assignments of the 1H and 13C spectra of erythromycin B and a description of the assignment process. 500 MHz NMR spectra of 4 mM erythromycin B in aqueous solution: ROESY spectrum connectivity table. Transferred NOESY spectrum of erythromycin B (4 mM) in the presence of E. coli deuteriated ribosomes (0.48 μM) mixing time 75 ms, connectivity table. Folded-out, 8-endo-folded-out and folded-in conformers of erythromycin B: full details. NOESY spectrum obtained when erythromycin B (4 mM) was incubated with 0.96 μM LiCl treated 50S ribosomal core particles. Broth microdilution Minimum Inhibitory Concentrations: Protocol. See DOI: 10.1039/c0md00251h |
| ‡ Present address: Pharmaceutics Research Centre, School of Pharmacy, Kerman University of Medical Sciences, Kerman, Iran. |
| § Present address: Centre for Drug Research, Universiti Sains Malaysia, 11800, Penang, Malaysia. |
| ¶ Present address: Ludwig-Maximilians-Universität München, Feodor-Lynen-Strasse 25, D-81377 München, Germany. |
| This journal is © The Royal Society of Chemistry 2011 |
