Alkynyl-farnesol reporters for detection of protein S-prenylation in cells†‡
Guillaume
Charron
,
Lun K.
Tsou
,
William
Maguire
,
Jacob S.
Yount
and
Howard C.
Hang
*
The Laboratory of Chemical Biology and Microbial Pathogenesis, The Rockefeller University, New York, NY, 10065. E-mail: hhang@mail.rockefeller.edu
First published on 25th November 2010
Abstract
Protein S-prenylation is a lipid modification that regulates membrane-protein and protein-protein interactions in cell signaling. Though sites of protein S-prenylation can be predicted based upon conserved C-terminal CaaX or CC/CXC motifs, biochemical detection of protein S-prenylation in cells is still challenging. Herein, we report an alkynyl-isoprenol chemical reporter (alk-FOH) as an efficient substrate for prenyltransferases in mammalian cells that enables sensitive detection of S-farnesylated and S-geranylgeranylated proteins using bioorthogonal ligation methods. Fluorescent detection alleviates the need to deplete cellular isoprenoids for biochemical analysis of S-prenylated proteins and enables robust characterization of S-prenylated proteins, such as effectors that are injected into host cells by bacterial pathogens. This alkynyl-prenylation reporter provides a sensitive tool for biochemical analysis and rapid profiling of prenylated proteins in cells.
Introduction
S-prenylation is a post-translational lipid modification that controls membrane targeting of many proteins underlying fundamental cellular signaling pathways1 and diseases such as cancer2 and microbial infections.3S-prenylation refers to the posttranslational installation of either farnesyl (15-carbon) or geranylgeranyl (20-carbon) isoprenoid lipid chains on proteins at one or two cysteine residues located at or near the C-terminus through a thioether linkage (Fig. 1A). The largest group of S-prenylated proteins are the small G proteins (small GTPases) that encompasses Ras, Rho and Rab families of proteins.4S-farnesylation controls H-Ras and N-Ras localization and signaling during cellular differentiation and is further regulated by dynamic fatty-acylation cycles,5 while K-Ras4B plasma membrane association and signaling are mediated by S-farnesylation and phosphorylation or calmodulin binding to an adjacent polybasic motif.6,7 In contrast, membrane targeting of Rho and Rab proteins is mediated by S-geranylgeranylation through transient association with lipid-binding proteins like Rho-GDI.8,9 Other proteins like heterotrimeric G protein γ subunits and nuclear lamins also require S-prenylation for proper signaling functions.10–13 Misregulation of protein S-prenylation on many substrates often results in major defects in cell signaling and disease progression.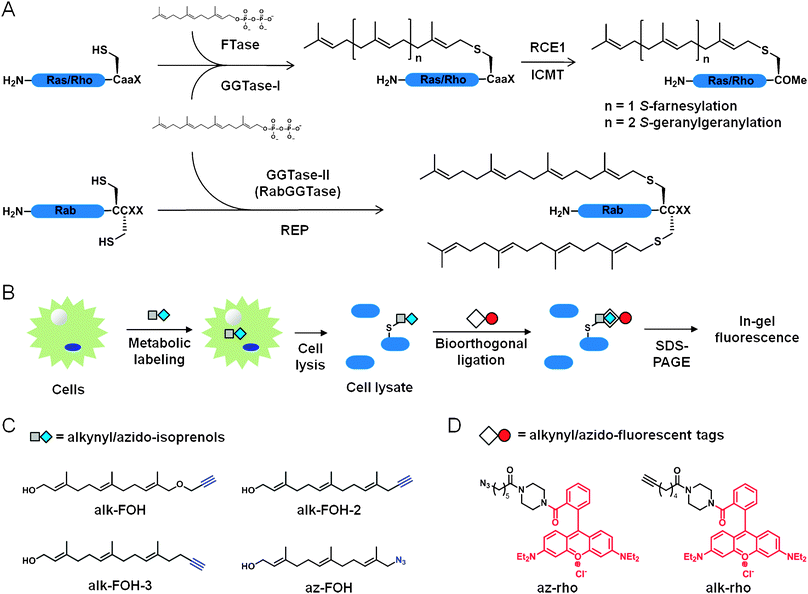 | ||
| Fig. 1 Bioorthogonal reporters of protein prenylation. (A) CaaX protein S-farnesylation and S-geranylgeranylation (top). Rab protein dual S-geranylgeranylation (bottom). (B) Scheme for metabolic labeling with prenylation reporters. (C) Prenylation reporters. (D) Detection tags. | ||
The significance of S-prenylated proteins has motivated the dissection of their biosynthetic pathways. The precursors for S-prenylation are derived from isoprenoid pyrophosphate substrates. Farnesyltransferase (FTase) catalyzes the transfer of farnesyl diphosphate (FPP) onto proteins while releasing a free diphosphate (Fig. 1A). Geranylgeranyltransferase type 1 and 2 (GGTase-I and GGTase-II) use geranylgeranyl diphosphate (GGPP) as a substrate (Fig. 1A). FTase and GGTase-I recognize a C-terminal CaaX motif where C is the modified cysteine, “a” is an aliphatic amino acid, and X determines the selectivity for either FTase (prefers Cys, Met, Ser, Ala and Gln) or GGTase-I (prefers Leu and Phe), although this rule is not stringent.14 For example, K-Ras (with CVIM or CIIM motifs) becomes geranylgeranylated in vivo upon farnesyltransferase inhibition15 and RhoB (with a CKVL motif) is known to be both farnesylated and geranylgeranylated in vivo.16 Upon prenylation in the cytosol, CaaX proteins are recruited to the endoplasmic reticulum where CaaX prenyl protease 2 (RCE1) cleaves the aaX tri-peptide and isoprenylcysteine carboxylmethyltransferase (ICMT) methylates the newly exposed C-terminal carboxylic acid of the cysteine. GGTase-II (also referred as RabGGTase) exclusively modifies Rab proteins and requires Rab escort proteins (REP) to present the C-terminal CC or CXC double cysteines of Rab proteins, which gets dually geranylgeranylated (Fig. 1A). Rab proteins bearing a C-terminal CXC motif also get carboxymethylated. Based on the conserved CaaX-box and CC/CXC motifs, bioinformatic analyses predict a few hundred S-prenylated proteins in mammalian proteomes.17 The biochemical dissection of protein prenylation pathways sparked significant interest in the development of robust detection methods to better understand the roles of S-prenylated proteins in cell signaling, but still remains challenging.
Prenylated proteins have been traditionally visualized by metabolic labeling with radioactive substrates such as [3H]mevalonic acid, [3H]FPP and [3H]GGPP, which have low sensitivity and require long exposure times for detection (weeks to months). To address these limitations, chemical reporters of protein prenylation have been developed to improve the detection of lipidated proteins.18,19 Fluorescent (NBD-GPP and NBD-FPP)20 and biotinylated (BGPP)21 analogs of isoprenoid diphosphates were successfully incorporated onto prenylated proteins in mammalian cells, but the bulk size of the fluorophore or biotin moieties may perturb membrane targeting of proteins and downstream signaling. Azido- and alkynyl-isoprenoid diphosphates coupled with bioorthogonal ligation22 provide improved methods for visualizing S-prenylated proteins (Fig. 1B).18,19 Azido- and alkynyl-geranyl diphosphates enabled the selective in vitro tagging by FTase of peptides and proteins bearing a CaaX motif,23–25 while azido-geranylgeranyl diphosphate afforded in vitro labeling by GGTase-I and GGTase-II of geranylgeranylated proteins.26 Further improvement was achieved with azido-farnesol (Az-FOH, Fig. 1C)27 and azido-geranylgeraniol28 metabolic incorporation in mammalian cells onto farnesylated and geranylgeranylated proteins, respectively.
Having previously demonstrated that alkyne-functionalized chemical reporters provide more sensitive detection than their azido-counterpart for fatty-acylated proteins,29 acetylated proteins30 as well as newly synthesized proteins,31 we synthesized and evaluated a panel of alkynyl-isoprenols to improve the detection of protein S-prenylation in cells. We demonstrate here that the alkynyl-farnesol (alk-FOH, Fig. 1C) chemical reporter is a readily accessible substrate for FTase and GGTases in cells and allows sensitive fluorescent detection of S-farnesylated and S-geranylgeranylated proteins using copper(I)-catalyzed azide-alkyne cycloaddition (CuAAC) methods.
Results and discussion
We designed alkynyl-isoprenol chemical reporters based on farnesol, since corresponding chemical reporters are substrates of FTase and GGTase-I in vitro.32 We utilized a synthetic scheme that would allow access to a diverse panel of alkynyl-farnesol derivatives (alk-FOH, alk-FOH-2 and alk-FOH-3), as well as an azide analog (az-FOH) for comparison purposes, from a common intermediate (Fig. 2). After THP protection, commercially available farnesol can be selectively oxidized at the terminal allylic position to give functionalized farnesol derivative 1.23 Allylic alcohol 1 was then alkylated with propargyl bromide and deprotected in acidic ethanol yielding alk-FOH, a precursor to previously reported alkynyl-farnesol diphosphate that can be incorporated onto peptide substrates by both FTase and GGTase-I in vitro.32 Corey-Kim like halogenation33 of allylic alcohol 1 enabled access to allylic bromide intermediate 3. CuI/K2CO3 mediated coupling with TMS-acetylene afforded enyne 4, which was subsequently deprotected in two steps to yield alk-FOH-2. Alternatively, allyl bromide 3 was alkylated with lithiated TMS-1-propyne to give enyne 5, which after deprotection afforded alk-FOH-3 that is one methylene unit longer than alk-FOH-2. Az-FOH was also obtained in a similar manner by displacement of the bromide 3 with sodium azide, followed by THP deprotection.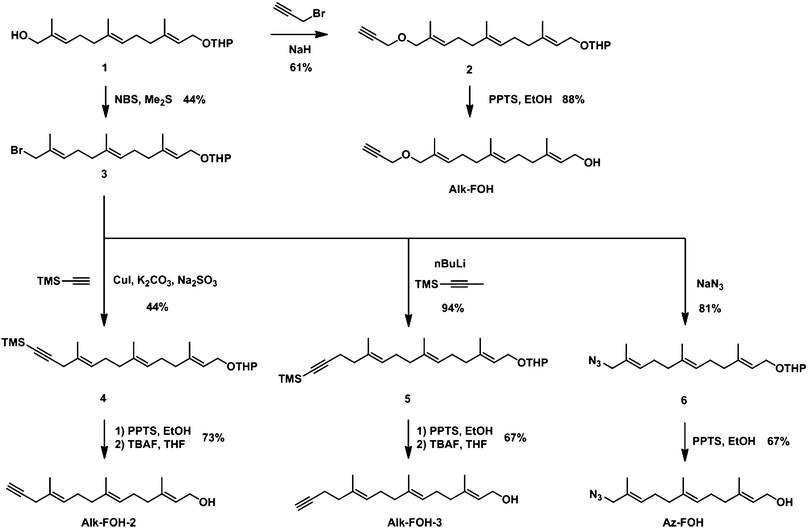 | ||
| Fig. 2 Synthesis of prenylation reporters. | ||
With all three alkynyl-farnesol reporters in hand, we assayed whether these compounds would be metabolically incorporated into mammalian cells and installed onto endogenous target proteins. Jurkat T cells were incubated with alk-FOH, alk-FOH-2, alk-FOH-3 or az-FOH at 50 μM for 4 h, lysed and conjugated to azido-rhodamine29 (in the case of alk-FOHs) or alk-rhodamine29 (for az-FOH) via CuAAC, separated by SDS-PAGE and analyzed by in-gel fluorescence scanning (Fig. 3). The banding pattern of proteins with the alk-FOH probes was similar to az-FOH labeling, suggesting that all three alk-FOHs were incorporated onto farnesylated proteins. Labeling was stronger with alk-FOH and alk-FOH-3 with markedly stronger signal in the 20–25 kDa molecular weight region of small GTPases. We also determined the effect of HMG-CoA reductase inhibitors (lovastatin) on the incorporation efficiency and detection sensitivity in comparison to az-FOH. Robust signal over the background was obtained without HMG-CoA reductase inhibitors using alkynyl-FOHs, whereas only few specific bands could be detected with previously reported az-FOH.28 Optimal concentration for alk-FOH was 50 μM, as judged by labeling of Jurkat cell lysates (Fig. S1‡). Addition of alk-FOH at 100 μM resulted in cellular toxicity that was not observed at lower concentrations (data not shown).
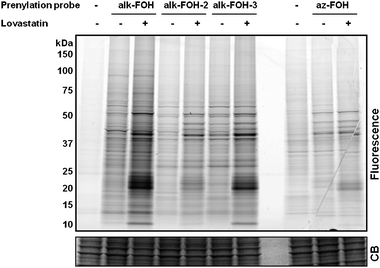 | ||
| Fig. 3 Comparative analysis of prenylation reporters. Jurkat cells were treated with either 20 μM lovastatin or with DMSO as a solvent control for 24 h before supplementing the media with prenylation reporters (50 μM, 4 h). Cell lysates labeled with alkynyl-isoprenols (alk-FOH, alk-FOH-2 and alk-FOH-3) or azido-isoprenol (az-FOH) were conjugated via CuAAC to azido-rhodamine (az-rho) or alkynyl-rhodamine (alk-rho), respectively. Lysates (20 μg) were separated by SDS-PAGE and scanned for fluorescence (top panel) or stained with Coomassie blue as a loading control (lower panel). | ||
To determine whether the alkynyl-farnesol reporters indeed targeted S-prenylated proteins, we evaluated the labeling of known farnesylated protein (H-Ras) and two different classes of geranylgeranylated proteins (RhoA and Rab7). All three reporters successfully visualized Ras farnesylation upon lovastatin treatment (Fig. 4A). Nonetheless, alk-FOH and alk-FOH-3 afforded better detection of Ras farnesylation than alk-FOH-2 and az-FOH, even at endogenous levels (without lovastatin pre-treatment, Fig. 4A). We focused on alk-FOH, as this chemical reporter can be readily synthesized and is stable compared to other alkynyl-farnesol reporters. Alk-FOH was then assayed with a known substrate protein of each prenyltransferase in HeLa cells transiently expressing Ras for FTase, RhoA for GGTase-I and Rab7 for GGTase-II. All three proteins were labeled by alk-FOH (Fig. 4B). Furthermore, selective depletion of the Ras signal was observed when incubating the cells with a FTase inhibitor (FTI-277, 10 μM), and selective diminution of the RhoA signal was observed with a GGTase-I inhibitor (GGTI-2133, 10 μM) while the signal was unaffected in the case of Rab7 (Fig. 4B). This demonstrates that alk-FOH can be incorporated as a substrate for all three prenyltransferases in cells and hence is a useful tool to detect protein S-prenylation. Alk-FOH was also incorporated by a variety of other cell types (fibroblasts: HeLa, 3T3; leukocytes: Jurkat, DC2.4 and Raw264.7) demonstrating the generality of this chemical reporter (Fig. S2‡). Each cell line exhibited a different banding pattern, highlighting the heterogeneity of S-prenylated proteins among unique cell types.
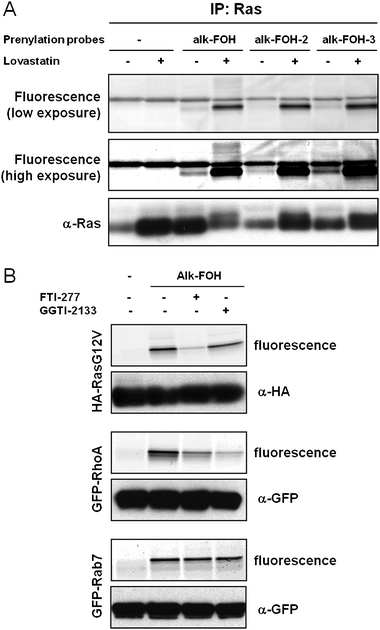 | ||
| Fig. 4 Fluorescent visualization of protein S-prenylation on known prenylated proteins. (A) Jurkat cells were treated with either 20 μM lovastatin or with DMSO as a solvent control for 24 h before supplementing the media with prenylation reporters (50 μM, 4 h). Immunopurified Ras labeled with alkynyl-isoprenols (alk-FOH, alk-FOH-2 and alk-FOH-3) was conjugated via CuAAC to azido-rhodamine (az-rho), followed by separation by SDS-PAGE and fluorescence detection (top panels) or immunoblotting as a loading control (lower panel). (B) HeLa cells transiently expressing HA-RasG12V, GFP-RhoA or GFP-Rab7 were treated with either 10 μM FTI-277, GGTI-2133, or with DMSO as a solvent control for 1 h before supplementing the media with prenylation reporter alk-FOH (50 μM, 4 h). Immunopurified proteins labeled with alk-FOH were conjugated via CuAAC to azido-rhodamine (az-rho), followed by separation by SDS-PAGE and fluorescence detection (top panels) or immunoblotting as a loading control (lower panel). | ||
Having demonstrated the efficient detection of S-prenylated proteins in mammalian cells with alk-FOH, we evaluated S-prenylation of Salmonella type III secretion system effector SifA. The bacterial protein effector SifA is translocated into the host cytoplasm after bacterial infection and localizes to membranes of the Salmonella-containing vacuoles (SCVs).34 SifA induces the formation of tubular membrane extensions of the SCV termed Salmonella-induced filaments (Sifs),34 which are necessary for maintaining the integrity of the SCV and Salmonella virulence.35 The C-terminal motif of SifA (331CLCCFL336) contains an unusual prenylation motif that can be interpreted either as a CaaX motif and/or a Rab protein motif (CCXX) and S. typhimurium lacking this SifA C-terminal hexapeptide exhibited a significant defect in bacterial replication.36 Although the S-prenylation prediction suite (PrePS)14 does not recognize SifA as a substrate for any prenyltransferase, SifA was labeled specifically at cysteine 333 by [3H]mevalonic acid in rabbit reticulocyte lysate in vitro.37 These data suggest that SifA is S-prenylated by the host cell machinery for recruitment to the SCV membrane,36,37 although SifA S-prenylation could not be detected by [3H]mevalonic acid labeling in cells. The precise roles of SifA S-palmitoylation and S-prenylation during Salmonella infection are unclear, as single point mutations of putative lipidation sites do not recapitulate the defect in Salmonella replication observed with the SifA mutant lacking the entire C-terminal hexapeptide.36,37
To provide more insight into SifA S-prenylation in cells, N-terminally HA-tagged SifA (HA-SifA) was transiently expressed in HeLa cells and metabolically labeled with alk-FOH. HA-SifA was then immunopurified, reacted with az-rho via CuAAC and analyzed by in-gel fluorescence scanning. Alk-FOH afforded robust visualization of SifA prenylation in cells for the first time (Fig. 5A). As opposed to previous in vitro radioactive labeling studies,37 we observed residual cellular HA-SifA prenylation (∼40%) in the presence of GGTase-I inhibitors (GGTI-298 or GGTI-2133, 10 μM). Furthermore, inhibition of FTase (with FTI-277, 10 μM) or dual inhibition of FTase and GGTase-I in cells retained residual SifA prenylation (∼20% and ∼50%, respectively) (Fig. 5A), suggesting that SifA is a substrate for more than GGTase-I. Single mutations of SifA C-terminal cysteines to serines (HA-SifAC333S, HA-SifAC334S) each retained about 30% of prenylation compared to wild-type, while dual mutations (HA-SifAC333,334S) or triple cysteine mutations (HA-SifAC331,333,334S) were not prenylated by alk-FOH in cells (Fig. 5B), suggesting that SifA is dually prenylated at cysteines 333 and 334. These results are in contrast with SifA in vitro radioactive labeling studies, where a single mutation (HA-SifAC333S) was sufficient to deplete SifA prenylation.37
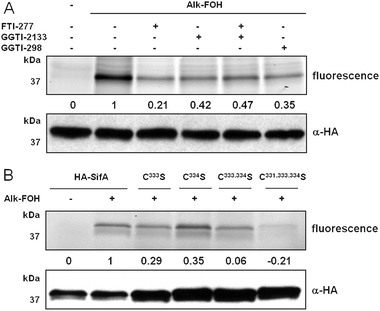 | ||
| Fig. 5 Alk-FOH analysis of SifA prenylation. (A) HeLa cells transiently expressing HA-SifA were treated with either 10 μM FTI-277, GGTI-298, GGTI-2133 or 10 μM of both FTI-277 and GGTI-2133, or with DMSO as a solvent control, for 1 h before supplementing the media with prenylation reporter alk-FOH (50 μM, 4 h). Immunopurified HA-SifA labeled with alk-FOH was conjugated via CuAAC to azido-rhodamine (az-rho), followed by separation by SDS-PAGE and fluorescence detection (top panels) or immunoblotting as a loading control (lower panel). (B) HeLa cells transiently expressing HA-SifA, HA-SifAC333S, HA-SifAC334S, HA-SifAC333,334S, or HA-SifAC331,333,334S were treated with prenylation reporter alk-FOH (50 μM, 4 h). Immunopurified HA-SifA labeled with alk-FOH was conjugated via CuAAC to azido-rhodamine (az-rho), followed by separation by SDS-PAGE and fluorescence detection (top panels) or immunoblotting as a loading control (lower panel). Fluorescence was quantified by mean fluorescence intensity adjusted for loading and normalized for background (0) and strongest signal (1). | ||
The differences in SifA prenylation between our results and previous studies may be due to cellular labeling with alk-FOH compared to in vitro labeling with [3H]mevalonic acid. The improved fluorescence detection with alk-FOH in cells may reveal low levels of S-prenylation not visualized by [3H]mevalonic acid in vitro. Prenylation analyses of proteins bearing the unusual double cysteine CCXX C-terminal motif similar to SifA have been described. Geranylgeranyl modification of Rab5A (CCSN) is catalyzed by an enzyme in brain cytosol but not by purified GGTase-I.38 Similarly, Rab5A prenylation with GGTase-II/REP-1 and [3H]GGPP followed by mass spectrometry analysis demonstrated that Rab5A is geranylgeranylated on both cysteines by GGTase-II.39 However, the CCXX motif is not always a marker for GGTase-II substrates. For example, in vitro translational analyses by radioactive labeling in rabbit reticulocyte lysate demonstrated that both Ral-A (CCIL) and Ral-B (CCLL) are geranylgeranylated exclusively at the cysteine in the fourth position from the carboxyl terminus and [3H]GGPP incorporation onto RalA/B could be blocked with GGTase-I inhibitors.40,41 Alternatively, Wrch-1, a Rho family GTPase bearing a CCFV C-terminal motif and a brain-specific isoform of Cdc42 with a CCIF motif were not found to be prenylated, but intead S-fatty acylated.42,43 CCXX can therefore be a motif for prenylation and S-palmitoylation. Nonetheless, our analyses of SifA prenylation with inhibitors of prenyltransferases and cysteine mutants suggest that SifA may be heterogeneously prenylated in host cells and single cysteine mutations do not completely abolish S-prenylation. While our cellular alk-FOH labeling of SifA differ from in vitro [3H]mevalonic acid labeling studies (Fig. 5), the robust and heterogeneous SifA prenylation from our alk-FOH cellular labeling is consistent with residual membrane partitioning of these SifA mutants as well as their activity during Salmonella infection.37
In conclusion, the alkynyl-farnesol, alk-FOH, provides an efficient chemical reporter of S-prenylated proteins in cells. Several improvements over previously reported detection methods were achieved. Alk-FOH is stable, readily accessible through chemical synthesis, and a substrate of all three prenyltransferases in mammalian cells. Importantly, HMG-CoA reductase inhibitors are not required for alk-FOH detection of endogenously prenylated proteins and transfer of hydrophobic proteins onto membranes is circumvented by in-gel fluorescence detection. Our approach enabled the detection of SifA prenylation in cells for the first time, which revealed robust and heterogeneous prenylation of the Salmonella T3SS effector SifA. Overall, the increased sensitivity achieved here should assist in biochemical and functional analysis of protein S-prenylation in cells.
Experimental
Plasmids construction
The human HA-RasG12V construct was a gift from Prof. Marilyn Resh (Memorial Sloan-Kettering Cancer Center), GFP-RhoA (plasmid 12965) and GFP-Rab7 (plasmid 12605) constructs were acquired from Addgene. DNA coding for SifA was amplified by PCR from Salmonella Typhimurium IR715 strain and inserted into the pCMV-HA (Clontech) vector for mammalian expression. The forward primer, 5′-GAATTCCC ATGCCGATTACTATAGGGAA-3′, added an EcoRI restriction site as well as nucleotides for in frame expression of the vector's HA tag. The reverse primer, 5′-GTCGAC TTATAAAAAACAACATAAACAGCCGCT-3′ added a SalI restriction site. DNA sequencing was performed and results were identical to previously reported SifA gene sequences from the LT2 strain of Salmonella Typhimurium. Mutagenesis was performed using the Stratagene Quikchange Multi Site-Directed Mutagenesis Kit. Primers used for individual mutants are as follows: SifAC333S: 5′-CAGAACAACAAAGCGGCTGTTTATCGTGTTTTTTATAAGTCGACGGCC-3′, SifAC334S: 5′-GAACAACAAAGCGGCTGTTTATGTTCGTTTTTATAAGTCGAC-3′, SifAC333,334S: 5′-GAACAACAAAGCGGCTGTTTATCGTCGTTTTTATAAGTCGAC-3′, SifAC331,333,334S: 5′-CTCAGAACAACAAAGCGGCTCGTTATCGTCGTTTTTATAAGTCGAC-3′.Cell culture, transfections and metabolic labeling
Jurkat cells were cultured in RPMI medium, while Raw264.7, HeLa, 3T3 and DC2.4 cells were cultured in DMEM, both supplemented with 10% fetal bovine serum (FBS), 100 U/mL penicillin and 100 μg mL−1 streptomycin. Cells were maintained in a humidified 37 °C incubator with 5% CO2. For transfections, HeLa cells were grown in 6-well plates to approximately 90% confluence, and transfected using Lipofectamine 2000 (Invitrogen) according to manufacturer's recommendation. Cells were treated with alk-FOH (50 μM, 50 mM stock solution in DMSO) for 4 h using the same volume of DMSO in the negative controls. For coincubation with inhibitors, HeLa cells were pretreated for 1 h with FTI-277 (10 μM, 20 mM stock solution in DMSO), GGTI-298 (10 μM, 10 mM stock solution in DMSO) or GGTI-2133 (10 μM, 10 mM stock solution in DMSO) prior to alk-FOH metabolic labeling. Cells were then harvested, washed twice with ice-cold PBS and pelleted at 1000 g for 5 min. Cells were directly lysed or flash frozen in liquid nitrogen and stored at −80 °C prior to lysis.Immunoprecipitation
Cell pellets obtained from 3 confluent wells of a 6-well plate were lysed with 150 μL of ice-cold Brij lysis buffer (1% Brij 97, 50 mM triethanolamine pH 7.4, 150 mM NaCl, 5 × EDTA-free Roche protease inhibitor cocktail, 10 mM PMSF). Cell lysates were collected after centrifuging at 1000 g for 5 min at 4 °C to remove cell debris. Protein concentration was determined by the BCA assay (Pierce). Proteins of interest were immunoprecipitated from 400 μg protein cell lysates in 250 μL ice-cold Brij lysis buffer using either 15 μL of packed anti-v-H-ras (Ab-1) rat mAb (Y13-259) agarose conjugate (Calbiochem), 25 μL of packed anti-HA (HA-7) mouse mAb agarose conjugate (Sigma), or 1 μL anti-GFP rabbit pAb (ab290, Abcam) and 50 μL of packed protein A-agarose beads (Roche) per sample. Upon incubation at 4 °C for 2 h on a nutating mixer (Labnet), the beads were washed (3 × 1 mL) with ice-cold modified RIPA lysis buffer (1% Triton X-100, 1% sodium deoxycholate, 0.1% SDS, 50 mM triethanolamine pH 7.4, 150 mM NaCl).CuI-catalyzed azide-alkyne cycloaddition (CuAAC)/click reaction
The purified protein bound to beads was suspended in 47 μL of ice-cold PBS, to which was added 3 μL freshly premixed click reaction cocktail [azido-rhodamine (100 μM, 10 mM stock solution in DMSO), tris(2-carboxyethyl)phosphine hydrochloride (TCEP) (1 mM, 50 mM freshly prepared stock solution in deionized water), tris[(1-benzyl-1H-1,2,3-triazol-4-yl)methyl]amine (TBTA) (100 μM, 10 mM stock solution in DMSO) and CuSO4·5H2O (1 mM, 50 mM freshly prepared stock solution in deionized water)] for 1 h at 4 °C on a nutating mixer. The beads were washed (3 × 1 mL) with ice-cold modified RIPA lysis buffer, resuspended in 40 μL loading buffer [27.5 μL (4% SDS, 50 mM triethanolamine pH 7.4, 150 mM NaCl), 10 μL 4 × SDS-loading buffer (40% glycerol, 200 mM Tris-HCl pH 6.8, 8% SDS, 0.4% bromophenol blue) and 2.5 μL 0.5 M Bond-Breaker TCEP Solution (Thermo Scientific)], heated for 5 min at 95 °C, and 20 μL of the supernatant was loaded on 2 separate SDS-PAGE gels (4–20% Bio-Rad Criterion Tris-HCl gel), one for fluorescence detection and the other for immunoblotting.For cell lysates, 50 μg proteins were clicked in 47 μl SDS-buffer (4% SDS, 50 mM triethanolamine pH 7.4, 150 mM NaCl) with 3 μL freshly premixed click reaction cocktail (same as above) for 1 h at room temperature. Proteins were precipitated by adding ice-cold methanol (1 mL), placing at −80 °C overnight, centrifuging at 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 g for 10 min at 4 °C and discarding the supernatant. The protein pellets were allowed to air-dry, resuspended in 50 μL loading buffer (same as above), heated for 5 min at 95 °C, and 20 μg of protein was loaded on 2 separate SDS-PAGE gels.
000 g for 10 min at 4 °C and discarding the supernatant. The protein pellets were allowed to air-dry, resuspended in 50 μL loading buffer (same as above), heated for 5 min at 95 °C, and 20 μg of protein was loaded on 2 separate SDS-PAGE gels.
In-gel fluorescence imaging
After SDS-PAGE separation, the gel was soaked in destaining solution (40% MeOH, 10% acetic acid, 50% H2O) overnight at 4 °C on an orbital shaker, rehydrated with deionized water and visualized by scanning the gel on an Amersham Biosciences Typhoon 9400 variable mode imager (excitation 532 nm, 580 nm filter, 30 nm band-pass).Immunoblotting
Proteins separated by SDS-PAGE were transferred (50 mM Tris, 50 mM glycine, 0.1% SDS, 10% MeOH in deionized water, Bio-Rad Trans-Blot Semi-Dry Cell, 20 V, 45 min) onto a nitrocellulose membrane (0.45 μm, Bio-Rad) which was subsequently blocked (10% non-fat dried milk, 2% BSA and 0.1% Tween-20 in PBS) overnight at 4 °C on an orbital shaker. The membrane was washed thrice with PBST (0.1% Tween-20 in PBS), and incubated with either anti-Ras (RAS10) mouse mAb (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4000, Upstate), anti-HA (HA-7) mouse mAb (1
4000, Upstate), anti-HA (HA-7) mouse mAb (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2000, Sigma), or anti-GFP (JL-8) mouse mAb (1
2000, Sigma), or anti-GFP (JL-8) mouse mAb (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000, Clontech) for 1 h at room temperature in PBST. The membrane was washed thrice with PBST, incubated with anti-mouse (H + L) donkey HRP-conjugated antibody (Jackson IR, 1
1000, Clontech) for 1 h at room temperature in PBST. The membrane was washed thrice with PBST, incubated with anti-mouse (H + L) donkey HRP-conjugated antibody (Jackson IR, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 15
15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000) for 1 h at room temperature in 5% non-fat dried milk and 1% BSA in PBST, washed thrice with PBST, and subsequently developed with ECL Western blotting detection reagents (Thermo Scientific).
000) for 1 h at room temperature in 5% non-fat dried milk and 1% BSA in PBST, washed thrice with PBST, and subsequently developed with ECL Western blotting detection reagents (Thermo Scientific).
Acknowledgements
G.C. thanks the Rockefeller/Sloan-Kettering/Cornell Tri-Institutional Program in Chemical Biology. J.S.Y. is supported by the Irving Institute Fellowship Program of the Cancer Research Institute. H.C.H. acknowledges support from Ellison Medical Foundation and NIH/NIGMS 1R01 GM087544-01A2.References
- M. D. Resh, Nat. Chem. Biol., 2006, 2, 584–590 CrossRef CAS.
- P. A. Konstantinopoulos, M. V. Karamouzis and A. G. Papavassiliou, Nat. Rev. Drug Discovery, 2007, 6, 541–555 CrossRef CAS.
- R. T. Eastman, F. S. Buckner, K. Yokoyama, M. H. Gelb and W. C. Van Voorhis, J. Lipid Res., 2006, 47, 233–240 CAS.
- R. B. Lobell, Adv. Immunol., 1998, 68, 145–189 Search PubMed.
- O. Rocks, A. Peyker, M. Kahms, P. J. Verveer, C. Koerner, M. Lumbierres, J. Kuhlmann, H. Waldmann, A. Wittinghofer and P. I. Bastiaens, Science, 2005, 307, 1746–1752 CrossRef CAS.
- M. Fivaz and T. Meyer, J. Cell Biol., 2005, 170, 429–441 CrossRef CAS.
- T. G. Bivona, S. E. Quatela, B. O. Bodemann, I. M. Ahearn, M. J. Soskis, A. Mor, J. Miura, H. H. Wiener, L. Wright, S. G. Saba, D. Yim, A. Fein, I. Perez de Castro, C. Li, C. B. Thompson, A. D. Cox and M. R. Philips, Mol. Cell, 2006, 21, 481–493 CrossRef CAS.
- G. R. Hoffman, N. Nassar and R. A. Cerione, Cell, 2000, 100, 345–356 CrossRef CAS.
- Y. An, Y. Shao, C. Alory, J. Matteson, T. Sakisaka, W. Chen, R. A. Gibbs, I. A. Wilson and W. E. Balch, Structure, 2003, 11, 347–357 CrossRef CAS.
- Y. Marrari, M. Crouthamel, R. Irannejad and P. B. Wedegaertner, Biochemistry, 2007, 46, 7665–7677 CrossRef CAS.
- T. Mulligan, H. Blaser, E. Raz and S. A. Farber, Cell. Signalling, 2010, 22, 221–233 CrossRef CAS.
- A. E. Rusinol and M. S. Sinensky, J. Cell Sci., 2006, 119, 3265–3272 CrossRef CAS.
- S. G. Young, M. Meta, S. H. Yang and L. G. Fong, J. Biol. Chem., 2006, 281, 39741–39745 CrossRef CAS.
- S. Maurer-Stroh and F. Eisenhaber, GenomeBiology, 2005, 6, R55 CrossRef.
- C. A. Rowell, J. J. Kowalczyk, M. D. Lewis and A. M. Garcia, J. Biol. Chem., 1997, 272, 14093–14097 CrossRef CAS.
- G. C. Prendergast, Nat. Rev. Cancer, 2001, 1, 162–168 CrossRef CAS.
- S. Maurer-Stroh, M. Koranda, W. Benetka, G. Schneider, F. L. Sirota and F. Eisenhaber, PLoS Comput. Biol., 2007, 3, e66 CrossRef.
- R. N. Hannoush and J. Sun, Nat. Chem. Biol., 2010, 6, 498–506 CrossRef CAS.
- G. Charron, J. Wilson and H. C. Hang, Curr. Opin. Chem. Biol., 2009, 13, 382–391 CrossRef CAS.
- B. Dursina, R. Reents, C. Delon, Y. Wu, M. Kulharia, M. Thutewohl, A. Veligodsky, A. Kalinin, V. Evstifeev, D. Ciobanu, S. E. Szedlacsek, H. Waldmann, R. S. Goody and K. Alexandrov, J. Am. Chem. Soc., 2006, 128, 2822–2835 CrossRef CAS.
- U. T. Nguyen, Z. Guo, C. Delon, Y. Wu, C. Deraeve, B. Franzel, R. S. Bon, W. Blankenfeldt, R. S. Goody, H. Waldmann, D. Wolters and K. Alexandrov, Nat. Chem. Biol., 2009, 5, 227–235 CrossRef CAS.
- J. A. Prescher and C. R. Bertozzi, Nat. Chem. Biol., 2005, 1, 13–21 CrossRef CAS.
- M. W. Rose, N. D. Rose, J. Boggs, S. Lenevich, J. Xu, G. Barany and M. D. Distefano, J. Pept. Res., 2005, 65, 529–537 CrossRef CAS.
- B. P. Duckworth, Z. Zhang, A. Hosokawa and M. D. Distefano, ChemBioChem, 2007, 8, 98–105 CrossRef CAS.
- M. W. Rose, J. Xu, T. A. Kale, G. O'Doherty, G. Barany and M. D. Distefano, Biopolymers, 2005, 80, 164–171 CrossRef CAS.
- A. F. Berry, W. P. Heal, A. K. Tarafder, T. Tolmachova, R. A. Baron, M. C. Seabra and E. W. Tate, ChemBioChem, 2010, 11, 771–773 CrossRef CAS.
- Y. Kho, S. C. Kim, C. Jiang, D. Barma, S. W. Kwon, J. Cheng, J. Jaunbergs, C. Weinbaum, F. Tamanoi, J. Falck and Y. Zhao, Proc. Natl. Acad. Sci. U. S. A., 2004, 101, 12479–12484 CrossRef CAS.
- L. N. Chan, C. Hart, L. Guo, T. Nyberg, B. S. Davies, L. G. Fong, S. G. Young, B. J. Agnew and F. Tamanoi, Electrophoresis, 2009, 30, 3598–3606 CrossRef CAS.
- G. Charron, M. M. Zhang, J. S. Yount, J. Wilson, A. S. Raghavan, E. Shamir and H. C. Hang, J. Am. Chem. Soc., 2009, 131, 4967–4975 CrossRef CAS.
- Y. Y. Yang, J. M. Ascano and H. C. Hang, J. Am. Chem. Soc., 2010, 132, 3640–3641 CrossRef CAS.
- M. Grammel, M. M. Zhang and H. C. Hang, Angew. Chem., Int. Ed., 2010, 49, 5970–5974 CAS.
- A. Hosokawa, J. W. Wollack, Z. Zhang, L. Chen, G. Barany and M. D. Distefano, Int. J. Pept. Res. Ther., 2007, 13, 345–354 Search PubMed.
- V. J. Davisson, A. B. Woodside, T. R. Neal, K. E. Stremler, M. Muehlbacher and C. D. Poulter, J. Org. Chem., 1986, 51, 4768–4779 CrossRef CAS.
- M. A. Stein, K. Y. Leung, M. Zwick, F. Garcia-del Portillo and B. B. Finlay, Mol. Microbiol., 1996, 20, 151–164 CrossRef CAS.
- C. R. Beuzon, S. Meresse, K. E. Unsworth, J. Ruiz-Albert, S. Garvis, S. R. Waterman, T. A. Ryder, E. Boucrot and D. W. Holden, EMBO J., 2000, 19, 3235–3249 CrossRef CAS.
- E. Boucrot, C. R. Beuzon, D. W. Holden, J. P. Gorvel and S. Meresse, J. Biol. Chem., 2003, 278, 14196–14202 CrossRef CAS.
- A. T. Reinicke, J. L. Hutchinson, A. I. Magee, P. Mastroeni, J. Trowsdale and A. P. Kelly, J. Biol. Chem., 2005, 280, 14620–14627 CrossRef CAS.
- B. T. Kinsella and W. A. Maltese, J. Biol. Chem., 1992, 267, 3940–3945 CAS.
- C. C. Farnsworth, M. C. Seabra, L. H. Ericsson, M. H. Gelb and J. A. Glomset, Proc. Natl. Acad. Sci. U. S. A., 1994, 91, 11963–11967 CAS.
- B. T. Kinsella, R. A. Erdman and W. A. Maltese, J. Biol. Chem., 1991, 266, 9786–9794 CAS.
- S. C. Falsetti, D. A. Wang, H. Peng, D. Carrico, A. D. Cox, C. J. Der, A. D. Hamilton and S. M. Sebti, Mol. Cell. Biol., 2007, 27, 8003–8014 CrossRef CAS.
- A. C. Berzat, J. E. Buss, E. J. Chenette, C. A. Weinbaum, A. Shutes, C. J. Der, A. Minden and A. D. Cox, J. Biol. Chem., 2005, 280, 33055–33065 CrossRef CAS.
- R. Kang, J. Wan, P. Arstikaitis, H. Takahashi, K. Huang, A. O. Bailey, J. X. Thompson, A. F. Roth, R. C. Drisdel, R. Mastro, W. N. Green, J. R. Yates 3rd, N. G. Davis and A. El-Husseini, Nature, 2008, 456, 904–909 CrossRef CAS.
Footnotes |
| † This article is part of a themed issue of Molecular BioSystems on Post-translational modifications. |
| ‡ Electronic supplementary information (ESI) available: Fig. S1 and S2; synthesis details. See DOI: 10.1039/c0mb00183j |
| This journal is © The Royal Society of Chemistry 2011 |
