DOI:
10.1039/C0MD00208A
(Concise Article)
Med. Chem. Commun., 2011,
2, 126-131
A class of novel AA-Trp-Trp-OBzl: synthesis, in vitro anti-proliferation, in vivo anti-tumor action, and intercalation mechanism†
Received
8th November 2010
, Accepted 25th November 2010
First published on 17th December 2010
Abstract
From the anti-tumoral active COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundN-tryptophanyl-β-carboline-3-carboxylic acid benzyl ester and β-carboline-3-carbonyltryptophan benzyl ester, an anti-tumoral pharmacophore, Trp-Trp-OBzl, was drawn. Into the N-terminus of Trp-Trp-OBzl, L-amino acids were introduced and twenty AA-Trp-Trp-OBzls were provided. The automated docking studies showed AA-Trp-Trp-OBzls to be desirable intercalators. The in vitro and in vivo assays explored that thirteen of twenty AA-Trp-Trp-OBzls were anti-tumoral active, and nine of twenty AA-Trp-Trp-OBzls were more active than COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundcytarabine. The acute toxicity, spleen index and increased body weight demonstrated that AA-Trp-Trp-OBzls did not damage the immunologic function of the treated mice and had a LD50 value of more than 500 mg kg−1. DNA intercalation was considered the action mechanism of Asn-Trp-Trp-OBzl.
1. Introduction
DNA recognition is critical for several anti-tumor agents including groove binders, alkylating compounds and intercalating agents to offer their anti-tumor activity. DNA intercalation is known to be a pivotal step in their action mechanism of several clinically used anti-tumor drugs, such as anthracyclines, acridines and anthraquinones.1 The discovery of new DNA intercalators has been a practical approach to push clinical cancer therapy. Recently, a number of intercalators have been reported,2,3 and some β-carbolines such as COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundharmine and its derivatives are involved.4 Intercalation of COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundβ-carboline towards DNA generally result in cytotoxicity.5,6 By intercalation β-carbolines are able to inhibit topoisomerases I/II and finally damage DNA. In our ongoing efforts a series of β-carboline-3-carbonyl- amino acid benzyl esters were presented as potent anti-tumor derivatives of natural products. It was demonstrated that their in vitro cytotoxicity depended on the building blocks, i.e.β-carboline-3-carboxylic acid, amino acid and benzyl moieties.7 These requirements were generally followed in the design of N-(3-carboxyl-9-benzyl carboline-1-yl)ethylamino acids, as well as their in vitro anti-proliferation and the in vivo anti-tumor activities were analyzed with DOCK score.8
COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundTrp-Trp is biologically important either as a peptide or as a fragment of peptides. In the intercalator design both 2-Trp and 3-Trp modified COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundβ-carboline-3-carboxylic acid benzyl ester was reported to be potential anti-tumor agents.8,9 The DOCK scores from the automated docking study suggest that Trp-Trp-OBzl could be the pharmacophore of these anti-tumor molecules (Fig. 1). In this context the present paper used Trp-Trp-OBzl as the lead compound and twenty tripeptide benzyl esters, AA-Trp-Trp-OBzl, were prepared to discover DNA intercalators.
2. Results and discussion
2.1. Docking of AA-Trp-Trp-OBzls towards d(CGATCG)2
The automated docking studies of AA-Trp-Trp-OBzls were performed by using the LigandFit/LigandScore in DS Modeling2.1 and d(CGATCG)2 (Protein Data Bank, 1D12).10–13 The DOCK scores of AA-Trp-Trp-OBzls are listed in Table 1. As seen, the DOCK scores of nine AA-Trp-Trp-OBzls are 49 to 94. While the DOCK scores of the intercalators with higher in vivo anti-tumor activities were usually 50 to 60.8,9 According to the DOCK scores these nine AA-Trp-Trp-OBzls should be DNA intercalators and may possess higher in vivo anti-tumor activity.
Table 1
DOCK scores of AA-Trp-Trp-OBzl intercalating towards d(CGATCG)2
2.2. Synthesis of AA-Trp-Trp-OBzl
AA-Trp-Trp-OBzls (4a-t, AA = twenty amino acid residues) were prepared according to the synthetic route depicted in Scheme 1. Using a standard procedure Boc-Trp was coupled with Trp-OBzl to form Boc-Trp-Trp-OBzl (1, 96% yield). In a solution of COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundhydrogen chloride in COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundethyl acetate the Boc group of 1 was removed and Trp-Trp-OBzl (2) was obtained in 98% yield. The coupling reaction of Boc-AA with Trp-Trp-OBzl provided Boc-AA-Trp-Trp-OBzls (3a-t, 41%–96% yields). In a solution of COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundhydrogen chloride in COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundethyl acetate the Boc groups of 3a-t were removed, AA-Trp-Trp-OBzls (4a-t) were obtained and the yields were 90% to 99%. To ensure the purity, 3a-t were purified on a chromatographic column, while 4a-t were purified repeatedly using ether-promoted solidification and their HPLC purities were more than 97%. These data demonstrate that the used procedures and conditions are suitable for the preparation of AA-Trp-Trp-OBzls in high quality and acceptable yield.
 |
| | Scheme 1 Synthetic route of AA-Trp-Trp-OBzl. i) DCC, COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundHOBt and NMM; ii) COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundHydrogen chloride in COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundethyl acetate (4 M). In 3a & 4a AA = Ala, 3b & 4b AA = Gly, 3c & 4c AA = Phe, 3d & 4d AA = Leu, 3e & 4e AA = COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundIle, 3f & 4f AA = Val, 3g & 4g AA = Ser, 3h & 4h AA = COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundThr, 3i & 4i AA = Tyr, 3j & 4j AA = Pro, 3k & 4k AA = Met, 3l & 4l AA = Glu(CBzl), 3m & 4m AA = Asp(OBzl), 3n & 4n AA = Cys(S-CH2C6H4-OCH3-p), 3o & 4o AA = NG-NO2-Arg, 3p & 4p AA = Nω-CBz-Lys, 3q & 4q AA = Gln, 3r & 4r AA = COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundAsn, 3s & 4s AA = His, 3t & 4t AA = Trp. | |
2.3. Anti-proliferation activities of 4a-t
To explore the tumoricidal indication of 4a-t in the in vitro anti-proliferation assay, the carcinoma cell lines, HepG2 (human hepatocellular liver carcinoma cell line), S180 (mouse sarcoma cell line), H22 (mouse hepatocellular carcinoma cell line), K562 (human immortalised myelogenous leukaemia line) and B16 (mouse melanoma cell line) were used. In the assays the cells were exposed to 4a-t or COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundcytarabine (positive control), underwent the standard MTT tests and gave their proliferation inhibitions. The IC50 values are summarized in Table 2. It can be seen that 4a-t possess distinct in vitro anti-proliferation activities, the IC50 values are 11.0 μM to 181.9 μM, most of them are able to inhibit the proliferation of the carcinoma cells. The IC50 values of 4c-g,i,k,l,n-q are significantly lower than that of COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundcytarabine. The IC50 values of 4e,k,l,r are less than 20 μM, of which the DOCK scores are 61–94. These suggest that the lead, Trp-Trp-OBzl, works well, inserting an amino acid residue into its N-terminus increases the in vitro efficacy, and the formed tripeptides can properly intercalate into DNA.
Table 2
IC50 (μM) of 4a-t inhibiting carcinoma cell linesa
| Compd. |
S180
|
K562
|
B16
|
H22
|
HepG2 |
n = 6.
Compared to COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundcytarabine p < 0.01.
Compared to COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundcytarabine p < 0.05.
Compared to COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundcytarabine p > 0.05.
|
COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundCytarabine
|
40.33 ± 3.19 |
43.61 ± 2.10 |
49.22 ± 3.31 |
37.82 ± 2.67 |
38.10 ± 2.14 |
|
4a
|
72.68 ± 7.53 |
116.16 ± 9.65 |
131.92 ± 10.75 |
84.93 ± 8.50 |
49.88 ± 4.01 |
|
4b
|
79.87 ± 8.60 |
88.56 ± 8.87 |
158.56 ± 12.80 |
51.13 ± 3.34d |
58.27 ± 3.49 |
|
4c
|
33.95 ± 3.00b |
43.69 ± 3.49d |
33.97 ± 3.53b |
37.57 ± 2.80d |
41.16 ± 3.31d |
|
4d
|
36.74 ± 2.59c |
37.93 ± 2.62b |
53.40 ± 4.00 |
36.04 ± 2.81d |
41.54 ± 3.41d |
|
4e
|
15.14 ± 1.10b |
16.25 ± 1.12b |
17.23 ± 1.21b |
16.15 ± 1.06b |
15.08 ± 0.89b |
|
4f
|
36.13 ± 2.98c |
40.44 ± 1.51c |
50.15 ± 3.69d |
35.99 ± 3.03d |
37.35 ± 2.98d |
|
4g
|
26.31 ± 1.89b |
25.63 ± 2.12 b |
48.41 ± 8.40d |
25.20 ± 1.89b |
25.92 ± 1.91 b |
|
4h
|
155.43 ± 10.41 |
172.80 ± 12.36 |
181.90 ± 12.84 |
153.49 ± 10.14 |
154.59 ± 10.80 |
|
4i
|
22.90 ± 1.91b |
21.26 ± 1.83b |
24.78 ± 1.63b |
22.96 ± 1.91b |
23.62 ± 1.85b |
|
4j
|
76.00 ± 5.22 |
91.83 ± 7.01 |
94.00 ± 4.35 |
91.48 ± 7.12 |
91.67 ± 7.35 |
|
4k
|
19.81 ± 0.98b |
16.99 ± 1.06b |
25.25 ± 1.64b |
17.52 ± 0.99b |
24.33 ± 1.56b |
|
4l
|
16.60 ± 1.01b |
14.34 ± 0.98b |
21.37 ± 1.04b |
14.74 ± 0.95b |
22.28 ± 1.55b |
|
4m
|
163.69 ± 12.34 |
170.50 ± 11.16 |
157.30 ± 12.30 |
107.12 ± 9.46 |
122.03 ± 12.21 |
|
4n
|
26.23 ± 1.79b |
20.63 ± 1.84b |
32.81 ± 2.91b |
24.65 ± 1.90b |
35.00 ± 1.89 |
|
4o
|
29.38 ± 1.88b |
27.22 ± 1.83 b |
37.41 ± 1.93 b |
27.46 ± 1.85b |
38.06 ± 1.92d |
|
4p
|
33.04 ± 2.95b |
34.47 ± 2.67b |
57.37 ± 3.90 |
33.01 ± 2.21b |
25.89 ± 2.04b |
|
4q
|
27.58 ± 2.04b |
24.61 ± 1.98 b |
31.83 ± 2.82 b |
25.11 ± 2.00b |
26.25 ± 9.96b |
|
4r
|
13.52 ± 0.95b |
10.99 ± 1.02b |
13.83 ± 0.98b |
11.00 ± 0.96b |
11.86 ± 1.01b |
|
4s
|
83.31 ± 6.20 |
101.04 ± 10.07 |
100.43 ± 9.01 |
95.52 ± 6.30 |
97.23 ± 7.86 |
|
4t
|
142.05 ± 10.84 |
164.36 ± 12.31 |
161.51 ± 13.00 |
105.84 ± 9.37 |
163.27 ± 12.01 |
|
6r
|
19.7 ± 2.97 |
57.4 ± 10.6 |
−23.0 ± 5.99 |
17.6 ± 8.45 |
20.9 ± 7.48 |
2.4. Tumor weights of 4a-t treated S180 mice
In the in vivo assays the tumor weights of 4a-t treated S180 mice were recorded and are listed in Table 3. It was noticed that when the mice were given a daily i.p. injection of 8.9 μmol kg−1 of 4a-t in 0.2 ml of normal saline (NS) for seven consecutive days, the tumor weights were 0.123 to 0.985 g (NS receiving mice, 0.827 g) and eleven tripeptide benzyl esters (4c,e,f,g,i,k,l,n,o,q,r) significantly decreased the tumor weight of the treated mice, four tripeptide benzyl esters (4d,j,p,s) possessed weak activity and five tripeptide benzyl esters (4a,b,h,j,m,t) had no effect on the tumor weight of the treated mice. The efficacies of nine tripeptide benzyl esters (4e,g,i,k,l,n,o,q,r, tumor weights ranging from 0.123 g to 0.472 g) are significantly higher than that of COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundcytarabine (positive control, tumor weight 0.563 g). The efficacies of four tripeptide benzyl esters (4c,d,f,p, tumor weights ranging from 0.550 g to 0.713 g) are essentially equal to that of COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundcytarabine. While 4e,k,l,r have the highest efficacies, which also have the highest DOCK scores. These suggest that the lead, Trp-Trp-OBzl, works well, inserting an amino acid residue into its N-terminus increases the in vivo efficacy, and the formed tripeptides can properly intercalate into DNA.
Table 3 Effects of 4a-t on tumor weights of S180 micea
| Compd. |
Tumor Weight |
Inhibition (%) |
Compd. |
Tumor Weight |
Inhibition (%) |
Dose = 8.9 μmol kg−1, n = 12.
Compared to NS p < 0.01.
Compared to NS p < 0.01, to COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundcytarabine p < 0.05.
Compared to NS and COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundcytarabine p < 0.01.
|
| NS |
0.827 ± 0.165 |
0.00 |
COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundCytarabine
|
0.563 ± 0.102b |
31.93 ± 5.59 |
|
4a
|
0.921 ± 0.204 |
−11.38 ± 2.29 |
4k
|
0.319 ± 0.077d |
61.46 ± 10.33 |
|
4b
|
0.881 ± 0.194 |
−6.51 ± 0.64 |
4l
|
0.175 ± 0.049d |
78.85 ± 8.12 |
|
4c
|
0.623 ± 0.107b |
24.69 ± 4.86 |
4m
|
0.900 ± 0.264 |
−8.85 ± 1.36 |
|
4d
|
0.713 ± 0.040b |
13.78 ± 1.87 |
4n
|
0.427 ± 0.122c |
48.39 ± 9.58 |
|
4e
|
0.164 ± 0.078d |
80.13 ± 8.71 |
4o
|
0.454 ± 0.111c |
45.10 ± 7.35 |
|
4f
|
0.550 ± 0.190b |
33.51 ± 6.64 |
4p
|
0.703 ± 0.102b |
15.01 ± 2.16 |
|
4g
|
0.429 ± 0.155c |
48.16 ± 9.12 |
4q
|
0.472 ± 0.102c |
42.99 ± 7.55 |
|
4h
|
0.891 ± 0.151 |
−7.66 ± 0.66 |
4r
|
0.123 ± 0.017d |
85.13 ± 13.51 |
|
4i
|
0.386 ± 0.101d |
53.37 ± 8.85 |
4s
|
0.729 ± 0.157 |
11.91 ± 2.20 |
|
4j
|
0.724 ± 0.185 |
12.47 ± 2.47 |
4t
|
0.985 ± 0.216 |
−19.07 ± 4.01 |
2.5. Effect of 4a-t on the organ and body weights of treated S180 mice
Immunologic function can be damaged in chemotherapy. The spleen index and the body weight are the important parameters of estimating immunologic function. To evaluate the effect of the treatment on the immunologic function, the spleen indexes and the increased body weights of 4a-t treated S180 mice were measured and are listed in Table 4. The data indicate that the liver, brain and kidney weights as well as the spleen indexes of 4a-t treated S180 mice are substantially equal to those of NS and COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundcytarabine treated S180 mice, but the increased body weights of 4a-t treated S180 mice are significantly higher than that of NS and COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundcytarabine treated S180 mice. The significant differences in the increased body weights demonstrate that the immunologic damage resulted from the therapy of 4a-t is smaller than that from the therapy of NS and COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundcytarabine. These suggest that the lead, Trp-Trp-OBzl, works well, inserting an amino acid residue into its N-terminus increases the safety of the administration.
Table 4 Effect of 4a-t on the spleen index and organ weights of S180 micea
| Compd. |
Liver |
Brain |
Kidney |
Spleen index |
Increased BW |
n = 12; BW = body weight.
Compared to NS and cytarabine p < 0.01.
Compared to COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundcytarabine p < 0.05.
|
| NS |
1.85 ± 0.23 |
0.27 ± 0.03 |
0.16 ± 0.02 |
7.26 ± 1.37 |
8.55 ± 1.22 |
COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundCytarabine
|
1.62 ± 0.11 |
0.29 ± 0.02 |
0.16 ± 0.03 |
7.50 ± 1.84 |
7.12 ± 1.29 |
|
4a
|
1.65 ± 0.19 |
0.28 ± 0.02 |
0.16 ± 0.02 |
7.69 ± 1.88 |
7.20 ± 0.76 |
|
4b
|
1.81 ± 0.22 |
0.29 ± 0.01 |
0.17 ± 0.02 |
8.01 ± 1.79 |
7.39 ± 1.26 |
|
4c
|
1.75 ± 0.18 |
0.28 ± 0.01 |
0.15 ± 0.02 |
7.66 ± 1.90 |
9.66 ± 1.21b |
|
4d
|
1.58 ± 0.23 |
0.28 ± 0.02 |
0.16 ± 0.02 |
7.02 ± 2.60 |
7.29 ± 1.06 |
|
4e
|
1.86 ± 0.21 |
0.27 ± 0.02 |
0.16 ± 0.02 |
8.25 ± 1.78 |
9.81 ± 1.19b |
|
4f
|
1.79 ± 0.20 |
0.27 ± 0.02 |
0.16 ± 0.02 |
8.12 ± 1.66 |
9.95 ± 1.21b |
|
4g
|
1.88 ± 0.11 |
0.27 ± 0.01 |
0.15 ± 0.02 |
7.96 ± 1.36 |
9.72 ± 1.13b |
|
4h
|
1.89 ± 0.17 |
0.27 ± 0.01 |
0.17 ± 0.01 |
7.77 ± 1.63 |
7.71 ± 1.15 |
|
4i
|
1.83 ± 0.19 |
0.27 ± 0.01 |
0.17 ± 0.02 |
7.99 ± 1.02 |
10.12 ± 1.30b |
|
4j
|
1.40 ± 0.27 |
0.27 ± 0.03 |
0.17 ± 0.02 |
7.49 ± 1.82 |
6.46 ± 1.04 |
|
4k
|
1.82 ± 0.11 |
0.30 ± 0.01 |
0.15 ± 0.01 |
7.98 ± 1.37 |
9.73 ± 1.25b |
|
4l
|
1.90 ± 0.20 |
0.27 ± 0.02 |
0.16 ± 0.03 |
8.21 ± 1.32 |
9.69 ± 1.04b |
|
4m
|
1.59 ± 0.23 |
0.29 ± 0.02 |
0.16 ± 0.02 |
8.21 ± 1.55 |
7.35 ± 1.01 |
|
4n
|
1.87 ± 0.21 |
0.28 ± 0.02 |
0.15 ± 0.02 |
7.92 ± 1.08 |
9.83 ± 1.12b |
|
4o
|
1.80 ± 0.25 |
0.27 ± 0.03 |
0.16 ± 0.02 |
7.66 ± 1.68 |
7.99 ± 1.13 |
|
4p
|
1.88 ± 0.27 |
0.27 ± 0.02 |
0.16 ± 0.02 |
8.22 ± 1.60 |
10.05 ± 1.31b |
|
4q
|
1.87 ± 0.06 |
0.27 ± 0.02 |
0.16 ± 0.01 |
7.89 ± 1.96 |
9.88 ± 1.29b |
|
4r
|
1.83 ± 0.06 |
0.28 ± 0.02 |
0.16 ± 0.03 |
8.36 ± 1.88 |
9.87 ± 1.19b |
|
4s
|
1.81 ± 0.27 |
0.27 ± 0.02 |
0.14 ± 0.01 |
8.21 ± 1.66 |
8.41 ± 1.21c |
|
4t
|
1.79 ± 0.25 |
0.30 ± 0.05 |
0.17 ± 0.02 |
7.45 ± 1.80 |
7.41 ± 1.26 |
2.6. Dose-dependent in vivo anti-tumor action of 4r
The most potent 4r was selected to explore the dose dependent action of 4a-t. At the doses of 8.9, 0.89 and 0.089 μmol kg−1, the inhibitions of 4r are 85.1%, 46.7%, and 26.6%, respectively (Table 5). Therefore, 4r possesses dose-dependently anti-tumoral action.
Table 5 Anti-tumor activity of 4r at different doses against S180 micea
2.7. Acute toxicity of mice receiving 4c,e,f,g,i,k,l,n,o,q,r
A series of intercalators may induce serious neurotoxicity.14 To evaluate the neurotoxicity an acute toxicity assay was performed for the anti-tumoral active 4c,e,f,g,i,k,l,n,o,q,r. The results indicate that even the single dose of 4c,e,f,g,i,k,l,n,o,q,r is up to 500 mg kg−1 (i.p. injection) the mice neither exhibited neurotoxic behavior, such as tremor, twitch, jumping, tetanus, and supination, nor occurred death. On the 7th day the necropsy findings in 4c,e,f,g,i,k,l,n,o,q,r receiving mice revealed that the therapy led no apparent changes in any organs. This suggests that 4c,e,f,g,i,k,l,n,o,q,r are comparatively non-toxic, and their LD50 values are more than 500 mg kg−1.
2.8. Action mechanism of 4a-t
The variation of DNA spectra induced by small molecule has been widely used to identify the intercalation.15–18 To explore the intercalation of 4a-t towards DNA, calf thymus DNA (CT DNA) was selected as the model DNA, 4r was selected as model tripeptide, and the UV (see ESI†), circular dichroic (CD, see ESI†), fluorescence, viscosity and melting temperature measurements were performed.
2.8.1.
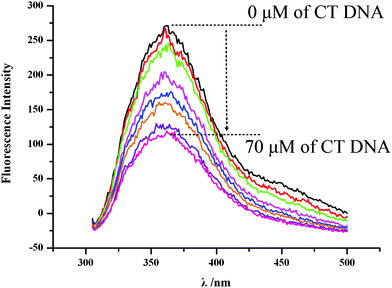
Fluorescence spectra of 4r without and with CT DNA.
The fluorescence spectra (Fig. 2) of 4r alone and it (final concentration 50 μM) plus CT DNA (final concentration 10, 20, 30, 40, 50, 60 and 70 μM) in PBS buffer (pH 7.4) were measured on a Shimadzu RF-5310PC spectrofluorometer (emission wavelength, 365 nm and excitation wavelength, 281 nm). The fluorescence spectra explain a phenomenon of fluorescence quenching, i.e. CT DNA concentration- dependently decreases the fluorescence intensity of 4r, and the intercalation of 4r toward CT DNA. As seen, when the final concentration of CT DNA is 70 μM the fluorescence intensity of 4r is decreased to the minima (corresponding a 35.28% decrease of the fluorescence intensity).
 |
| | Fig. 2
Fluorescence spectra of 4r in PBS (concentration 50 μM, pH = 7.0, λem = 365 nm, λex = 281 nm) explain the fluorescence quenching induced by 10 μl of the solution of a series concentrations (0, 10, 20, 30, 40, 50, 60 and 70 μM) of CT DNA in PBS. | |
2.8.2. Viscosity of CT DNA without and with 4r17,18.
During the intercalation, the base pairs of DNA will be pushed apart to accommodate and bind small molecule and consequently results in the increase of the viscosity of DNA. To reveal the intercalation the relative viscosity of the sample solutions of [4r]/[CT DNA] in the ratios of 0 – 0.36 was measured. Fig. 3 indicates that the relative viscosity of CT DNA is gradually increased with the increase of the concentration of 4r, which is the result of the intercalation of 4r towards CT DNA.
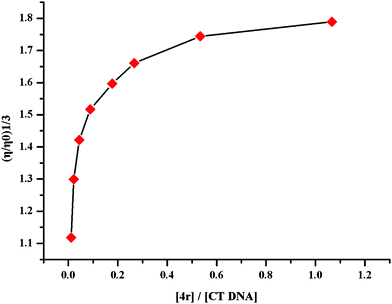 |
| | Fig. 3 Effect of 4r on the relative viscosity of CT DNA (200 μM). | |
2.8.3. Melting temperature of CT DNA without and with 4r19,20.
The melting temperature (Tm) of DNA is widely used to define the thermostability of the double helix. Once the temperature of DNA solution is high enough the double-stranded DNA will dissociate to single-stranded one and results in the increase of the absorbance. The temperature that induces the absorbance a 50% increase is termed as Tm. Here the temperature of the PBS (pH 7.4) solution of CT DNA alone (100 μM) and it plus 4r (10 μM) rose at a rate of 0.1. The melting curves of CT DNA alone and it plus 4r were shown with Fig. 4. As seen, the Tm of CT DNA plus 4r is 20.8 ± 0.6 °C higher than that of it alone. This Tm indicates that the addition of 4r increases the thermostability of the double-stranded DNA, which should be attributed to the intercalation of 4r toward CT DNA.
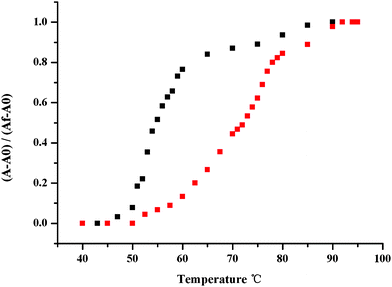 |
| | Fig. 4 Thermal denaturation curves of CT-DNA in the absence and presence of 4r. Tm measurements were performed in PBS at pH 7.4 with a 4r/DNA ratio of 0.10. | |
3. Conclusions
From a similar ring-opening modification of the anti-tumoral active N-trypto- phanyl-β-carboline-3-carboxylic acid benzyl ester and β-carboline-3-carbonyl- tryptophan benzyl ester a pharmacophore, Trp-Trp-OBzl, could be obtained. The N-terminal modification results in 20 anti-tumoral AA-Trp- Trp-OBzls. In the in vivo assay nine AA-Trp-Trp-OBzls were more potential than COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundcytarabine. In the acute toxicity assay AA-Trp-Trp-OBzls had a LD50 value of more than 500 mg kg−1. The automated docking, UV, CD, fluorescence, viscosity and melting temperature studies supported that the in vivo anti-tumor action was the result of the intercalation of AA-Trp-Trp-OBzls towards DNA.
Acknowledgements
This work was completed in the Beijing area major laboratory of peptide and small molecular drugs, supported by PHR (IHLB, KM 200910025009), the National Natural Scientific Foundation of China (30801426, 81072552), and Special Project (2008ZX09401-002) of China.
References
- G. Momekov, A. Bakalova and M. Karavanova, Novel approaches towards development of non-classical platinum-based antineoplastic agents: Design of platinum complexes characterized by an alternative DNA-binding pattern and/or tumor-targeted cytotoxicity, Curr. Med. Chem., 2005, 12, 2177–2191 CrossRef CAS.
- A. Bolognese, G. Correale, M. Manfra, A. Lavecchia, O. Mazzoni, E. Novellino, P. La Colla, G. Sanna and R. Loddo, Antitumor agents 3. Design, synthesis, and biological evaluation of new pyridoisoquinolindion and dihydrothienoquinolin- dione derivatives with potent cytotoxic activity, J. Med. Chem., 2004, 47, 849–858 CrossRef CAS.
- M. A. Lynch, O. Duval, A. Sukhanova, J. Devy, S. P. Mackay, R. D. Waigh and I. Nabiev, Synthesis, biological activity and comparative analysis of DNA binding affinities and human DNA topoisomerase I inhibitory activities of novel 12-alkoxy-benzo[c]phenanthridinium salts, Bioorg. Med. Chem. Lett., 2001, 11, 2643–2646 CrossRef CAS.
- R. Cao, W. Peng, H. Chen, Y. Ma, X. Liu, X. Hou, H. Guan and A. Xu, DNA binding properties of 9-substituted harmine derivatives, Biochem. Biophys. Res. Commun., 2005, 338, 1557–1563 CrossRef CAS.
- M. Laronze, M. Boisbrun, S. Leonce, B. Pfeiffer, P. Renard, O. Lozach, L. Meijer, A. Lansiaux, C. Bailly, J. Sapi and J. Y. Laronze, Synthesis and anticancer activity of new pyrrolocarbazoles and pyrrolo-beta-carbolines, Bioorg. Med. Chem., 2005, 13, 2263–2283 CrossRef CAS.
- A. M. Sobhani, S. A. Ebrahimi and M. J. Mahmoudian, An in vitro evaluation of human DNA topoisomerase I inhibition by Peganum harmala L. seeds extract and its beta-carboline alkaloids, Pharm Pharm Sci, 2002, 5, 19–23 Search PubMed.
- M. Zhao, L. Bi, W. Wang, C. Wang, M. Baudy-Floc, J. Ju and S. Peng, Synthesis and cytotoxic activities of beta-carboline amino acid ester conjugates, Bioorg. Med. Chem., 2006, 14, 6998–7010 CrossRef CAS.
- J. H. Wu, M. Zhao, K. D. Qian, K. H. Lee, S. M. Natschke and S. Q. Peng, Novel N-(3-carboxyl-9-benzyl-beta-carboline-1-yl)ethylamino acids: Synthesis, anti- tumor evaluation, intercalating determination, 3D QSAR analysis and docking investigation, Eur. J. Med. Chem., 2009, 44, 4153–4161 CrossRef CAS.
- J. H. Wu, C. Y. Li, M. Zhao, W. J. Wang, Y. J. Wang and S. Q. Peng, A class of novel carboline intercalators: Their synthesis, in vitro anti-proliferation, in vivo anti- tumor action and 3D QSAR analysis, Bioorg. Med. Chem., 2010, 18, 6220–6229 CrossRef CAS.
- A. M. Sobhani, S. R. Amini, J. D. A. Tyndall, E. Azizi, M. Daneshtalab and A. Khalaj, A theory of mode of action of azolylalkylquinolines as DNA binding agents using automated flexible ligand docking J Mol Graphics Model, 2006, 25, 459–469 Search PubMed.
- A. Marco, C. Loza-MejíaRafael and L. R. Alfonso, Molecular modeling of tricyclic compounds with anilino substituents and their intercalation complexes with DNA sequences, J. Mol. Graphics Modell., 2009, 27, 900–907 CrossRef.
- C. M. A. Alves, S. Naik and P. J. G. Coutinho, Novel long alkyl side chain benzo[a]phenoxazinium chlorides: synthesis, photophysical behaviour and DNA interaction, Tetrahedron, 2009, 65, 10441–10452 CrossRef CAS.
- C. M. Venkatachalam, X. Jiang, T. Oldfield and M. Waldman, LigandFit: a novel method for the shape-directed rapid docking of ligands to protein active sites, J. Mol. Graphics Modell., 2003, 21, 289–307 CrossRef CAS.
- A. M. Sobhani, S. R. Amini, J. D. A. Tyndall, E. Azizi, M. Daneshtalab and A. Khalaj, A theory of mode of action of azolylalkylquinolines as DNA binding agents using automated flexible ligand docking, J. Mol. Graphics Modell., 2006, 25, 459–469 CrossRef CAS.
- Z. Rezaei, H. Firouzabadi, N. Iranpoor, A. Ghaderi, M. R. Jafari, A. A. Jafari and H. R. Zare, Design and one-pot synthesis of α-aminophosphonates and bis(α- aminophosphonates) by iron(III) chloride and cytotoxic activity, Eur. J. Med. Chem., 2009, 44, 4266–4275 CrossRef CAS.
- N. Li, Y. Ma, C. Yang, L. P. Guo and X. R. Yang, Interaction of anticancer drug mitoxantrone with DNA analyzed by electrochemical and spectroscopic methods, Biophys. Chem., 2005, 116, 199–205 CrossRef CAS.
- U. Varadarajan, C. Alfonso and U. N. Balachandran, Copper(II) complexes of N4 tetradentate ligands with flexible alkyl spacers: Crystal structure, DNA binding and cleavage studies, Polyhedron, 2007, 26, 3008–3016 CrossRef CAS.
- P. U. Maheswari and M. Palaniandavar, DNA binding and cleavage properties of certain tetrammine ruthenium(II) complexes of modified 1,10-phenanthrolines- effect of hydrogen-bonding on DNA-binding affinity, J. Inorg. Biochem., 2004, 98, 219–230 CrossRef.
- T. S. Pitchumony, H. S. Evans and M. Palaniandavar, Synthesis, structure and DNA interaction of cobalt(III) bis-complexes of 1,3-bis(2-pyridylimino)isoindoline and 1,4,7-triazacyclononane, J. Inorg. Biochem., 2005, 99, 2110–2118 CrossRef.
- F. Arjmand and M. Aziz, Synthesis and characterization of dinuclear macrocyclic cobalt(II), copper(II) and zinc(II) complexes derived from 2,2,2′,2′-S, S[bis(bis- N, N-2-thiobenzimidazolyloxalato-1,2-ethane)]: DNA binding and cleavage studies, Eur. J. Med. Chem., 2009, 44, 834–844 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c0md00208a |
|
| This journal is © The Royal Society of Chemistry 2011 |
Click here to see how this site uses Cookies. View our privacy policy here. 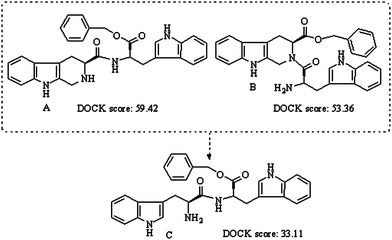

![[x with combining macron]](https://www.rsc.org/images/entities/i_char_0078_0304.gif) ± SD g.
b Compared to 0.89 μmol kg−1 of cytarabine p < 0.05.
c Compared to 0.089 μmol kg−1 of cytarabine p < 0.05.
d Compared to 0.89 μmol kg−1 of 4r p < 0.01.
e Compared to 0.089 μmol kg−1 of 4r p < 0.01; f) Compared to NS p < 0.01.
f Compared to NS p < 0.01.
± SD g.
b Compared to 0.89 μmol kg−1 of cytarabine p < 0.05.
c Compared to 0.089 μmol kg−1 of cytarabine p < 0.05.
d Compared to 0.89 μmol kg−1 of 4r p < 0.01.
e Compared to 0.089 μmol kg−1 of 4r p < 0.01; f) Compared to NS p < 0.01.
f Compared to NS p < 0.01.