Proteolytic Activity Matrix Analysis (PrAMA) for simultaneous determination of multiple protease activities†‡
Miles A.
Miller
a,
Layla
Barkal
a,
Karen
Jeng
a,
Andreas
Herrlich
b,
Marcia
Moss
c,
Linda G.
Griffith
a and
Douglas A.
Lauffenburger
*a
aDepartment of Biological Engineering, Massachusetts Institute of Technology, Cambridge, MA 02139. E-mail: lauffen@mit.edu
bWhitehead Institute for Biomedical Research, Cambridge, MA 02139
cBiozyme, Inc., USA
First published on 23rd December 2010
Abstract
Matrix metalloproteinases (MMPs) and A Disintegrin and Metalloproteinases (ADAMs) are two related protease families that play key roles in matrix remodeling and growth factor ligand shedding. Directly ascertaining the proteolytic activities of particular MMPs and ADAMs in physiological environments in a non-invasive, real-time, multiplex manner remains a challenge. This work describes Proteolytic Activity Matrix Analysis (PrAMA), an integrated experimental measurement and mathematical analysis framework for simultaneously determining the activities of particular enzymes in complex mixtures of MMPs and ADAMs. The PrAMA method interprets dynamic signals from panels of moderately specific FRET-based polypeptide protease substrates to deduce a profile of specific MMP and ADAM proteolytic activities. Deconvolution of signals from complex mixtures of proteases is accomplished using prior data on individual MMP/ADAM cleavage signatures for the substrate panel measured with purified enzymes. We first validate PrAMA inference using a compendium of roughly 4000 measurements involving known mixtures of purified enzymes and substrates, and then demonstrate application to the live-cell response of wildtype, ADAM10−/−, and ADAM17−/− fibroblasts to phorbol ester and ionomycin stimulation. Results indicate PrAMA can distinguish closely related enzymes from each other with high accuracy, even in the presence of unknown background proteolytic activity. PrAMA offers a valuable tool for applications ranging from live-cellin vitro assays to high-throughput inhibitor screening with complex enzyme mixtures. Moreover, our approach may extend to other families of proteases, such as caspases and cathepsins, that also can lack highly-specific substrates.
Insight, innovation, integrationMetalloproteinases (MPs) impact diverse biological processes, and extensive post-translational regulations coupled with a complex repertoire of MP substrate preferences have hindered understanding of MP biology. Although methods exist to study MP activity, they typically face a trade-off between invasiveness and specificity. Here, we present a combined mathematical and experimental framework to measure real-time MP activity non-invasively, demonstrating capability for discerning specific MP activities from complex mixtures and biological samples. We develop and validate the approach by analyzing an extensive compendium of measurements using purified MPs, and apply it to several well-studied cell lines. This approach can adapt to a broad range of applications, and we show how one can a priori design studies of this type to meet particular objectives. |
Introduction
Matrix metalloproteinases (MMPs) comprise a family of 23 zinc-dependent endopeptidases that are part of the Metzincin family of enzymes and are generally active on or near the cell surface.1,2 As central regulators of extracellular microenvironments throughout the human body, MMPs play key roles in normal physiological processes including development,3angiogenesis,4,5tissue remodeling,6 wound repair, and inflammation.7 On the other hand, they are also implicated in a wide array of pathologies, ranging from cancer, tumor invasion, and metastasis,8 to respiratory diseases such as asthma and chronic obstructive pulmonary disease (COPD).9MMP proteolytic activities are tightly controlled. Once active, certain MMPs (e.g., MMP2) have been demonstrated to act on hundreds of endogenous substrates.10,11MMP substrates include signaling molecules (e.g., cytokines, chemokines, growth factors, GCPRs, growth factor receptors, and cytokine receptors), extracellular matrix components (e.g., collagen, laminin, and fibronectin), cell adhesion molecules, clotting and complement cascade proteins, and proteases themselves.12–15Closely related to MMPs, ADAM (A Disintegrin and Metalloproteinase) enzymes are metalloproteinases (MPs) within the Metzincin family that are mostly bound at the cell surface.16 At least 13 ADAMs existing in humans have intact metalloproteinase domains and proteolytic activity.14 ADAMs mediate various cellular behaviors including migration, adhesion,17 proliferation,15 and apoptosis.16,18 Similar to MMPs, ADAM family enzymes are found throughout the body and support diverse physiological processes such as development15 and angiogenesis.5,16 Likewise, ADAMs can become dysregulated in a variety of diseases and play roles in pathologies including cancer,1,18 inflammatory bowel disease, and asthma.16 Current research suggests that ADAMs have a narrower repertoire of substrates compared to MMPs, and principally function to shed the ectodomain of surface-bound proteins such as growth factor ligands, growth factor receptors, cell adhesion molecules, and cytokine receptors.14,15
Three key properties of MP biology have created the need for methods that directly observe protease activity in a specific, non-invasive, real-time, and multiplex manner. First, the extensive amount of post-translational modification and regulatory mechanisms controlling MP proteolytic activity make direct activity measurements a valuable and complementary addition to common methods of assessing protein function, such as western blotting, immunohistochemistry, and genetic manipulation.19–22 Second, the plethora of endogenous substrates cleaved by certain MPs, the context dependency of endogenous substrate cleavage, and the overlapping endogenous substrate specificity of closely related MPs make it difficult to quantitatively match the contributions of specific proteases to global observations of endogenous substrate degradation.11,19,23 Quantitative determination of selected protease activities would complement measurements that focus on endogenous substrate cleavage, thereby facilitating attempts to match particular proteolytic activities to patterns of substrate degradation. Third, cyclical feedback interactions and compensatory mechanisms among closely related MPs can severely complicate the interpretation of protease network interactions.15,24,25 Non-invasive and multiplexed measurement of MP activity would allow for the assessment of protease network interactions without artificially biasing the underlying network structure.
While many useful methodologies currently exist to study MPs, unfortunately none simultaneously allow for direct, non-invasive, multiplex, real-time measurements of specific protease activity. In general, existing methods such as zymography, activity based probes, and mass-spectrometry based methods all must choose between invasiveness, specificity, and throughput.10,26–30 Synthetic polypeptide protease substrates have been extensively developed to directly assess MP activity in a non-invasive and real-time manner.20,31–34 These substrates typically consist of a fluorescence resonance energy transfer (FRET) donor and quencher fluorophore that are separated by a 3–10 amino acid linker containing a protease cleavage motif. Upon cleavage of the polypeptide linker, the donor fluorophore separates from the quencher and fluorescence increases. Protease activity dynamics can then be tracked by observing the change in fluorescence over time. Similarly to many endogenous MP substrates, MP FRET-substrates are generally cleaved by multiple closely related proteases.32,35,36 Several strategies, including positional scanning of synthetic combinatorial libraries35,37 and directed evolution using phage display38 have attempted to optimize substrate sequences such that they are more selectively cleaved by a specific protease. Combinations of multiple substrates and inhibitors have also been implemented to increase specificity.39 These strategies often succeed at distinguishing between two or a few proteases, but cross-reactivity nevertheless remains problematic in more complex mixtures.35,40,41
This work describes an approach we term ‘Proteolytic Activity Matrix Analysis’ (PrAMA) as a method of using panels of FRET-substrates to infer a dynamic, quantitative, and specific profile of MMP and ADAM proteolytic activities. PrAMA ascertains specific protease activity by deconvoluting from measurements derived from relatively non-specific FRET-substrates, employing prior knowledge of individual MMP/ADAM cleavage signatures ascertained using purified enzymes. This approach allows PrAMA to elucidate particular enzyme activities from cleavage signatures obtained in complex samples containing multiple proteases. The integrated experimental measurement and mathematical analysis framework exploits the advantages of FRET-substrates, which support non-invasive real-time measurements of live-cell activity, while addressing their problems of limited specificity and multiplexing. Peptide library microarrays have been previously implemented to assess global patterns of protease activity and infer specific protease activity.42 Nevertheless, PrAMA's novel combination of mathematical and experimental methodologies allows for greater quantification of protease activity, lower expense, and higher throughput compared to microarray-based approaches. Ultimately, PrAMA fills a niche that complements many other current methods of assaying MP activity and substrate degradation. This niche is especially important for multivariate network-level analysis, where the ability to simultaneously measure multiple MP activities in a non-invasive, real-time, and relatively high-throughput manner confers the greatest benefits. We anticipate that such network-level approaches will be valuable for designing clinical trials focused on MMPs and for illuminating unintended consequences of the many trials that have failed in the last decade.11,43
We present a compendium on the order of 4000 measurements involving mixtures of FRET-substrates and purified recombinant MPs, and use these measurements to both construct the PrAMA inference parameters and test the limits of PrAMA inference accuracy. A priori determination of the PrAMA inference parameters can predict optimal subsets of substrates for distinguishing particular MPs from each other. We demonstrate PrAMA as being capable of accurately inferring MP activity even in the presence of background protease activities. Finally, we apply PrAMA to assess the live-cell proteolytic response of wildtype, ADAM10−/−, and ADAM17−/− mouse embryonic fibroblasts (MEFs) to phorbol ester and ionomycin stimulation. Overall, this work presents the foundation, validation, and theoretical analysis of a general methodology that has potential applications ranging from systems biology to in vitroinhibitor screening.
Materials and methods
Materials
Recombinant human ADAMs 8, 9, 10, and 17 were purchased from R & D Systems. The catalytic domains of the following recombinant human enzymes were purchased from Enzo Life Sciences: ADAM12 and MMPs 1, 2, 3, 7, 8, 9, 10, 12, 13, and 14. MMP9 Inhibitor I (Cat. No. 444278) was purchased from Calbiochem. GM6001 was obtained from Enzo Life Sciences. Recombinant human TNFα and EGF were obtained from Millipore (Billerica, MA). 18 FRET-substrates were obtained from BioZyme, Inc. In this work, we refer to substrates as numbers 1–18, and these reference numbers correspond to polypeptide sequences listed in Table S1.‡ We performed time-lapse fluorimetry using 384-well OptiPlates from Perkin-Elmer and the Spectromax M3 and M2e fluorimeters (Molecular Devices). We used excitation and emission wavelengths of 485 nm and 530 nm, respectively, for all experiments.Substrate assays with purified enzymes
For all experiments, substrates were diluted from 5 mM stock in dimethyl sulfoxide (DMSO) to a final concentration of 10 μM in the appropriate assay buffer. We conducted experiments in four different assay buffers. ‘ADAM buffer’ consists of 20 mM Tris, pH 8.0, and 6 × 10−4% Brij-35. ADAM buffer also includes 10 mM CaCl2 for experiments involving ADAM8. ‘MMP buffer’ consists of 50 mM Tris, pH 7.5, 150 mM NaCl, 2 mM CaCl2, 5 μM ZnSO4, and 0.01% Brij-35. We obtained the third buffer, Clonetics™ Mammary Epithelial Cell Basal Media (‘MEBM’), as a phenol-red and serum free solution from Lonza, pH 7.4. Final active concentration of MMPs and ADAMs in activity assays ranged from 0.01 nM to 7.5 nM.As a fourth buffer, we spiked purified MMP into the conditioned media from the MDA-MB-231 cell line, which is an estrogen receptor negative (ER−) breast cancer cell line derived from the pleural effusion of a breast cancer patient. We obtained these cells from the American Type Culture Collection (ATCC, Manassas, VA) and routinely cultured them at 37 °C, 5% CO2, in DMEM supplemented with 10% foetal calf serum, 100 U ml−1penicillin, 100 μg ml−1streptomycin, 4 mM L-glutamine, and 4.5 g L−1D-glucose. We collected cell supernatant under the following conditions: cells were grown to 80% confluency in 10 cm tissue-culture treated polystyrene plates obtained from Corning Life Sciences (Lowell, MA), serum-starved for 4 h in basal media consisting of DMEM supplemented with penicillin/streptomycin, and stimulated with basal media supplemented with either 10 ng ml−1TNFα or 10 ng ml−1epidermal growth factor (EGF). Supernatant was collected at 12 h, spun down at 200 g for 5 min to remove debris, and immediately flash-frozen. For FRET-substrate assays involving this supernatant, final reactions were composed of a 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 mixture of 20 μM substrate diluted from 5 mM DMSO stock into phosphate buffered saline, 4 nM active MMP7 diluted in ‘MMP buffer’, and thawed supernatant.
1 mixture of 20 μM substrate diluted from 5 mM DMSO stock into phosphate buffered saline, 4 nM active MMP7 diluted in ‘MMP buffer’, and thawed supernatant.
We determined active site concentrations by comparing observed cleavage rates to previously published catalytic efficiencies for the same substrates in either ‘MMP buffer’ or ‘ADAM buffer’.32,44 In some cases we performed active site titration with GM6001 to either confirm this comparison or to substitute it when comparison was unavailable. Activity data for active site titrations were fit to the Morrison equation using non-linear least squares curve-fitting (see below). We normalized substrate concentration to a positive control, comprised of 10 μM substrate incubated with 0.5% trypsin and 0.2% EDTA (Sigma). Almost all experiments were performed in technical triplicate, except for the MMP7 dilution series and the experiments involving cell supernatant, which both were performed in technical duplicate. For the experiments in triplicate, we excluded clear outliers in a few cases (<10% of all triplicates) using Dixon's Q-test with a 90% threshold. We performed all experiments at 37 °C. In general, readings measured fluorescence approximately every half-hour for roughly five hours. All experimental data are provided in the ESI.‡ We conducted all computational work using Matlab (2009a, MathWorks, Natick, MA).
Live-cell substrate assays
Mouse embryonic fibroblasts (MEF cells) were a kind gift from Dr Carl Blobel (Hospital for Special Surgery, New York, USA) and Dr Paul Saftig (University of Kiel, Germany). ADAM1045 and ADAM1719 knockout fibroblasts were derived from E9.5 and E13.5 embryos, respectively. Cells were maintained at 37 °C, 5% CO2, in DMEM supplemented with 10% foetal calf serum, 100 U ml−1penicillin, 100 μg ml−1streptomycin, 4 mM L-glutamine, and 4.5 g L−1D-glucose. For PrAMA, we plate 5000 cells per well in clear-bottom 384-well polystyrene plates from Thermo Scientific (roughly 70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells cm−2). The following day, we change media and add one of seven FRET substrates (substrates 1, 2, 5, 6, 7, 9, and 15) at 10 μM to each well, along with either 1 μM phorbol 12-myristate 13-acetate (PMA), 10 μM ionomycin (IM), or a DMSO control. In all cases media contained <1% DMSO. Following addition of substrate, cells were imaged at 20–40 min intervals and at 37 °C for 2 h. We performed experiments in biological quadruplicate and excluded data lying more than 1.5 standard deviations from the mean (at most one sample per quadruplicate).
000 cells cm−2). The following day, we change media and add one of seven FRET substrates (substrates 1, 2, 5, 6, 7, 9, and 15) at 10 μM to each well, along with either 1 μM phorbol 12-myristate 13-acetate (PMA), 10 μM ionomycin (IM), or a DMSO control. In all cases media contained <1% DMSO. Following addition of substrate, cells were imaged at 20–40 min intervals and at 37 °C for 2 h. We performed experiments in biological quadruplicate and excluded data lying more than 1.5 standard deviations from the mean (at most one sample per quadruplicate).
Enzyme kinetics modeling
We model enzyme kinetics as an extension of the classical Michaelis–Menten (M–M) model, where the initial rate of cleavage, V0, is assumed to be constant and is defined as the following: V0 = Ci,j[Si]0[Ej]. Ci,j describes the catalytic efficiency, kcat/Km, with which the jth enzyme [Ej] cleaves the ith substrate [Si]. This model assumes Km ≫ [S], which has been experimentally confirmed for several substrates.46 We assume minimal inner-filter effect, although this can become significant at substrate concentrations above those used in this work (>20 μM).47 In all experiments we aim to infer either the initial rate of substrate cleavage (V0) or the catalytic efficiency (Ci,j) in a reaction, depending on whether [E] is known or unknown (i.e., ‘self-blinded’ in this work), respectively. We infer these parameters by fitting a kinetic model to the time-lapse fluorimetry data, where fluorescence Fp(t) indicates product formation. Typically, inference using M–M kinetics involves fitting early time-points to the linear M–M model Fp(t) = (V0t + B)F0/[S]0, where B is the background signal and F0 is the peak fluorescence from the positive control (described above). We extend the linear M–M description to a non-linear ‘depletion-decay’ model. We let [S]/[S]0 = 1 − Fp/F0, and describe the observed fluorescence, Fobs, in the following equations:| dFobs/dt = V0(F0 − Fp)/[S]0 − kdFobs | (1) |
| dFp/dt = V0(F0 − Fp)/[S]0 | (2) |
| Fobs(t) = F0V0(e−V0t/[S]0)(kd[S]0 − V0)−1 + Ae−kdt | (3) |
| A = ekdT0F0[(1 − e−V0T0/[S]0) − V0(e−V0T0/[S]0)(kd[S]0 − V0)−1] | (4) |
We fit model parameters in several steps. First, we subtract the signal of a negative control (FRET-substrate only) from all other signals. The maximum fluorescence in the positive control, which is generally at the first time point, indicates F0. We determine kd from the negative slope of the log-transformed positive control 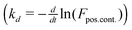 . We obtain the remaining two parameters (V0 and T0) by non-linear curve-fitting. In several cases, we explicitly measured T0 and compared model fitting with and without explicitly defining that parameter. Results indicate that V0 inference remains consistent regardless of whether T0 is inferred or measured. In cases where [E] is known (i.e. not blinded), we calculate Ci,j by the following relation: Ci,j = V0/([Si]0[Ej]).
. We obtain the remaining two parameters (V0 and T0) by non-linear curve-fitting. In several cases, we explicitly measured T0 and compared model fitting with and without explicitly defining that parameter. Results indicate that V0 inference remains consistent regardless of whether T0 is inferred or measured. In cases where [E] is known (i.e. not blinded), we calculate Ci,j by the following relation: Ci,j = V0/([Si]0[Ej]).
PrAMA inference
PrAMA inference uses panels of FRET-substrate cleavage measurements, coupled with known catalytic efficiencies, Ci,j, for all relevant i substrates and j enzymes, to infer specific protease activity from a complex mixture of unknown enzymes. PrAMA operates under the assumption that total observed cleavage for the ith substrate V0,i in a mixture of enzymes is the summation of cleavages from each individual protease in that mixture:| V0,i = [Si]0∑Ci,j[Ej] | (5) |
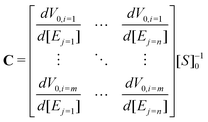 | (6) |
| VT0 = [S]0CET | (7) |
Various methods can be applied to solve eqn (7). In this work, we implement a non-negative least squares algorithm combined with inference sensitivity analysis. We employ sensitivity analysis to quantitatively determine robustness to experimental error and to tune inference sensitivity and specificity. PrAMA inference involves three main procedures once V0 and C have been measured. First, we perform a bootstrapping scheme of randomly generating an ensemble of 1000 cleavage vectors Vs0. Sampling is from a log-normal distribution with μ = V0 and a standard deviation representative of the average variance between the experimentally observed and PrAMA expected cleavage rates obtained from PrAMA validation sets of data. Second, we use least squares to solve eqn (7) for every Vs0 in the sampling ensemble, and compute the mean and standard deviation of the ensemble inference results E. In some cases, we added artificial error to C for each iteration of the bootstrapping scheme, representative of experimentally observed parameter uncertainty. This additional process did not significantly improve PrAMA inference, however, and was excluded unless stated otherwise.
As the third step, we apply a robustness filter to the inference results to tune specificity. This filter, termed the σT threshold, roughly defines specific protease activities as significant if they are inferred in more than a certain percentage of the ensemble of inferences. We scale σT as a fraction of the inference standard deviation that is subtracted from the mean inference value. For example, setting σT = 1.0 roughly defines protease activity as significant if observed in at least 84% of the ensemble inference results.
Results
Characterization of the non-linear kinetic model
PrAMA infers specific protease activity levels from panels of kinetic cleavage measurements using FRET-based polypeptide substrates (see Fig. 1 for a schematic & illustration of the procedure). The first steps of PrAMA involve determining the kinetic parameters V0 and C from time-lapse fluorimetry data (see Methods). To establish these two parameters, we employ a ‘depletion-decay’ kinetic model of substrate cleavage that elaborates on linear M–M kinetics to account for first-order photobleaching decay and substrate depletion. Typical raw time-course fluorimetry output for a mixture of enzymes and substrates can be fit by both the linear and decay-depletion models reasonably well (Fig. 2A). However, the often subtle non-linearity of the data can reveal significant differences in the underlying kinetics. For the time-course in Fig. 2A, the cleavage rate V0 inferred from the decay-depletion model is roughly 50% greater than that inferred when using the linear approximation. For a systematic comparison of the two models, we inferred trypsin activity from time-lapse fluorimetry measurements across 2–3 orders of magnitude in enzyme concentration, using both the linear and decay-depletion models (Fig. 2B). With log-log scaling, the R2 coefficient of determination for the decay-depletion model in Fig. 2B is 0.9, compared to only R2 = 0.25 for the linear model. Disparities in inference accuracy lie almost entirely at the extremely high and low enzyme concentration range, where substrate depletion and photobleaching, respectively, become most significant. In effect, the decay-depletion model increases the range over which protease activities can be quantitatively and accurately measured by over an order of magnitude.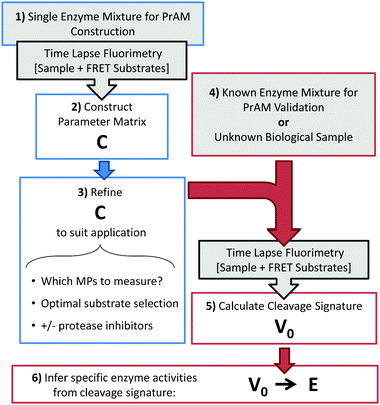 | ||
| Fig. 1 PrAMA overview. Blue indicates PrAMA development & construction, red indicates PrAMA implementation, and grey indicates experimental preparation & procedure. | ||
![Modeling
protease
cleavage kinetics and specificity signatures. (A) A typical time-lapse fluorimetry output for a single enzyme (0.5 nM MMP10) and substrate 13 in MMP buffer. The data is fit with both linear and decay-depletion kinetic models. The decay-depletion model almost perfectly overlays the data. (B) Inferred kinetic rates (V0/[S]0) of trypsin cleaving substrate 7 over a range of concentrations. (C) Decay-depletion model simulations of substrate cleavage using the following parameters: F0 = 104FLU, kcat/Km = 105 M−1s−1, [E] = 1 nM, [S] = 10 μM. (D) Hierarchical biclustering of observed cleavage efficiencies for various ADAMs and MMPs. Cleavage parameters were averaged over several concentrations. (E,F) PrAMA inference of individual MMPs at different concentrations (E, ∼0.5 nM vs. ∼0.05 nM) and buffers (F, MMP minimal media vs.MEBM). Each column represents an individual PrAMA experiment. The abscissa indicates the actual enzyme present, and the ordinate indicates the inferred MMP present. Each PrAMA output (each column) is normalized to have a total signal of 1.](/image/article/2011/IB/c0ib00083c/c0ib00083c-f2.gif) | ||
| Fig. 2 Modeling protease cleavage kinetics and specificity signatures. (A) A typical time-lapse fluorimetry output for a single enzyme (0.5 nM MMP10) and substrate 13 in MMP buffer. The data is fit with both linear and decay-depletion kinetic models. The decay-depletion model almost perfectly overlays the data. (B) Inferred kinetic rates (V0/[S]0) of trypsin cleaving substrate 7 over a range of concentrations. (C) Decay-depletion model simulations of substrate cleavage using the following parameters: F0 = 104FLU, kcat/Km = 105 M−1s−1, [E] = 1 nM, [S] = 10 μM. (D) Hierarchical biclustering of observed cleavage efficiencies for various ADAMs and MMPs. Cleavage parameters were averaged over several concentrations. (E,F) PrAMA inference of individual MMPs at different concentrations (E, ∼0.5 nM vs. ∼0.05 nM) and buffers (F, MMP minimal media vs.MEBM). Each column represents an individual PrAMA experiment. The abscissa indicates the actual enzyme present, and the ordinate indicates the inferred MMP present. Each PrAMA output (each column) is normalized to have a total signal of 1. | ||
Several factors make photobleaching a significant issue in this work. We conduct protease activity assays using fluorescein-based FRET substrates over long (often >5 h) time scales. Fluorescein is relatively sensitive to photobleaching, and long time scales further amplify photo-sensitivity effects. In this application we typically read fluorescence every 15–30 min and observe resultant first-order decay constants (kd values) as high as 10−4 s−1. Computational simulations using the decay-depletion model demonstrate how significantly decay can influence the observed fluorescence (Fig. 2C). Our results indicate that even the small amount of photobleaching incurred with infrequent plate-reader measurements may result in a several-fold decrease in fluorescence after hours have elapsed.
PrAMA construction
PrAMA requires the explicit measurements of the entries of matrix C, which characterize the catalytic efficiencies with which individual enzymes cleave FRET-substrates, before specific enzyme activities can be deconvolved from complex reaction mixtures. The dimensions of C can be customized depending on the experimental application, and Fig. 2D depicts one possible configuration involving 18 FRET-substrates, 4 ADAMs, and 10 MMPs. In this figure, catalytic efficiencies range from 103 M−1 s−1 to 106 M−1s−1. Although we successfully measured cleavage rates below this range, such low signals typically have high error and ultimately have low impact on PrAMA inference. We hierarchically biclustered the elements of C with a Euclidean distance metric and average linkage, without mean-centering or variance normalizing, and with optimal leaf ordering. Several clear patterns emerge from this clustering, as indicated by the dendrograms flanking the array (Fig. 2D). MP clustering somewhat recapitulates DNA sequence based phylogenetic profiling of the enzyme families. For example, ADAMs partition from the MMPs, and closely related MMPs such as the gelatinases (MMP2 & MMP9) cluster together. The substrates form three distinct clusters: substrates with greater specificity towards ADAMs (cyan on the dendrogram); substrates with greater specificity towards MMPs (red); and substrates cleavable by both MMPs and ADAMs (blue, green to a lesser extent). More than anything, however, C explicitly indicates the lack of selectivity among these substrates. Hence, this parameter matrix underscores the need for a deconvolution process in extracting specific protease activities from FRET-substrate measurements, and C ultimately becomes integral to this task.A compendium of cleavage signature measurements
We performed roughly 4000 experiments using a variety of enzyme combinations, buffers, and substrates to measure, validate, and test the parameters and principles of PrAMA inference. We conducted a wide array of PrAMA experiments whereby panels of FRET-substrates were used to measure the cleavage signature V0 of various enzyme mixtures, including roughly 50 single enzyme mixtures, 30 double enzyme mixtures, and 10 triple enzyme mixtures. We tested several buffers, enzyme concentrations, enzyme combinations, and have presented the V0 values corresponding to these reaction conditions in Fig. 3. All experiments and inferred cleavage signatures V0 are available in the ESI.‡ We hierarchically biclustered the reaction conditions according to their mean-centered and variance-standardized cleavage signatures V0, using Euclidean distance, average linkage, and optimal leaf ordering. Clustering can often serve as an effective tool to group and/or classify objects. In this application, hierarchical clustering successfully groups over 75% of individual MMPs with themselves, based on their V0 observed at different concentrations and buffers (Fig. 3A). Clustering analysis becomes more difficult to interpret in multi-enzyme mixtures, yet some patterns still emerge (Fig. 3B–C). For example, V0 signatures from mixtures containing ADAM enzymes form two main clusters, corresponding to reactions with and without MMP present (Fig. 3C). Nevertheless, simple hierarchical clustering inadequately classifies such complicated enzyme mixtures, motivating a more effective inference methodology to ascertain individual enzyme activities.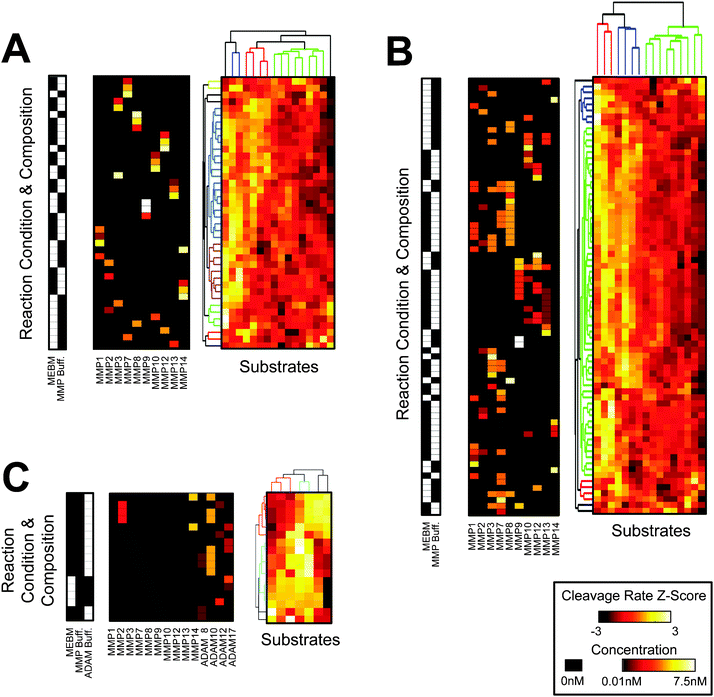 | ||
| Fig. 3 A compendium of cleavage signatures from purified proteases and protease mixtures. Hierarchical biclustering organizes reaction mixtures by their corresponding cleavage signatures, which consist of cleavage rates (s−1) of substrates (shown in columns) by various protease mixtures (shown in rows). Within each signature V0, the Z-score indicates deviation from the mean cleavage rate, following variance-standardization. Three arrays accompany each subplot A–C. The left array describes the buffer (indicated by white) that corresponds to each cleavage signature, aligned by row with the other two arrays. The middle array describes MMP concentrations, and corresponds by row with the adjacent cleavage signatures. Experiment groupings are as follows: (A) all single MMP experiments, (B) all single and mixture MMP experiments, and (C) all single and mixture experiments involving ADAMs. | ||
PrAMA classifies individual MPs
We first tested the ability of PrAMA to identify an individual enzyme based on its cleavage signature observed at one concentration and/or buffer compared to another. To demonstrate, we constructed a 16 × 5 parameter matrix C describing the cleavage of substrates 1–16 by MMPs 1, 2, 3, 7, and 8 at {0.4, 0.1, 0.9, 0.7, 1.0} nM, respectively, in MMP buffer. We used these parameters to analyze cleavage signatures V0 from the same enzymes at an order of magnitude lower concentration. Results indicate PrAMA accurately infers the specific MMP based on its cleavage signature, with an average total cross-reactivity of 11% (Fig. 2E). As a second demonstration, we used the same parameter matrix C to infer cleavage signatures corresponding to MMPs 1–8 in MEBM buffer rather than MMP buffer, again at different concentrations. Results for this analysis are even better: 4/5 PrAMA experiments show zero cross-reactivity, and one has 13% cross-reactivity between MMP3 and MMP7 (Fig. 2F).PrAMA inference strengths and weaknesses
We analyzed several properties of the parameter matrix C in order to a priori predict which MMPs PrAMA can accurately infer with high specificity. We transform C into a model covariance error matrix RM that describes inference uncertainty as a function of data uncertainty, RD, which we directly measure from the variance of replicate experiments. RM is mathematically defined as the following:| RM = (CTRDC)−1 | (8) |
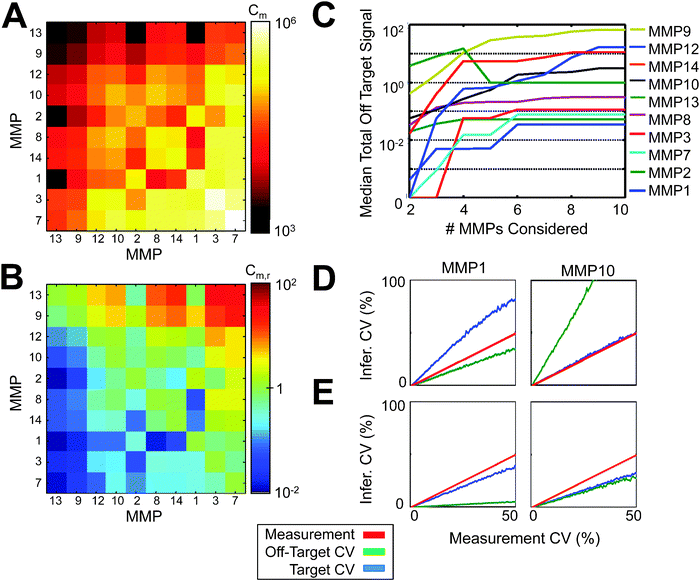 | ||
| Fig. 4 Parameter matrix error analysis. (A) Model error covariance matrix RM. (B) Relative model error matrix, RrM. (C) Median off-target PrAMA inference error as a function of the number of MMPs considered in the parameter matrix, averaged over all possible combinations of MMP subsets. (D,E) Average target and off-target inference error as functions of the synthetic measurement error, when all MMPs (D) or only MMPs 1 & 10 (E) are considered in the parameter matrix. (C–E) Parameter matrix C was constructed using enzymes at ∼0.5 nM. σT = 0 for all results here. (C) Cleavage signatures were obtained at ∼0.05 nM. | ||
The diagonal elements of RM represent on-target model uncertainties, while off-diagonal elements indicate off-target model error. To emphasize the relative amounts of on- and off-target uncertainty, we subtract the diagonal elements of RM from their respective rows to produce a “relative” model uncertainty matrix, RrM (Fig. 4B). Large positive values in RrM indicate the potential for high cross-reactivity in PrAMA inference. For example, the RrM rows for MMPs 9, 12, and 13 have large elements corresponding to MMPs 3 and 7. This suggests that signals from MMPs 9, 12, and 13 are likely to be mistakenly inferred as MMPs 3 and 7. We experimentally tested such cross-reactivity by performing PrAMA inference on MMP signals using different C configurations. We tested all combinations of MMPs considered by C, and performed PrAMA to infer MMP activity from individual enzyme mixtures (Fig. 4C). Results indicate that indeed MMPs 9, 12, and 13 have high cross-reactivity with other MMPs. MMPs 1, 2, 3, and 7 are inferred with the greatest specificity. Encouragingly, results suggest that inference cross-reactivity is relatively independent of the number of MMPs considered, past a certain point. For most MMPs, there is hardly any increase in average cross-reactivity when increasing the number of MMPs considered from 6 to 10. For MMP13, the effects of high pairwise uncertainty become diluted when more MMPs are considered, and total cross reactivity actually decreases. To test cross-reactivity as a function of experimental variability, we simulated PrAMA by inferring MMP activity from cleavage signatures generated from the columns of C, but with increasing artificial amounts of multiplicative error added to the simulated cleavage signatures. Fig. 4D shows the average results from 1000 iterations of these simulations for MMPs 1 and 10, when all MMPs are considered in the C parameter matrix. In agreement with RM and RrM, MMP1 has higher on-target error, while MMP10 has high cross-reactivity. A priori analysis of RM can suggest potential protease inhibitors to add or biophysical separation techniques to apply in order to eliminate the number of MMPs considered in a given sample. Both on- and off-target error significantly decrease when MMPs 1 and 10, which according to RM have relatively low cross-reactivity, are the only two proteases considered in PrAMA inference. Ultimately this analysis (a) reveals the potential need for additional FRET-substrates with certain specificities, (b) suggests which MMPs can be accurately and simultaneously measured in a given sample, and (c) suggests the potential use of inhibitors or supplementary experimental methods to achieve greater inference resolution. For example, RM analysis suggests the potential need for additional substrates that better distinguish MMP9 from MMP7. Although general non-specificity may be difficult to eliminate, substrates could be designed to minimize particular cross-reactivity through a bioinformatic analysis of cleavage motifs52,53 or through targeted directed evolution methods.54,55
Inference sensitivity analysis improves PrAMA accuracy
Experimental error propagates through PrAMA inference in a complex manner. Consequently, we perform a bootstrapping sensitivity analysis to directly account for observed experimental error, gauge its effect on PrAMA inference, and to tune PrAMA specificity/sensitivity (see Methods). Experimental replicates of C and V0 have an average standard deviation of 20%. Experimental variance is a mix of both additive and multiplicative error, and standard deviation drops to roughly 10% at protease activity levels above 104 M−1s−1. In general, our results suggest that correct PrAMA inference is more robust to experimental and/or artificial noise than incorrectly inferred enzyme activities (i.e. false positive results). As an example, we inferred MMP activity from a cleavage signature V0 corresponding to a reaction that contained MMP8, but with increasing synthetic multiplicative sampling error applied to the observed V0 (Fig. 5A). In this instance, MMP8 does not have the highest average inferred MP activity when synthetic sampling error is high. Nonetheless, PrAMA infers MMP8 activity with the greatest consistency compared to the other MPs considered. We developed a robustness threshold, termed the σT threshold, to take advantage of this general observation. Total cross-reactivity of the inferred MMP activity is a function of (a) the experimental and/or synthetic sampling error of the parameters V0 and C, and (b) the σT threshold. For PrAMA inference of the single-enzyme mixtures involving MMPs 1–8, total cross-reactivity can be totally eliminated by applying the appropriate σT threshold, even when the standard deviation of the applied Gaussian multiplicative error approaches 100% (Fig. 5B). As another example, the σT threshold can completely eliminate as much as 170% cross-reactivity in the inference of individual MMPs 9, 10, 12, and 13 (Fig. 5C–D).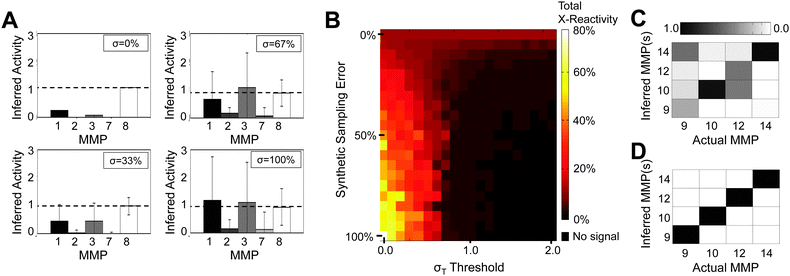 | ||
| Fig. 5 Robustness thresholds filter off-target inference. (A) Inference results for cleavage signatures obtained using MMP 8 at ∼0.05 nM and parameters obtained at ∼0.5 nM. Increasing amounts of multiplicative error was added to cleavage signatures, randomly sampled from a normal distribution with standard deviations of σ = {33, 67, 100%}. (B) Total cross-reactivity in inference results for cleavage signatures obtained using MMPs 1,2,3,7, and 8 at ∼0.5 nM and parameters obtained at ∼0.05 nM. Cross-reactivity is undefined at high sampling error and σT thresholds, when no signal falls above σT. (C,D) PrAMA inference of MMPs 9, 10, 12, and 13 at different concentrations (∼0.5 nM and ∼0.05 nM) before (C) and after (D) applying 30% sampling error and threshold σT = 2. Each column represents an individual PrAMA experiment. The abscissa indicates the actual enzyme present, and the ordinate indicates the inferred MMP present. Each PrAMA output (each column) is normalized to have a total signal of 1. | ||
PrAMA inference of MP mixtures
We tested the ability of PrAMA to infer specific protease activities from cleavage signatures V0 observed from 92 total mixtures of one, two, three, and four MPs. We analyzed 48 single-enzyme mixtures, 30 two-enzyme mixtures, 12 triple-enzyme, and 2 four-enzyme mixtures involving 14 different MPs (see Fig. 6A, and supplemental data in the ESI‡). For these mixtures, we did not consistently observe statistically significant deviation between the observed cleavage signatures V0 and the expected cleavage patterns based on PrAMA assumptions (e.g., see eqn (7)). Fig. S2–S12 show raw time-lapse fluorimetry data, inferred & expected cleavage signatures V0, and PrAMA inference results for several of these enzyme combinations.‡ | ||
| Fig. 6 PrAMA inference of enzyme mixtures. (A) Heat maps indicating PrAMA inference results using mixtures of purified enzymes. Each column corresponds to a different enzyme mixture, and rows indicate which enzymes are considered in the parameter matrix C. The top heat map shows actual enzyme activity concentrations, and the bottom two indicate inference results obtained using different σT thresholds. Each PrAMA output (each column) is normalized to have a total signal of 1. (B) ROC curve describing PrAMA accuracy for inference shown in A. True positive rate (TPR) and false positive rate (FPR) describe accuracies with which PrAMA infers whether or not an enzyme is present in the mixture. (C–D) Heat maps and ROC curves describing PrAMA inference results for mixtures involving ADAMs 10, 17, and MMP 2, 14. (E) Maximum accuracies for inference of individual enzymes in mixtures shown in A. Although 14 enzymes are considered in the parameter matrix as in A–B, here we present inference accuracy for each enzyme individually. (F) PrAMA inference results (ordinate) of MMP7 at different actual concentrations (abscissa). Inference was performed using substrates 1–16 in MMP buffer (black) and conditioned media (red). | ||
For each mixture, we define MP activity as significant if inferred at levels above a defined σT threshold. If that enzyme is actually present in the reaction mixture, then we label the inference for that specific MP as ‘true positive’. Receiver-operator characteristic (ROC) curves then summarize the total PrAMA inference results (Fig. 6A). Tuning the σT threshold moves inference results along the ROC curve to adjust the true positive and false positive rates. Inference accuracy, defined as the ratio (true positives + true negatives)/(total positives + total negatives), is maximally ∼90% for all mixtures, using a parameter matrix that considers the presence of all 14 MPs used in this study. Maximum accuracy for single-enzyme mixtures is slightly above 90%, while more complex triple and four-enzyme mixtures have inference accuracy of roughly 80%. We explored an alternative bootstrapping method in this work (see Methods), but found PrAMA results to be robust to the algorithm variation (Fig. S13‡). Fig. 6A shows PrAMA inference results with increasing levels of σT. Previous information regarding MP interactions, co-expression, and cellular localization partly informed the selection of MP mixture compositions. For example, mixtures involving membrane-bound ADAM enzymes also included MMP14, which is the only MMP we analyzed that is membrane-bound with a transmembrane domain. We also included MMP2 in these mixtures, as previous work indicates MMP14 activates MMP2 at the cell surface.56 We analyzed 5 double-enzyme and 10 single-enzyme mixtures involving ADAM10, ADAM17, MMP2, and MMP14. Fig. 6D presents PrAMA results using a parameter matrix that focuses on just these four enzymes. Maximum PrAMA inference accuracies for both the single and double enzyme mixtures are roughly 90%.
Using PrAMA results from the 92 enzyme mixtures shown in Fig. 6A–B, we analyzed which particular proteases PrAMA most reliably measures. We calculated inference accuracy for each of the 14 MPs as a function of the significance threshold σT. ROC curves (Fig. S14‡) and corresponding maximum accuracies (Fig. 6E) for each enzyme reveal ADAM17 to be the most reliably identified MP. Results indicate MMP7 and MMP3 are the most difficult enzymes to measure, and this agrees with previously discussed RM analysis (Fig. 4A). As a caveat, these results may be somewhat skewed by the non-random selection of protease mixture compositions. Nevertheless, results suggest that even enzymes with relatively low catalytic efficiencies for the substrates, such as ADAM12, can be assessed with high accuracy.
We performed PrAMA inference on mixtures containing various concentrations of MMP7 to ascertain PrAMA's ability to quantitatively infer MMP activity, in addition to simply inferring whether or not an enzyme is present (Fig. 6F). We used 16 substrates and considered 10 MMPs (i.e., constructed a 16 × 10 matrix C) in this analysis, and tested 7 concentrations ranging from 0.01 nM to 1 nM. In all cases, PrAMA inferred MMP7 activity with 100% specificity. Furthermore, PrAMA detected quantitative differences in protease activity with high accuracy. R2 = 0.98 for a log-log plot that describes inferred MMP7 activity as a function of its actual concentration. PrAMA has less success in quantifying absolute differences in activity among multiple MPs (Fig. S15A–B‡), in part due to the fact that the relationship between MMP concentration and observed protease activityV0 is enzyme-specific and can deviate from linearity (Fig. S15C and S16‡). In general, we observe the recombinant MMPs employed in this work to be less efficient at higher concentrations. Enzyme concentration effects on proteolytic activity may be due to issues such as non-specific protein adsorption and aggregation. To test this hypothesis, we added increasing concentrations of Brij 35 to the reaction buffer (Fig. S17A‡). Although Brij can decrease proteolytic efficiency, our results suggest that Brij improves assay linearity perhaps by decreasing non-specific aggregation at higher enzyme concentrations (Fig. S17B‡), which has been observed for other secreted proteins.57 Even when nonlinear relationships between MMP concentration and observed protease activityV0 exist, PrAMA inference does not seem to distort these relationships (Fig. S15C‡). Consequently, quantitative comparisons of individual protease activities from one experimental sample to another can still be accurately made.
PrAMA with unknown background protease activity
In many potential applications, PrAMA will not be able to explicitly account for all protease activities in the parameter matrix C. In this work we account for up to 14 MPs simultaneously, and more substrates and enzymes can be potentially included in the PrAMA for future applications. Nevertheless, some biological samples may contain unknown proteinases that are also capable of cleaving the FRET-substrates. Robustness to these unknown proteinases is a crucial property of PrAMA. To test this, we applied PrAMA inference to enzyme mixtures with known “background” protease activity that is not explicitly accounted for in the parameter matrix C. For example, we constructed a 16 × 2 parameter matrix C to infer MMP9 & MMP10 activities. We tested PrAMA inference on 5 enzyme mixtures that contained MMP9 and/or MMP10, as well as at least one additional MP that was unaccounted for in C (Fig. 7A). We repeated this process for MMPs 1–8 (3 mixtures) and ADAMs 10 & 17 (3 mixtures). All three sets of analyses performed roughly as well as the PrAMA inference results where all protease activity was explicitly accounted for in C. PrAMA inference of ADAMs 10 & 17 yielded a maximum accuracy of 100%, although statistical significance was modest (p = 0.17) due to the small sample size (3 mixtures) and low inference dimensionality (only 2 enzymes). Statistical significances of the other two PrAMA results were greater (MMP9 & 10, p < 0.1; MMPs 1–8, p < 0.001).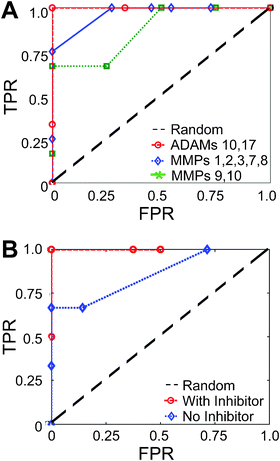 | ||
| Fig. 7 Using PrAMA with background protease activity and protease inhibitors . (A) The figure legend indicates which MMPs are considered in the parameter matrix. The corresponding ROC curves describe PrAMA inference accuracy for mixtures that contain MMPs and ADAMs that are both considered and ignored in the parameter matrix. (B) Inference for a triple-enzyme mixture involving MMP3, MMP7, and MMP9, with or without invoking information from an MMP9 inhibitor. For inference with inhibitor, p < 0.001. | ||
We also used PrAMA to infer protease activity over a background of conditioned media from the breast cancer cell line MDA-MB-231 (Fig. 6F). We added recombinant, active MMP7 to supernatant collected 12 h after stimulating cells with EGF and the inflammatory cytokine TNFα. We considered 10 MMPs in the parameter matrix C, and ultimately were able to identify MMP7 protease activity with 100% specificity. PrAMA did not detect any additional MMP activity in these samples.
Augmenting PrAMA with specific protease inhibitors
The accuracy and specificity of PrAMA can be bolstered by using protease inhibitors in conjunction with traditional PrAMA methods. Adding inhibitors to solutions of active proteases may be appropriate when specificity and accuracy are considered more important than non-invasiveness. In this work, we present an example of how PrAMA can be combined with inhibitors. First, we measured the cleavage profile V0 of a mixture containing 0.5 nM MMP3 & MMP7 and 0.05 nM MMP9 in MMP buffer using substrates 1–16. Second, we measured the cleavage rate V0 of substrate-1 when the mixture had an added 100 nM MMP-9 inhibitor (IC50 = 5 nM). The decrease in observed cleavage rate caused by adding the inhibitor, divided by the previously known catalytic efficiency Ci,j and substrate concentration, produced an inferred MMP9 concentration within 20% of the actual concentration: [E] = V0/([S]Ci,j) ≈ 0.04 nM. Based on this, we subtracted the expected MMP9 component of the cleavage signature from the total signature V0 observed with no inhibitor and performed PrAMA inference on the remaining non-MMP9 component. For this example, PrAMA achieves correct inference of all three enzymes only when incorporating information gleaned from using the inhibitor (Fig. 7B). Without this information, PrAMA fails to infer MMP9 without including several false-positives.PrAMA inference of live-cell response to PMA & IM stimulation
We applied PrAMA to a well-studied set of wildtype, ADAM10−/−, and ADAM17−/− MEF cell lines in order to validate the approach in a live-cell context. Fig. 8A depicts the increased substrate cleavage observed in response to PMA stimulation for four of the total seven substrates used in this example. Within 2 h, PMA causes a statistically significant increase in substrate cleavage for at least one substrate in all three cell lines (Fig. 8B): 4/7 substrates significantly increased cleavage with WT cells; 3/7 increased with ADAM10−/− cells; and substrate 15 increased cleavage with ADAM17−/− cells. In some cases, we observe more statistically significant changes in cleavage when including measurements at later time points. Substrates 1, 9, and 15 significantly increase cleavage in ADAM17−/− cells when assessing cleavage 8 h after PMA stimulation (p < 0.05). From these measurements it would be difficult to attribute changes in substrate cleavage to particular MPs without PrAMA. We assess protease activity by using a parameter matrix C that considers the presence of five MPs (Fig. 8C). We define the observed cleavage vector V0 as the change of cleavage rate in response to PMA stimulation, using all seven substrates. In this proof of principle, we limit PrAMA inference to the first 2 h following cellular stimulation, with the anticipation that later time-point measurements may face greater issues relating to off-target substrate cleavage. PrAMA inference indicates that PMA stimulates significant ADAM17 activity (Fig. 8D). In all three cell lines, PrAMA did not detect a significant increase in MMP2, MMP14, or ADAM10 activity. Results indicate that substrate cleavage and subsequent PrAMA are sensitive to treatment with the metalloproteinase inhibitor GM6001 (Fig. S18‡). As further validation, PrAMA infers ADAM17−/− cells to have 90% less ADAM17 activity than WT. Remaining ADAM17 signal in ADAM17−/− cells likely arises from other proteases (e.g., ADAM9) with similar substrate selectivity.![Live
cell
inference of PMA-stimulated
MP
activity. PrAMA was conducted using three cell lines (WT, ADAM10−/−, and ADAM17−/− MEFs) and 7 total substrates, tracking substrate cleavage up to two hours after adding substrate. (A) Time-lapse fluorimetry for 4 of the 7 total substrates used in this experiment. (B) Inferred substrate cleavage rates (V0/[S]0), corresponding by row to the time-courses shown in A and using all four time-points in A for the inference. Stars indicate p < 0.05, comparing between cleavage rates for the control and stimulated conditions. (C) Parameter matrix used in this experiment, with each column divided by its Euclidean norm. (D) PrAMA inference results for the increased activity caused by PMA stimulation, using significance threshold σT = 1.4. No significant increase in MMP2, MMP14, or ADAM10 activity was detected at this threshold. For MMP9 and ADAM17, all inferred differences were statistically significant (p < 0.05). For all subplots, error bars indicate standard deviation of three biological replicates.](/image/article/2011/IB/c0ib00083c/c0ib00083c-f8.gif) | ||
| Fig. 8 Live cell inference of PMA-stimulated MP activity. PrAMA was conducted using three cell lines (WT, ADAM10−/−, and ADAM17−/− MEFs) and 7 total substrates, tracking substrate cleavage up to two hours after adding substrate. (A) Time-lapse fluorimetry for 4 of the 7 total substrates used in this experiment. (B) Inferred substrate cleavage rates (V0/[S]0), corresponding by row to the time-courses shown in A and using all four time-points in A for the inference. Stars indicate p < 0.05, comparing between cleavage rates for the control and stimulated conditions. (C) Parameter matrix used in this experiment, with each column divided by its Euclidean norm. (D) PrAMA inference results for the increased activity caused by PMA stimulation, using significance threshold σT = 1.4. No significant increase in MMP2, MMP14, or ADAM10 activity was detected at this threshold. For MMP9 and ADAM17, all inferred differences were statistically significant (p < 0.05). For all subplots, error bars indicate standard deviation of three biological replicates. | ||
We also applied PrAMA to assess the protease-activity response to ionomycin treatment. We used six substrates to ascertain activities of the same five proteases considered in the PMA example, again using wildtype, ADAM10−/−, and ADAM17−/− MEF cells (Fig. 9). All substrates show a statistically significant increase in cleavage upon IM stimulation within 2 h (p < 0.05), even in the mutant cell lines. In contrast to PMA stimulation, PrAMA results suggest that IM stimulates MMP9 & ADAM10 activity. In all three cell lines, PrAMA did not detect an increase in MMP2, MMP14, or ADAM17 activity at the same level of significance as detected for ADAM10 and MMP9 (zero at the significance level corresponding to results in Fig. 9D). Encouragingly, ADAM10−/− cells show a >90% decrease in inferred ADAM10 activity compared to wildtype cells. Again, remaining ADAM10 signal may be attributed to other proteases with similar substrate preference. Interestingly, the knockout cell lines seem to exhibit heightened general proteolytic response to IM stimulation compared to the wildtype cells (Fig. 9A–B). However, PrAMA analysis suggests that this increased activity likely arises from proteases other than ADAM10 or ADAM17. Fig. 9C shows that wildtype cells cleave the good ADAM substrates (6 & 9) at a greater relative rate compared to the knockout cell lines, even if the absolute cleavage rate is lower. Consequently, PrAMA infers the knockout cells to exhibit stronger MMP9 rather than ADAM activity.
![Live
cell
inference of
ionomycin
stimulated
MP
activity. PrAMA was conducted using three cell lines (WT, ADAM10−/−, and ADAM17−/− MEFs) and 6 total substrates, tracking substrate cleavage up to two hours after adding substrate. (A) Time-lapse fluorimetry for 3 of the 6 total substrates used in this experiment. (B) Inferred substrate cleavage rates (V0/[S]0), corresponding by row to the time-courses shown in A and using all time-points in A for the inference. Stars indicate p < 0.05, comparing between cleavage rates for the control and stimulated conditions. (C) Observed cleavage vector V0 (columns), normalized to have total signal of 1, for each of the three cell lines. (D) PrAMA inference results for the increased activity caused by IM stimulation, using significance threshold σT = 1.4. No significant increase in MMP2, MMP14, or ADAM17 activity was detected at this threshold. For MMP9 and ADAM10, all inferred differences were statistically significant (p < 0.05). For all subplots, error bars indicate standard deviation of three biological replicates.](/image/article/2011/IB/c0ib00083c/c0ib00083c-f9.gif) | ||
| Fig. 9 Live cell inference of ionomycin stimulated MP activity. PrAMA was conducted using three cell lines (WT, ADAM10−/−, and ADAM17−/− MEFs) and 6 total substrates, tracking substrate cleavage up to two hours after adding substrate. (A) Time-lapse fluorimetry for 3 of the 6 total substrates used in this experiment. (B) Inferred substrate cleavage rates (V0/[S]0), corresponding by row to the time-courses shown in A and using all time-points in A for the inference. Stars indicate p < 0.05, comparing between cleavage rates for the control and stimulated conditions. (C) Observed cleavage vector V0 (columns), normalized to have total signal of 1, for each of the three cell lines. (D) PrAMA inference results for the increased activity caused by IM stimulation, using significance threshold σT = 1.4. No significant increase in MMP2, MMP14, or ADAM17 activity was detected at this threshold. For MMP9 and ADAM10, all inferred differences were statistically significant (p < 0.05). For all subplots, error bars indicate standard deviation of three biological replicates. | ||
Ultimately, PrAMA results that show ADAM17 to be activated in response to PMA agree with multiple other reports in the literature.19,20,25 Furthermore, PrAMA indications that IM stimulates ADAM10 activity also agree with previous literature.19 This work complements these previous reports by observing specific ADAM17 and ADAM10 activity in a non-invasive, real-time manner, without resorting to pharmacological or genetic perturbations.
Optimal substrate selection
Various logistical constraints may exist that limit the number of substrates available to be used for PrAMA in certain applications. To address this issue, we implemented a common optimal design criterion for selecting substrates so as to maximize PrAMA accuracy. The determinant of the covariance error matrix RM is one metric describing the volume of inference uncertainty. Minimizing this volume, which is equivalent to maximizing the determinant of its inverse and minimizing the condition number, ultimately reduces model uncertainty and optimizes PrAMA inference accuracy.58 Using the following function, we optimally selected subsets of the total 18 FRET-substrates to perform various PrAMA inferences:| min det(RM) = max det(CTRDC) | (9) |
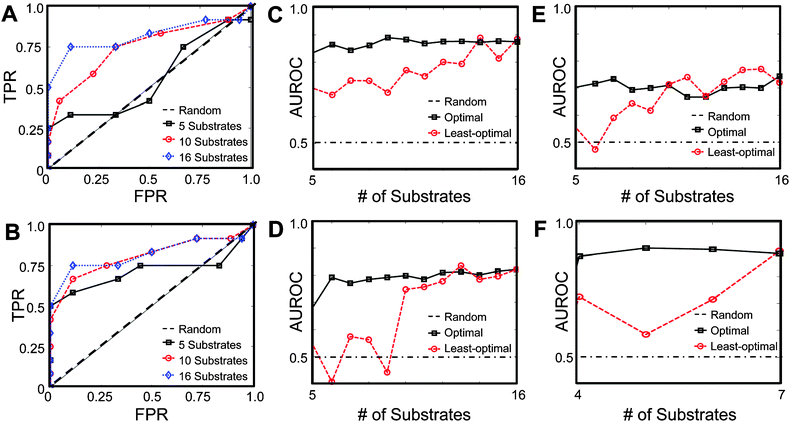 | ||
| Fig. 10 Optimal substrates selection improves PrAMA accuracy. (A,B) ROC curves describing PrAMA accuracy for double-enzyme experiments involving MMPs 1, 2, 3, 7, and 8, where PrAMA uses a worst (A) or best (B) subset of the substrates, as defined in the text. (C–F) Area under the ROC curve (AUROC) as a function of the number of best (black) or worst (red) substrates, for various sets of PrAMA experiments: single (C), double (D), and triple (E) enzyme mixtures involving MMPs 1, 2, 3, 7, and 8; (F) double-enzyme mixtures involving ADAMs 10 and 17. | ||
Discussion
Existing techniques used to study MPs each have advantages and disadvantages. Zymography is one of the oldest and most common MP activity assays, and a variety of zymographic techniques exist to measure the activities of diverse proteases and their inhibitors.28,59,60 Most zymographic techniques involve SDS-PAGE electrophoresis, which prevents continuous real-time measurement (and generally disrupts non-covalent MP complexes). In situ zymography, often applied to frozen tissue sections, allows for the observation of localized protease activity.27 However, in most cases the substrates used for in situ zymography (e.g., gelatin) are readily cleaved by a variety of MPs and measurements consequently lack specificity. Recently a variety of methods have been developed to observe the proteolytic degradation of endogenous MP substrates. For example, mass spectrometric techniques can quantify hundreds of proteins that have freshly cleaved amide bonds within complex biological samples.26 Problems involving the complex relationship between proteases and their substrates make it difficult to accurately and non-invasively infer the contributions of specific MPs to the global patterns of endogenous substrate degradation. Yet another recently developed method, activity based probes (ABPs), support the direct and specific measurement of diverse protease activities.29,30,61 Like typical zymography substrates, however, ABPs can act on a broad range of related proteases. Therefore, assays involving ABPs face a trade-off between invasiveness and specificity. For specific protease identities to be ascertained, biological samples analyzed with ABPs can be resolved by size through electrophoresis.Synthetic polypeptide protease substrates have been developed for an increasingly wide range of enzymes. Within the last few years several FRET-substrates have been designed with some specificity, thereby supporting their application in complex biological samples.20,31,32 Nevertheless, cross-reactivity with closely related MPs and distantly-related, but much more non-specific, proteases can still complicate the interpretation of FRET-substrate activity assays. As an example, several FRET-substrates with some specificity for ADAM17 have recently been developed with a sequence based on the ADAM17 cleavage site on pro-TNFα.31,32,62,63 Multiple recent reports employ these ADAM17 FRET-substrates, even though they have documented cross-reactivity with related MPs.35,64 At least six MPs have been recognized to cleave endogenous pro-TNFα, in some cases at the same site.32,65,66 Such non-specificity complicates interpretation of the observed FRET-substrate cleavage, especially when comparing multiple correlated MP activities in the same biological sample.64 Several MPs cleave many of the same synthetic and endogenous substrates that ADAM17 cleaves, suggesting the repertoire of ADAM17 substrates could be a subset of the repertoire for more promiscuous MPs (e.g., MMP14) or non-specific proteases like plasmin.67 At least to some degree, this situation is conceivable not just for ADAM17 but for a variety of MPs, and would make identifying truly specific substrates impossible.
Although MPs have been extensively studied for decades, no method yet exists to assay multiple protease activities in real-time with high specificity and non-invasiveness. One explanation partly accounting for this fact is that the ubiquitous regulatory interactions, diverse substrates, and distinct roles played by closely related MPs have only recently become fully appreciated. Both MMPs and ADAMs engage in regulatory networks controlled by cyclical feedback interactions. For example, ADAMs participate in an autocrine positive feedback loop in mammary epithelial cells: EGFR transactivation stimulates Erk activity, which in turn stimulates ADAM shedding of EGF ligands, further activating EGFR.69 In this situation, common methods of ascertaining the influence of ADAM activity on EGFR signaling, such as by applying a protease inhibitor or siRNA treatment, can both disrupt the underlying feedback interactions and potentially create compensatory reactions whereby closely related ADAMs modify their activity to accommodate perturbations.15,24,25,68 As another example, many MMPs activate themselves and one another. Such interactions can create positive feedback interactions that allow, for example, an initiating MMP activation event to trigger further protease activation.70 We predict non-invasive, multiplexed, real-time, and specific measurements of MP activity will be critical towards understanding the complex regulatory mechanisms underlying MP networks.
We anticipate that PrAMA should have broad applicability in protease biology. FRET-substrates have been extensively used for high-throughput inhibitor screening with individual purified enzymes. PrAMA would allow inhibitor screening to be performed in more complex enzyme mixtures and biological samples, and could be adapted for high-throughput in vitro functional assays of inhibitor activity. As discussed above, PrAMA is well suited for network-level analysis of in vitroprotease activity, and PrAMA can scale up and down in scope depending on the particular application. At the most basic level, PrAMA could use multiple FRET-substrates in tandem to bolster the specificity of an activity measurement for even a single protease. In other words, the parameter matrix C could be as small as (2 substrates × 1 enzyme). PrAMA can capture protease activity on a variety of time-scales, depending on the particular application. We demonstrate high sensitivity measurements that are made over the course of >5 h, and live-cell measurements can detect significant differences in cleavage within 30 min. Dynamic measurements on this short time-scale can be relevant for detecting rapid post-translational protease activation, while longer time-scale measurements have relevance, for example, to phenotypic responses that are downstream of transcriptional changes. Soluble FRET-substrates can be directly applied to both live-cells and cell lysate for protease activity measurement.31,64 Our initial experiments show that PrAMA can operate by adding individual yet distinct FRET-substrates to live-cells in a multi-well format. Furthermore, FRET-substrates with distinctive excitation/emission spectra may be simultaneously combined in the same solution for PrAMA of a single biological sample. FRET-substrates have been tethered directly to 3D substrata such as collagen,71 providing localized measurement of protease activity. For simultaneously analyzing many protease activities, the mathematical framework behind PrAMA can be applied to microarrays of peptides, for instance, that contain hundreds or thousands of FRET-peptide substrates. Previous work with peptide microarrays has demonstrated how patterns of peptidase activity can be deconvoluted using non-linear least squares in a similar manner to PrAMA, ultimately to infer the presence of specific proteases within complex biological samples.42PrAMA builds upon this previous work by using non-invasive dynamic measurements of peptide cleavage rather than static snapshots of cleavage patterns to quantitatively infer kinetic parameters (i.e., V0). The advantages of PrAMA's mathematical framework include explicitly accounting for substrate depletion & photobleaching effects, as well as readily extending to dynamic measurements of protease activity where V0 is not constant. Strategies for selecting optimal substrates depend on the application. Most commonly, optimal substrates are individually chosen by the combination of their specificity and cleavage efficiency.35,38,42 In this work, however, we employ a global optimization strategy to identify the set of substrates that combine to yield the greatest specificity and the least inference uncertainty. The principles behind PrAMA, including strategies for optimally selecting substrates, are readily extendable to other classes of enzymes, such as caspases and cathepsins. FRET-based protease substrates have been successfully applied to measuring in vitrocaspase activation. Like MPs, however, individual caspases have overlapping substrate specificity and it can be difficult to interpret which specific caspase has become activated.72 Lastly, PrAMA inference has many potential uses involving clinical samples. For example, simultaneous measurement of multiple protease activities in patient fluid samples or biopsies could reveal mechanistic insight and/or identify activity-based markers of disease state for diagnostic/prognostic use. Ultimately, this work presents an integrated mathematical and experimental framework that can be adapted and extended to a broad range of applications. We have demonstrated various methods of a priori analyzing how best to design PrAMA experiments, whether it be through choosing optimal substrates, identifying which proteases can be specifically measured with the available substrates, or understanding how to account for experimental variability.
Abbreviations
| ABP | activity based probe |
| ADAM | a disintegrin and metalloproteinase |
| B | background signal |
| DMEM | Dulbecco's Modified Eagle Medium |
| DMSO | dimethyl sulfoxide |
| C i,j | catalytic efficiency for the ith substrate and jth enzyme |
| C | catalytic efficiency parameter matrix |
| Cha | cyclohexylalanyl |
| Dabcyl | 4-(4-dimethylaminophenylazo)benzoyl |
| [E] | enzyme concentration |
| E | vector of enzyme activities |
| EGF | epidermal growth factor |
| F 0 | peak fluorescence from positive control |
| F obs | observed fluorescence from product formation |
| F p | fluorescence from product formation |
| Fam | 5-carboxyfluorescein |
| FRET | fluorescence resonance energy transfer |
| GABA | γ-aminobutyric acid |
| Homophe | homophenylalanyl |
| IM | ionomycin |
| k cat | turnover number |
| k cat/Km | catalytic efficiency |
| k d | photobleaching decay constant |
| K m | Michaelis–Menten constant |
| M–M | Michaelis–Menten |
| MEBM | Mammary Epithelial Basal Medium |
| MMP | matrix metalloproteinase |
| MP | metalloproteinase |
| PrAMA | Proteolytic Activity Matrix Analysis |
| R M | model covariance error matrix |
| R r M | relative model covariance error matrix |
| R D | data uncertainty covariance matrix |
| [S]0 | initial substrate concentration |
| [S] | substrate |
| σ T | significance of inference threshold |
| T 0 | lag time |
| TIMP | tissue inhibitor of metalloproteinase |
| TNFα | tumor necrosis factor-alpha |
| V 0 | initial rate of substrate cleavage |
| V0 | vector of V0's for all substrates |
| V s 0 | bootstrapping sample ensemble of multiple V0 |
Acknowledgements
This work was partially supported by NIH grants U54-CA112967, R01-EB010246 and R01-GM081336, and an NSF Graduate Fellowship to M.A.M. The authors thank Marcia Moss, Fred Rasmussen, Carl Blobel, Harvey Lodish, H. Steven Wiley, Barbara Imperiali, and members of the Lauffenburger and Griffith labs for useful discussions and materials.References
- C. López-Otín and L. M. Matrisian, Nat. Rev. Cancer, 2007, 7, 800–808 CrossRef CAS.
- K. Kessenbrock, V. Plaks and Z. Werb, Cell, 2010, 141, 52–67 CrossRef CAS.
- T. Vu and Z. Werb, Genes Dev., 2000, 14, 2123–2133 CrossRef CAS.
- J. Rundhaug, J. Cell. Mol. Med., 2005, 9, 267–285 CrossRef CAS.
- M. Handsley and D. Edwards, Int. J. Cancer, 2005, 115, 849–860 CrossRef CAS.
- A. Page-McCaw, A. Ewald and Z. Werb, Nat. Rev. Mol. Cell Biol., 2007, 8, 221–233 CrossRef CAS.
- S. Eming, T. Krieg and J. Davidson, J. Invest. Dermatol., 2007, 127, 514–525 CrossRef CAS.
- E. Deryugina and J. Quigley, Cancer Metastasis Rev., 2006, 25, 9–34 CrossRef CAS.
- M. Gueders, J. Foidart, A. Noel and D. Cataldo, Eur. J. Pharmacol., 2006, 533, 133–144 CrossRef CAS.
- R. Dean and C. Overall, Mol. Cell. Proteomics, 2007, 6, 611–623 CrossRef CAS.
- C. Morrison, G. Butler, D. Rodríguez and C. Overall, Curr. Opin. Cell Biol., 2009, 21, 645–653 CrossRef CAS.
- M. Egeblad and Z. Werb, Nat. Rev. Cancer, 2002, 2, 163–176 CrossRef.
- I. Stamenkovic, J. Pathol., 2003, 200, 448–464 CrossRef CAS.
- G. Murphy, Nat. Rev. Cancer, 2008, 8, 932–941 CrossRef.
- C. P. Blobel, Nat. Rev. Mol. Cell Biol., 2005, 6, 32–43 CrossRef CAS.
- N. Rocks, G. Paulissen, M. El Hour, F. Quesada, C. Crahay, M. Gueders, J. M. Foidart, A. Noel and D. Cataldo, Biochimie, 2008, 90, 369–379 CrossRef CAS.
- G. Kirfel, A. Rigort, B. Borm and V. Herzog, Eur. J. Cell Biol., 2004, 83, 717–724 CrossRef.
- M. Duffy, E. McKiernan, N. O'Donovan and P. McGowan, Clin. Cancer Res., 2009, 15, 1140–1144 CrossRef CAS.
- K. Horiuchi, S. Le Gall, M. Schulte, T. Yamaguchi, K. Reiss, G. Murphy, Y. Toyama, D. Hartmann, P. Saftig and C. P. Blobel, Mol. Biol. Cell, 2007, 18, 176–188 CAS.
- P. Xu and R. Derynck, Mol. Cell, 2010, 37, 551–566 CrossRef CAS.
- M. Pillinger, N. Marjanovic, S. Kim, J. Scher, P. Izmirly, S. Tolani, V. Dinsell, Y. C. Lee, M. J. Blaser and S. B. Abramson, J. Biol. Chem., 2005, 280, 9973–9979 CrossRef CAS.
- A. Murthy, W. Cruz-Munoz and R. Khokha, Springer Verlag, 2008, pp. 373–400.
- A. Herrlich, E. Klinman, J. Fu, C. Sadegh and H. Lodish, FASEB J., 2008, 22, 4281–4295 CrossRef CAS.
- E. Joslin, H. Shankaran, L. Opresko, N. Bollinger, D. Lauffenburger and H. S. Wiley, Mol. BioSyst., 2010, 6, 1293–1306 RSC.
- U. Sahin, G. Weskamp, K. Kelly, H. Zhou, S. Higashiyama, J. Peschon, D. Hartmann, P. Saftig and C. P. Blobel, J. Cell Biol., 2004, 164, 769–779 CrossRef CAS.
- G. Butler, R. Dean, C. Morrison and C. Overall, Methods Mol. Biol., 2010, 622, 451–470 CrossRef CAS.
- Z. Galis, G. Sukhova and P. Libby, FASEB J., 1995, 9, 974–980 CAS.
- D. Kleiner and W. Stetler-Stevenson, Anal. Biochem., 1994, 218, 325–329 CrossRef CAS.
- A. Saghatelian, N. Jessani, A. Joseph, M. Humphrey and B. Cravatt, Proc. Natl. Acad. Sci. U. S. A., 2004, 101, 10000–10005 CrossRef CAS.
- G. Blum, S. Mullins, K. Keren, M. Fonovi
![[c with combining breve]](https://www.rsc.org/images/entities/char_0063_0306.gif) , C. Jedeszko, M. J. Rice, B. F. Sloane and M. Bogyo, Nat. Chem. Biol., 2005, 1, 203–209 CrossRef CAS.
, C. Jedeszko, M. J. Rice, B. F. Sloane and M. Bogyo, Nat. Chem. Biol., 2005, 1, 203–209 CrossRef CAS. - M. Alvarez-Iglesias, G. Wayne, K. O'Dea, A. Amour and M. Takata, Lab. Invest., 2005, 85, 1440–1448 Search PubMed.
- M. Moss and F. Rasmussen, Anal. Biochem., 2007, 366, 144–148 CrossRef CAS.
- J. R. Doedens, R. M. Mahimkar and R. A. Black, Biochem. Biophys. Res. Commun., 2003, 308, 331–338 CrossRef CAS.
- E. L. Hassemer, S. M. Le Gall, R. Liegel, M. McNally, B. Chang, C. J. Zeiss, R. D. Dubielzig, K. Horiuchi, T. Kimura, Y. Okada, C. P. Blobel and D. J. Sidjanin, Genetics, 2010, 185, 245–255 CrossRef CAS.
- C. Caescu, G. Jeschke and B. Turk, Biochem. J., 2009, 424, 79–88 CrossRef CAS.
- H. Nagase and G. Fields, Biopolymers, 1996, 40, 399–416 CrossRef CAS.
- M. Lim and C. Craik, Bioorg. Med. Chem., 2009, 17, 1094–1100 CrossRef CAS.
- E. Chen, S. Kridel, E. Howard, W. Li, A. Godzik and J. W. Smith, J. Biol. Chem., 2002, 277, 4485–4492 CrossRef CAS.
- F. Rasmussen, N. Yeung, L. Kiefer, G. Murphy, C. Lopez-Otin, M. P. Vitek and M. L. Moss, Biochemistry, 2004, 43, 2987–2995 CrossRef CAS.
- M. Drag, M. Bogyo, J. Ellman and G. Salvesen, J. Biol. Chem., 2010, 285, 3310–3318 CrossRef CAS.
- D. Gosalia, C. Salisbury, D. Maly, J. Ellman and S. Diamond, Proteomics, 2005, 5, 1292–1298 CrossRef CAS.
- D. Gosalia, W. Denney, C. Salisbury, J. Ellman and S. Diamond, Biotechnol. Bioeng., 2006, 94, 1099–1110 CrossRef CAS.
- G. Dorman, S. Cseh, I. Hajdu, L. Barna, D. Konya, K. Kupai, L. Kovacs and P. Ferdinandy, Drugs, 2010, 70, 949–964 CrossRef CAS.
- M. Mohan, T. Seaton, J. Mitchell, A. Howe, K. Blackburn, W. Burkhart, M. Moyer, I. Patel, G. M. Waitt, J. D. Becherer, M. L. Moss and M. E. Milla, Biochemistry, 2002, 41, 9462–9469 CrossRef CAS.
- D. Hartmann, B. De Strooper, L. Serneels, K. Craessaerts, A. Herreman, W. Annaert, L. Umans, T. Lubke, A. Lena Illert, K. von Figura and P. Saftig, Hum. Mol. Genet., 2002, 11, 2615–2624 CrossRef CAS.
- M. Moss, F. Rasmussen, R. Nudelman, P. Dempsey and J. Williams, Comb. Chem. High Throughput Screening, 2010, 13, 358–365 CrossRef CAS.
- M. O. Palmier and S. R. Van Doren, Anal. Biochem., 2007, 371, 43–51 CrossRef CAS.
- A. Cornish-Bowden, Fundamentals of Enzyme Kinetics, Portland, London, 3rd edn, 1995 Search PubMed.
- J. A. Robinson and W. G. Characklis, Microb. Ecol., 1984, 10, 165–178 CrossRef.
- R. S. Obach and A. E. Reed-Hagen, Drug Metab. Dispos., 2002, 30, 831–837 CrossRef CAS.
- A. Nath and W. M. Atkins, Drug Metab. Dispos., 2006, 34, 1433–1435 CrossRef CAS.
- N. D. Rawlings, A. J. Barrett and A. Bateman, Nucleic Acids Res., 2009, 38, D227–D233.
- J. Verspurten, K. Gevaert, W. Declercq and P. Vandenabeele, Trends Biochem. Sci., 2009, 34, 319–323 CrossRef CAS.
- S. Ohkubo, K. Miyadera, Y. Sugimoto, M. Ki, K. Wierzba and Y. Yamada, Comb. Chem. High Throughput Screening, 2001, 4, 573–583 CAS.
- O. Schilling and C. M. Overall, Curr. Opin. Chem. Biol., 2007, 11, 36–45 CrossRef CAS.
- S. Hernandez-Barrantes, M. Toth, M. Bernardo, M. Yurkova, D. Gervasi, Y. Raz, Q. X. A. Sang and R. Fridman, J. Biol. Chem., 2000, 275, 12080–12089 CrossRef CAS.
- V. Sluzky, A. M. Klibanov and R. Langer, Biotechnol. Bioeng., 1992, 40, 895–903 CrossRef CAS.
- D. Belsley, Wiley, New York, 1991.
- J. Fowlkes, K. Thrailkill, D. Serra and H. Nagase, Endocrine, 1997, 7, 33–36 CrossRef CAS.
- G. Oliver, J. Leferson, W. Stetler-Stevenson and D. Kleiner, Anal. Biochem., 1997, 244, 161–166 CrossRef CAS.
- S. Sieber, S. Niessen, H. Hoover and B. Cravatt, Nat. Chem. Biol., 2006, 2, 274–281 CrossRef CAS.
- Y. Hiraoka, K. Yoshida, M. Ohno, T. Matsuoka, T. Kita and E. Nishi, Biochem. Biophys. Res. Commun., 2008, 370, 154–158 CrossRef CAS.
- F. Mohammed, D. Smookler, S. Taylor, B. Fingleton, Z. Kassiri, O. H. Sanchez, J. L. English, L. M. Matrisian, B. Au, W. C. Yeh and R. Khokha, Nat. Genet., 2004, 36, 969–977 CrossRef CAS.
- E. Walker and G. Rosenberg, Exp. Neurol., 2009, 216, 122–131 CrossRef CAS.
- A. Amour, C. Knight, W. English, A. Webster, P. Slocombe, V. Knauper, A. J. P. Docherty, J. D. Becherer, C. P. Blobel and G. Murphy, FEBS Lett., 2002, 524, 154–158 CrossRef CAS.
- H. Haro, H. Crawford, B. Fingleton, K. Shinomiya, D. Spengler and L. M. Matrisian, J. Clin. Invest., 2000, 105, 143–150 CrossRef CAS.
- E. Tam, C. Morrison, Y. Wu, M. Stack and C. Overall, Proc. Natl. Acad. Sci. U. S. A., 2004, 101, 6917–6922 CrossRef CAS.
- S. Le Gall, P. Bobe, K. Reiss, K. Horiuchi, X. Niu, D. Lundell, D. R. Gibb, D. Conrad, P. Saftig and C. P. Blobel, Mol. Biol. Cell, 2009, 20, 1785–1794 CrossRef CAS.
- E. Joslin, L. Opresko, A. Wells, H. S. Wiley and D. Lauffenburger, J. Cell Sci., 2007, 120, 3688–3699 CrossRef CAS.
- M. Sternlicht and Z. Werb, Annu. Rev. Cell Dev. Biol., 2001, 17, 463–416 CrossRef CAS.
- B. Z. Packard, V. V. Artym, A. Komoriya and K. M. Yamada, Matrix Biol., 2009, 28, 3–10 CrossRef CAS.
- L. Bouchier-Hayes, C. Muñoz-Pinedo, S. Connell and D. Green, Methods, 2008, 44, 222–228 CrossRef CAS.
Footnotes |
| † Published as part of an Integrative Biology themed issue in honour of Mina J. Bissell: Guest Editor Mary Helen Barcellos-Hoff. |
| ‡ Electronic supplementary information (ESI) available: Table S1; Text S1; Fig. S1–S18; supplemental data. See DOI: 10.1039/c0ib00083c |
| This journal is © The Royal Society of Chemistry 2011 |
