Recent development in computer simulations of lipid bilayers
Alexander P.
Lyubartsev
*a and
Alexander L.
Rabinovich
b
aPhysical Chemistry, Department of Materials and Environmental Chemistry, Stockholm University, Stockholm, 10691, Sweden. E-mail: alexander.lyubartsev@mmk.su.se; Fax: +46-8152187; Tel: +46-8161193
bLaboratory of Biochemistry, Institute of Biology, Karelian Research Center, Russian Academy of Sciences, Petrozavodsk, 185910, Russia
First published on 7th September 2010
Abstract
Rapid development of computer power during the last decade has made molecular simulations of lipid bilayers feasible for many research groups, which, together with the growing general interest in investigations of these very important biological systems has lead to tremendous increase of the number of research on the computational modeling of lipid bilayers. In this review, we give account of the recent progress in computer simulations of lipid bilayers covering mainly the period of the last 5 years, and covering several selected subjects: development of the force fields for lipid bilayer simulations, studies of the role of lipid unsaturation, the effect of cholesterol and other inclusions on properties of the bilayer, and use of coarse-grained models.
1 Introduction
Biomembranes surround cells: a membrane separates the interior of a cell from the outside environment. Being selectively permeable, membranes participate in control of the movement of various compounds (substances) into and out of cells. Biomembranes are very complex heterogeneous systems consisting of many different types of lipids, sterols, proteins, carbohydrates and various membrane associated molecules which are involved in a variety of cellular processes; consequently, membranes play an active part in the life of the cell, they exist as dynamic structures. Lipid molecules differ with respect to the type of hydrophilic head-group and occur with a wide variety of hydrophobic hydrocarbon chains of fatty acids (FAs). Usually the most abundant phospholipid in animal and plants is phosphatidylcholine (PC), its trivial name is “lecithin”: it is the key building block of membrane bilayers. Cholesterol (CHOL) molecules are essential component of mammalian cell membranes playing an important role in formation of heterogeneites (known also as rafts) which are supposed to be responsible for cell signaling.1 Knowledge of physical-chemical properties of lipid bilayers is a key element of our general understanding of biomembrane functioning, which is one of the greatest challenging problems in biophysical and biomedical sciences.A characteristic feature of lipid bilayers is that, in a physiological form, they exist in a liquid crystalline (fluid) state which implies a relatively high degree of disorder. Experimental measurements of structural and dynamical properties are obtained as averages over a large number of lipids and over a certain time interval, which not always can give an unambiguous picture of individual lipids and their interactions.
During the last decades computer simulations have become a well established tool of modern investigations of molecular structure. Monte Carlo (MC) or molecular dynamics (MD) can provide three-dimensional real-time imaging of the system with atomistic-level resolution, and hence can give essential structural and dynamical information which otherwise is hardly accessible by any experimental method. The first attempts of computer simulations of model bilayers composed of amphiphilic molecules with atomistic resolution were made by almost 30 years ago,2–4 which was followed in 90-ties by simulations of fully hydrated bilayers built by lipids typical for biological membranes.5–7 This list may be supplemented with some other early theoretical works8–12 (more detailed situation is described in the reviews presented below). The rapid development of the accessible computer power has made simulations of more and more complicated systems feasible, and allowed also increase the size of the simulated systems. Now simulation of about hundred fully hydrated lipids during 50–100 ns can be considered as a routine. Besides, some computer simulations of lipid bilayers have been performed for a much longer simulation time (e.g. 800 ns13 and 1 μs).14 The amount of works on simulations of lipid membrane systems has increased tremendously, and a number of reviews appeared accounting for this in the past decade15–26 and more recently.27–38 In this review, we give account of the recent development in computer simulations of lipid bilayers covering mainly the period of the last 5 years. More than two hundred papers have been chosen but this is a moderate part of the simulation studies performed in a given time in this active area of research. It is beyond the scope of this review to touch upon other topics and types of membrane systems. Unfortunately, as a result a number of important areas are not represented here sufficiently (or even mentioned), e.g. membrane – protein simulations, simulations of other membrane inclusions, etc. Some reviews can be enumerated here in this respect, e.g. reviews devoted to computer simulation studies of protein - nucleic acid complexes,39 membrane proteins,40 large conformational changes in proteins,41 infrared spectra in peptides and proteins,42 blood coagulation proteins,43etc. The absence of some references in our review is related with an existence of a quantity of excellent above-mentioned and similar reviews.
Throughout this review, the notation of N:k(n – j)cis for describing the structure of each hydrocarbon chain of lipids will be used, where N refers to the total number of carbon atoms in the chain, k is the number of the methylene-interrupted double bonds (i.e., one methylene group is localized between each pair of double bonds), j denotes the number of carbons between the chain terminal CH3 group and the nearest double bond, whereas cis refers to the conformation around the double bonds. For brevity, the fragment (n – j)cis in the notation is frequently omitted.
Some of the commonly occurring types of FA chains and PC molecules discussed in the text are listed below: systematic name, trivial name in parentheses (if it exists), and shorthand designation are presented.
Saturated FAs:
• dodecanoic (lauric, 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0)
0)
• tetradecanoic (myristic, 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0)
0)
• hexadecanoic (palmitic, 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0)
0)
• octadecanoic (stearic, 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0)
0)
• eicosanoic (arachidic, 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0)
0)
Monounsaturated FAs:
• cis-9-hexadecenoic (palmitoleic, 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1(n-7)cis)
1(n-7)cis)
• cis-9-octadecenoic (oleic, 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1(n-9)cis)
1(n-9)cis)
Polyunsaturated FAs with methylene-interrupted double bonds:
• cis-9,12-octadecadienoic (linoleic, 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2(n-6)cis)
2(n-6)cis)
• cis-9,12,15-octadecatrienoic (alpha-linolenic, 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3(n-3)cis)
3(n-3)cis)
• cis-5,8,11,14-eicosatetraenoic (arachidonic, 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4(n-6)cis)
4(n-6)cis)
• cis-5,8,11,14,17-eicosapentaenoic (20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5(n-3)cis)
5(n-3)cis)
• cis-4,7,10,13,16,19-docosahexaenoic (22![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6(n-3)cis)
6(n-3)cis)
PC molecules:
• 1,2-dilauroyl-sn-glycero-3-phosphatidylcholine (DLPC), 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/12
0/12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0 PC
0 PC
• 1,2-dimyristoyl-sn-glycero-3-phosphatidylcholine (DMPC), 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/14
0/14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0 PC
0 PC
• 1,2-dipalmitoyl-sn-glycero-3-phosphatidylcholine (DPPC), 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/16
0/16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0 PC
0 PC
• 1,2-distearoyl-sn-glycero-3-phosphatidylcholine (DSPC), 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/18
0/18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0 PC
0 PC
• 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphatidylcholine (POPC), 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/18
0/18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1(n-9)cis PC
1(n-9)cis PC
• 1-stearoyl-2-oleoyl-sn-glycero-3-phosphatidylcholine (SOPC), 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/18
0/18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1(n-9)cis PC
1(n-9)cis PC
• 1,2-dioleoyl-sn-glycero-3-phosphatidylcholine (DOPC), 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1(n-9)cis/18
1(n-9)cis/18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1(n-9)cis PC
1(n-9)cis PC
• 1-palmitoyl-2-linoleoyl-sn-glycero-3-phosphatidylcholine, 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/18
0/18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2(n-6)cis PC
2(n-6)cis PC
• 1-stearoyl-2-linoleoyl-sn-glycero-3-phosphatidylcholine, 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/18
0/18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2(n-6)cis PC
2(n-6)cis PC
• 1-palmitoyl-2-linolenoyl-sn-glycero-3-phosphatidylcholine, 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/18
0/18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3(n-3)cis PC
3(n-3)cis PC
• 1-stearoyl-2-linolenoyl-sn-glycero-3-phosphatidylcholine, 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/18
0/18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3(n-3)cis PC
3(n-3)cis PC
• 1-palmitoyl-2-arahidonoyl-sn-glycero-3-phosphatidylcholine (PAPC), 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/20
0/20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4(n-6)cis PC
4(n-6)cis PC
• 1-stearoyl-2-arahidonoyl-sn-glycero-3-phosphatidylcholine (SAPC), 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/20
0/20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4(n-6)cis PC
4(n-6)cis PC
• 1-palmitoyl-2-eicosapentaenoyl-sn-glycero-3-phosphatidylcholine (PEPC), 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/20
0/20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5(n-3)cis PC
5(n-3)cis PC
• 1-stearoyl-2-eicosapentaenoyl-sn-glycero-3-phosphatidylcholine (SEPC), 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/20
0/20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5(n-3)cis PC
5(n-3)cis PC
• 1-palmitoyl-2-docosahexaenoyl-sn-glycero-3-phosphatidylcholine (PDPC), 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/22
0/22![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6(n-3)cis PC
6(n-3)cis PC
• 1-stearoyl-2-docosahexaenoyl-sn-glycero-3-phosphatidylcholine (SDPC), 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/22
0/22![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6(n-3)cis PC
6(n-3)cis PC
Besides, structures of 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/22
0/22![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6(n-3)cis PC molecule and four FA chains, 18
6(n-3)cis PC molecule and four FA chains, 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1(n-9)cis, 18
1(n-9)cis, 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2(n-6) cis, 18
2(n-6) cis, 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3(n-3)cis, 20
3(n-3)cis, 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4(n-6)cis, are presented in Fig. 1.
4(n-6)cis, are presented in Fig. 1.
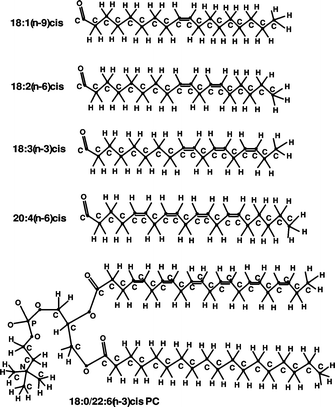 | ||
Fig. 1 Structures of phosphatidylcholine molecule of 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/22 0/22![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6(n-3)cis PC and hydrocarbon (FA) chains of 18 6(n-3)cis PC and hydrocarbon (FA) chains of 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1(n-9)cis, 18 1(n-9)cis, 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2(n-6)cis, 18 2(n-6)cis, 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3(n-3)cis and 20 3(n-3)cis and 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4(n-6)cis. 4(n-6)cis. | ||
Some other types of lipids abundant in living cells and discussed in this review are phosphatidylethanolamine (PE), sphingomyelin (SM), and phosphatidylserine (PS).
A typical snapshot of a lipid bilayer composed of 128 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/18
0/18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2(n-6)cis PC lipids and hydrated by 3840 water molecules, simulated by atomistic MD in rectangular periodic cell of size 64 × 64 × 66Å, is shown in Fig. 2.
2(n-6)cis PC lipids and hydrated by 3840 water molecules, simulated by atomistic MD in rectangular periodic cell of size 64 × 64 × 66Å, is shown in Fig. 2.
 | ||
Fig. 2 Snapshot of an equilibrium configuration of the hydrated bilayer system consisting of 128 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/18 0/18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2(n-6)cis PC lipids and 3840 water molecules as taken out from the computer simulations. Color-coding is the following: single bonded carbon atoms are green, double bonded carbons are yellow, water oxygens are red, lipid oxygens are dark red, hydrogens are grey, phosphorus atoms are black, and nitrogens are blue. 2(n-6)cis PC lipids and 3840 water molecules as taken out from the computer simulations. Color-coding is the following: single bonded carbon atoms are green, double bonded carbons are yellow, water oxygens are red, lipid oxygens are dark red, hydrogens are grey, phosphorus atoms are black, and nitrogens are blue. | ||
2 Force field development
Proper parametrization of the force field defining molecular interactions is ongoing problem in molecular simulations. A good force field should provide agreement with all available experimental data, within the simulation and experimental uncertainty. As simulations becoming longer, uncertainties caused by the equilibration stage and statistical error are decreasing. Experimental techniques are also improving. At some point, the force field which earlier provided satisfactory agreement with experimental data, may begin to show discrepancies. This in turn may initiate further improvements of the force field leading to better description of the molecular interactions and better agreement between computer simulations and experimental results.In simulation of lipid bilayers, two families of force fields were typically used in recent years: GROMOS44–46 and CHARMM.47,48 GROMOS employs united atoms approach representing each of non-polar CH, CH2 and CH3 groups of hydrocarbons as a single particle which allows to reach about 3-fold speedup comparing to all-atomic simulations. There exists several versions of the GROMOS force field which essentially fall into two groups, one with original GROMOS non-bonded parameters (for example, 45A3 and similar parameter sets46), and Berger modification49 which is the most frequently used. In the latter one, besides modification of non-bonded interaction parameters, the Ryckaert-Bellemans potential is implemented to describe torsion rotations of the hydrocarbon chains of lipids. GROMOS force field is fully supported in GROMACS simulation package.50
The CHARMM force field48 describes all hydrogens explicitly. Additionally, it has a more detailed description of intramolecular interactions, including Urey-Bradley term for covalent angles and a richer variety of parameters for dihedral angles. CHARMM parameters for lipids were introduced first in ref. 51 (within the Charmm22 parameter set, often denoted also as C22) and then were updated in ref. 24 (Charmm27, or C27 parameter set) and again in ref. 52 (C27r parameter set). The most recent C36 parameter set for lipids has been published in ref. 53. Besides the original CHARMM software, the CHARMM force field is native in the NAMD simulation package.54 It is also implemented in a number of other simulation packages.
Recently, another frequently used for biomolecular simulations AMBER force field (known also as GAFF, or Generalized Amber Force Field55) was also extended to include lipid parameters.56
Several methodological studies were devoted to validation of the different force fields used in bilayer simulations. The average area per lipid defined in constant pressure - zero tension simulations, is a parameter which is most often used to define the quality of the force field. Area per lipid is one of the most fundamental properties of a lipid bilayer and one of the most common ways to determine whether the bilayer system has reached equilibrium. When the area per lipid reaches a stable value, other structural properties (density distributions, NMR order parameters) do not change either. Simulated area per lipid can be also compared with experimental values available from X-ray or neutron diffraction and volumetric data. A collection of average lipid areas for several bilayers composed from different lipids and computed from different force fields, as well as experimental areas, is available in Table 1 of ref. 57. More reliable validation of a force field can be done by comparison of simulated and experimental structure factors as it was shown in ref. 58. Additional important source of data for validation of a force field used in lipid bilayer simulations is NMR bond order parameters.
In ref. 46, the 45A3 GROMOS parameter set, as well as few other versions of the GROMOS force field, were tested in simulations of 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/16
0/16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0 PC bilayer at 323 K, by comparison with experimentally known average membrane area per lipid, NMR bond order parameters and lipid lateral diffusion. In ref. 58, comparison of simulated and measured in X-ray or neutron diffraction structure factor was made for 18
0 PC bilayer at 323 K, by comparison with experimentally known average membrane area per lipid, NMR bond order parameters and lipid lateral diffusion. In ref. 58, comparison of simulated and measured in X-ray or neutron diffraction structure factor was made for 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1(n-9)/18
1(n-9)/18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1(n-9) PC lipids. In ref. 59, 14
1(n-9) PC lipids. In ref. 59, 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/14
0/14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0 PC bilayer was simulated using Berger parameter set49 at 30 and 50 °C and comparison with similar set of experimental data has been made. A common conclusion from these as well as some other studies60–62 can be made that while giving a fair representation of the bilayer structure and dynamics, the GROMOS force field still has some small, but going beyond possible computational or experimental error differences for the electron density profile (or the structure factor), area per lipid and some other properties.
0 PC bilayer was simulated using Berger parameter set49 at 30 and 50 °C and comparison with similar set of experimental data has been made. A common conclusion from these as well as some other studies60–62 can be made that while giving a fair representation of the bilayer structure and dynamics, the GROMOS force field still has some small, but going beyond possible computational or experimental error differences for the electron density profile (or the structure factor), area per lipid and some other properties.
Recently two new updates of GROMOS parameter set have been proposed. In the first one (called 43A1-S3 parameter set) some additional revision of parameters was made on the basis of ab initio computations and fitting to thermodynamical data for liquid alkanes.63 These corrections improved agreement with experiment for the area per lipid for a number of lipids in comparison with other versions of the GROMOS force field. Finally, the latest version of the GROMOS force field (the G53A6 parameter set) which greatly improved the fluidity of 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/16
0/16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0 PC lipid bilayers was recently reported.64 Specifically, the repulsion between the choline methyl groups and the non-ester phosphate oxygens was enhanced by increasing the van der Waals radius for this particular interaction. The structural properties of 16
0 PC lipid bilayers was recently reported.64 Specifically, the repulsion between the choline methyl groups and the non-ester phosphate oxygens was enhanced by increasing the van der Waals radius for this particular interaction. The structural properties of 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/16
0/16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0 PC bilayers (area and volume per lipid, electron density profiles, bilayer thickness and hydration, ordering and conformation of acyl chains) were in very good agreement with experiment.64 The ability of this parameter set64 to reproduce the structural and hydration properties of common phospholipids of varying length and degree of unsaturation of the acyl chains, i.e., pure bilayers of 12
0 PC bilayers (area and volume per lipid, electron density profiles, bilayer thickness and hydration, ordering and conformation of acyl chains) were in very good agreement with experiment.64 The ability of this parameter set64 to reproduce the structural and hydration properties of common phospholipids of varying length and degree of unsaturation of the acyl chains, i.e., pure bilayers of 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/12
0/12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0 PC, 14
0 PC, 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/14
0/14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0 PC, 18
0 PC, 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1(n-9)cis/18
1(n-9)cis/18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1(n-9)cis PC, and 16
1(n-9)cis PC, and 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/18
0/18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1(n-9)cis PC in a liquid crystal phase was examined in ref. 57. The simulations demonstrated that the set64 is well suited for the simulation of PC bilayers in the biologically relevant liquid-crystalline phase. The structural properties of the bilayers were validated using a broad range of experimental data for each lipid. Critically, the extent of hydration of the lipid headgroups was found to be in agreement with NMR, X-ray, and neutron diffraction as well as infrared spectroscopic data. The work in ref. 57 underlines the fact that to validate simulation models, especially those used to model lipid bilayers, there is a critical need to examine a range of experimental data as opposed to focusing on a single parameter, such as the area per lipid alone.
1(n-9)cis PC in a liquid crystal phase was examined in ref. 57. The simulations demonstrated that the set64 is well suited for the simulation of PC bilayers in the biologically relevant liquid-crystalline phase. The structural properties of the bilayers were validated using a broad range of experimental data for each lipid. Critically, the extent of hydration of the lipid headgroups was found to be in agreement with NMR, X-ray, and neutron diffraction as well as infrared spectroscopic data. The work in ref. 57 underlines the fact that to validate simulation models, especially those used to model lipid bilayers, there is a critical need to examine a range of experimental data as opposed to focusing on a single parameter, such as the area per lipid alone.
The CHARMM force field, describing all hydrogens explicitly, and having a richer variety of parameters for dihedral angles, many of which being developed on the basis of quantum-chemical calculations, may seem to have advantages in accurate description of lipid bilayers. However, recent detailed investigations have shown that the CHARMM force field, including its newer CHARMM27 version,24 have also non-negligible disagreements with experiment.58 Moreover, it was found that such fundamental parameter as the average area per lipid, is underestimated in constant-pressure simulations, and in some cases the bilayer goes to the gel phase at conditions corresponding to the liquid crystalline phase.65–68 This was the reason that many recent bilayer simulations employing CHARMM27 force field were done either in the NVT ensemble, or with a fixed area per lipid, or under non-zero surface tension.58,69–72 One of the reasons of such behavior can be traced to too strong preference for “trans” conformations in the saturated alkane chains of lipid tails described by the CHARMM27 force field.52,62,68
Recently, further update of CHARMM torsion parameters for alkane chains has been suggested, known as C27r parameter set.52 Revision of parameters was made on the basis of ab initio recomputation of the torsion potential energies for short alkanes, which lead to some decrease of the energy difference between trans- and gauche- conformations. Later studies have demonstrated however that C27r parameter set still does not reproduce the correct area per lipid in simulations at zero surface tension.70 Another way to improve parameters was to recalculate charges of the lipid headgroup, which was suggested by Sonne in ref. 67 and 73. In these works, the charges were computed within the ab initio Hartree–Fock approach for an ensemble of typical lipid conformations taken from a molecular dynamics trajectory, and then averaged. Though recalculation of charges has brought result closer to the experiment, the resulting area per lipid was still about 4–5 Å2 too low, both for 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/16
0/16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0 PC67 and 14
0 PC67 and 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/14
0/14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0 PC74 lipids.
0 PC74 lipids.
In ref. 68, an empirical way of gradual change of the energy difference between trans- and gauche- conformations of alkane chains, by scaling of the 1–4 electrostatic interactions (between atoms separated by exact three covalent bonds) was suggested. It was shown however that the parameter, which most closely reproduces the experimentally known gauche-trans ratio in liquid alkanes, still provides a too low area per lipid for 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/14
0/14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0 PC bilayer. However, after recomputation of atomic charges in the same manner as in ref. 67, a 100 ns simulation of 14
0 PC bilayer. However, after recomputation of atomic charges in the same manner as in ref. 67, a 100 ns simulation of 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/14
0/14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0 PC bilayer at 303 K has shown an area per lipid in perfect agreement with experiment, both in the case of using TIP3P and SPC water models. Very good agreement was also demonstrated for other experimentally measurable bilayer properties as bond order parameters, electron density and the structure factor.68 In ref. 75 simulations of bilayers composed of 16
0 PC bilayer at 303 K has shown an area per lipid in perfect agreement with experiment, both in the case of using TIP3P and SPC water models. Very good agreement was also demonstrated for other experimentally measurable bilayer properties as bond order parameters, electron density and the structure factor.68 In ref. 75 simulations of bilayers composed of 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/16
0/16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0 PC, 16
0 PC, 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/18
0/18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1(n-9)cis PC and 16
1(n-9)cis PC and 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/22
0/22![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6(n-3)cis PC lipids, with atomic charges derived in ref. 67 and with alkane torsion parameters described by C27r parameter set, also provided good agreement with experimental data for these types of lipids.
6(n-3)cis PC lipids, with atomic charges derived in ref. 67 and with alkane torsion parameters described by C27r parameter set, also provided good agreement with experimental data for these types of lipids.
The reason by which modification of charges based on ab initio computations for the whole lipid headgroup67,68 provides a significant improvement for the simulation results, can be rationalized from the following. In the original derivation of force field parameters, the charges of individual atomic groups constituting a lipid molecule were fixed as +1 for the choline group, −1 for the phosphate, and 0 for the rest. When these groups are gathered in a single molecule, some redistribution of charges occurs, leaving charge +0.76 on the choline group, −0.89 for the phosphate and +0.13 for the esters. Such redistribution leads also to decrease of “in-plane” lipid dipole moments and increase of the dipole moment normal to the membrane surface. Both factors favor to repulsion between the lipid headgroups and thus to increase of the area, bringing it in agreement with the experiment.
As another line of modification of the CHARMM force field for lipids, it was suggested to use a united atom description of hydrocarbons in lipid tails similar to the Ryckaert-Bellemans torsion potential but with some other parameters.76 However simulations using this model were carried out at constant area per lipid and comment was made that behavior of the average area and surface tension is similar to that for the unmodified all-atom CHARMM force field.
The most recent update of the CHARMM force field, the C36 parameter set, was presented in a recent paper53 and validated on six lipid types: 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/12
0/12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0 PC, 14
0 PC, 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/14
0/14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0 PC, 16
0 PC, 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/16
0/16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0 PC, 18
0 PC, 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1/18
1/18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 PC, 16
1 PC, 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/18
0/18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 PC and 16
1 PC and 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/18
0/18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 PE. The changes included reparameterization of partial atom charges and torsion potentials on the basis of ab initio computations, as well as revision of some Lennard-Jones parameters. Properties as average area per lipid at zero tension, structure factors, NMR order parameters, dipole electrostatic potential, showed certain improvements relative to the previous C27r parameter set.
1 PE. The changes included reparameterization of partial atom charges and torsion potentials on the basis of ab initio computations, as well as revision of some Lennard-Jones parameters. Properties as average area per lipid at zero tension, structure factors, NMR order parameters, dipole electrostatic potential, showed certain improvements relative to the previous C27r parameter set.
The Amber force field was used for description of atomistic models of lipid membranes less frequently, though some earlier simulations of lipid bilayers were performed just with Amber force field.77,78 Simulations carried out within the standard Amber94 set of force field parameters,79,80 as well as using its newer version known as GAFF, provided average lipid area below the experimental value for 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/14
0/14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0 PC and 18
0 PC and 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1/18
1/18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 PC lipids.81 Some additional modifications of the GAFF force field, including recomputations of atomic charges, were made in ref. 61. However, the average area per lipid in constant pressure simulations of 18
1 PC lipids.81 Some additional modifications of the GAFF force field, including recomputations of atomic charges, were made in ref. 61. However, the average area per lipid in constant pressure simulations of 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1/18
1/18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 PC bilayer still remained below experimental, and additional surface tension should be applied to maintain the correct area. The GAFF force field seems to need further optimization to reproduce correct bilayer structure for a tensionless membrane.
1 PC bilayer still remained below experimental, and additional surface tension should be applied to maintain the correct area. The GAFF force field seems to need further optimization to reproduce correct bilayer structure for a tensionless membrane.
There exist also other than force field factors affecting simulation results. Finite system-size effects in MD simulations of lipid bilayers are subject to much discussion in the membrane simulation community. In ref. 82, system-size effects on the structure of a 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1(n-9)cis/18
1(n-9)cis/18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1(n-9)cis PC bilayer are investigated by performing MD simulations of small and large single bilayer patches (72 and 288 lipids, respectively), as well as an explicitly multilamellar system consisting of a stack of five 72-lipid bilayers, all replicated in three dimensions by using periodic boundary conditions. The analysis82 demonstrates that finite-size effects are negligible in simulations of 18
1(n-9)cis PC bilayer are investigated by performing MD simulations of small and large single bilayer patches (72 and 288 lipids, respectively), as well as an explicitly multilamellar system consisting of a stack of five 72-lipid bilayers, all replicated in three dimensions by using periodic boundary conditions. The analysis82 demonstrates that finite-size effects are negligible in simulations of 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1(n-9)cis/18
1(n-9)cis/18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1(n-9)cis PC bilayers at low hydration. A similar study was performed for a saturated bilayer: MD simulations of 16
1(n-9)cis PC bilayers at low hydration. A similar study was performed for a saturated bilayer: MD simulations of 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/16
0/16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0 PC bilayers composed of 72 and 288 lipids were used in ref. 83 to examine system size dependence on dynamical properties.
0 PC bilayers composed of 72 and 288 lipids were used in ref. 83 to examine system size dependence on dynamical properties.
Concluding discussion on methodological issues in lipid bilayer simulation, it might be constructive to bring attention to the treatment of long-range corrections to the Lennard-Jones potential. While importance of correct treatment of the long-range electrostatic forces is well recognized,84,85 and vast majority of lipid bilayer simulations implement Ewald summation method, the role of long-range corrections to the Lennard-Jones forces is less appreciated. Most of molecular dynamics simulations employ a force cutoff distance of 10–14 Å, out of which van-der-Waals interactions are neglected. Though the attractive part of the Lennard-Jones potential may seem to be small at such distances, its total contributions to the energy and especially pressure are not negligible. They can be evaluated by assuming that the pair correlation function g(r) is equal to 1 beyond the cutoff distance r > Rcut86 for all atom pairs. Application of these expressions to a 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/14
0/14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0 lipid bilayer described by CHARMM27 force field results in correction for pressure of −360 bar for Rcut = 10 Å, which decreases to −130 bar for Rcut = 14 Å. Using of a transition region for the Lennard-Jones potential modifies the formulas but does not remove the need for correction, which can still be of order of hundred atmospheres. There exists also a more accurate isotropic periodic sum (IPS) approach87,88 which takes into account long-range corrections to the Lennard-Jones potential for inhomogeneous systems. Still, using of in principle more correct methods to compute intermolecular interactions not necessarily leads to improvement of the results, since “infinite” cutoff of the Lennard-Jones forces may occur not consistent with older force fields. Taking or not taking into account the long-range corrections (known also as dispersion corrections) in simulations of lipids may change the surface tension by several dyn/cm per leaflet85 which may lead to noticeable difference in computed average areas per lipid. Different treatments of out-cut-off corrections may also explain some differences in the results for simulated bilayers computed in different works implementing the same force field.
0 lipid bilayer described by CHARMM27 force field results in correction for pressure of −360 bar for Rcut = 10 Å, which decreases to −130 bar for Rcut = 14 Å. Using of a transition region for the Lennard-Jones potential modifies the formulas but does not remove the need for correction, which can still be of order of hundred atmospheres. There exists also a more accurate isotropic periodic sum (IPS) approach87,88 which takes into account long-range corrections to the Lennard-Jones potential for inhomogeneous systems. Still, using of in principle more correct methods to compute intermolecular interactions not necessarily leads to improvement of the results, since “infinite” cutoff of the Lennard-Jones forces may occur not consistent with older force fields. Taking or not taking into account the long-range corrections (known also as dispersion corrections) in simulations of lipids may change the surface tension by several dyn/cm per leaflet85 which may lead to noticeable difference in computed average areas per lipid. Different treatments of out-cut-off corrections may also explain some differences in the results for simulated bilayers computed in different works implementing the same force field.
3 Bilayers: Lipids with saturated, unsaturated, and polyunsaturated chains
It was mentioned that a typical biological membrane contains many species of lipid molecules, with different head groups and hydrocarbon tails. The most commonly occurring FA chains may contain 1–6 carbon - carbon double bonds of the cis configuration in different positions. In most cases, at least half of the FA chains are unsaturated. The double bonds of polyunsaturated (PU) chains are, as a rule, methylene-interrupted.The PUFA tails of lipids are of great importance for the structure and functioning of biomembranes.89–98 Docosahexaenoic acid, 22![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6(n-3)cis, is the longest and most unsaturated FA commonly found in nature.
6(n-3)cis, is the longest and most unsaturated FA commonly found in nature.
A number of animal and plant species, tissues or organs may be cited which contain membranes with one or several unsaturated lipid chains as their main component. PUFAs play a key role in membrane metabolism and the control of gene expression. It has been observed that membranes that are active metabolically, have high levels of PUFAs; 22![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6(n-3)cis chain and other PUFAs have been linked to the great number of biochemical processes, to an enormous variety of human afflictions, chronic diseases. Key transcription factors are regulated by (n-3) PUFAs, which in turn control levels of proteins involved in lipid and carbohydrate metabolism. 22
6(n-3)cis chain and other PUFAs have been linked to the great number of biochemical processes, to an enormous variety of human afflictions, chronic diseases. Key transcription factors are regulated by (n-3) PUFAs, which in turn control levels of proteins involved in lipid and carbohydrate metabolism. 22![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6(n-3)cis acid has been established as a key controller of hepatic lipid synthesis,99 22
6(n-3)cis acid has been established as a key controller of hepatic lipid synthesis,99 22![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6(n-3)cis acid and related FAs reduce colon cancer risk and inflammatory disorders of the intestine.100 PUFAs of the (n-3) series have immunosuppressive effects which make these molecules candidates for treating inflammatory symptoms associated with cardiovascular disease, obesity, arthritis, and asthma.101 22
6(n-3)cis acid and related FAs reduce colon cancer risk and inflammatory disorders of the intestine.100 PUFAs of the (n-3) series have immunosuppressive effects which make these molecules candidates for treating inflammatory symptoms associated with cardiovascular disease, obesity, arthritis, and asthma.101 22![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6(n-3)cis FA is highly concentrated in the central nervous system and is essential for proper neuronal and retinal function.102 The potential role of many oxidation products of 22
6(n-3)cis FA is highly concentrated in the central nervous system and is essential for proper neuronal and retinal function.102 The potential role of many oxidation products of 22![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6(n-3)cis FA on induction of apoptosis in cancer cells is reviewed in ref. 103.
6(n-3)cis FA on induction of apoptosis in cancer cells is reviewed in ref. 103.
Evidently the basis (and primary cause) of these and similar phenomena is the specific chemical structure of PUFA chains (in particular, 22![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6(n-3)cis) having methylene-interrupted cis double bonds, which results in their specific physical properties, which are in its turn cause their specific functioning in living organisms. Nevertheless, full understanding of the effects of lipid unsaturation on various physical properties of membranes at the molecular level, affecting their functioning, is not yet achieved. The mechanisms of many biological functions of PUFAs remain a subject of much debate.
6(n-3)cis) having methylene-interrupted cis double bonds, which results in their specific physical properties, which are in its turn cause their specific functioning in living organisms. Nevertheless, full understanding of the effects of lipid unsaturation on various physical properties of membranes at the molecular level, affecting their functioning, is not yet achieved. The mechanisms of many biological functions of PUFAs remain a subject of much debate.
Many theoretical investigations (during the period of last 7 years) were devoted to the various properties of unsaturated and PUFA chains of different lipid molecules in bilayers; several FA chains most frequently studied by molecular simulations and corresponding references are enumerated below: 22![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6(n-3)cis,93,104–116 22
6(n-3)cis,93,104–116 22![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5(n-3)cis,104 20
5(n-3)cis,104 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4(n-6)cis,66,107,110,112–115,117,118 18
4(n-6)cis,66,107,110,112–115,117,118 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3(n-3)cis107,110,112,113,115 18
3(n-3)cis107,110,112,113,115 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2(n-6)cis,66,107,110,112–115,119 18
2(n-6)cis,66,107,110,112–115,119 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1(n-9)cis.56,57,61,66,81,93,106–108,110,112–115,117,120–141
1(n-9)cis.56,57,61,66,81,93,106–108,110,112–115,117,120–141
MD simulations were also used to study the properties of mixed 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1(n-9)cis/18
1(n-9)cis/18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1(n-9)cis PC – 18
1(n-9)cis PC – 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1(n-9)cis/18
1(n-9)cis/18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1(n-9)cis PE bilayers as a function of PC/PE headgroup composition.120 The molecular organization in model membranes composed of 18
1(n-9)cis PE bilayers as a function of PC/PE headgroup composition.120 The molecular organization in model membranes composed of 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1(n-9)trans/18
1(n-9)trans/18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0 PC, 18
0 PC, 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1(n-9)cis/18
1(n-9)cis/18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0 PC, 18
0 PC, 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/18
0/18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0 PC was compared by MD simulations.140 It is shown that acyl chain order in 18
0 PC was compared by MD simulations.140 It is shown that acyl chain order in 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1(n-9)trans/18
1(n-9)trans/18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0 PC in the liquid crystalline state is much closer to that of 18
0 PC in the liquid crystalline state is much closer to that of 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1(n-9)cis/18
1(n-9)cis/18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0 PC than that of the substantially more ordered 18
0 PC than that of the substantially more ordered 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/18
0/18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0 PC, which is attributed to the reduced energy barrier to rotation about the C–C single bonds next to either a trans or cis carbon double bond.
0 PC, which is attributed to the reduced energy barrier to rotation about the C–C single bonds next to either a trans or cis carbon double bond.
Cardiolipin is a key lipid component in the inner mitochondrial membrane, where the lipid is involved in energy production and mechanisms in the apoptotic pathway. Cardiolipin has a unique dimeric structure with two negatively charged phosphatidyl moieties attached to a glycerol group and a total of four acyl chains. Three cardiolipin –16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/18
0/18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1(n-9)cis PC bilayers with different lipid compositions were simulated by MD in ref. 133 to investigate cardiolipin and its effect on the structure of lipid bilayers.
1(n-9)cis PC bilayers with different lipid compositions were simulated by MD in ref. 133 to investigate cardiolipin and its effect on the structure of lipid bilayers.
A large number of researches is traditionally involved in the computer simulation studies of saturated lipid bilayer systems. A series of simulations of hydrated 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/16
0/16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0 PC bilayers were presented in ref. 142. A number of properties and parameters (the cutoff methods, system sizes, and hydration) was varied in these simulations. In ref. 143, 16
0 PC bilayers were presented in ref. 142. A number of properties and parameters (the cutoff methods, system sizes, and hydration) was varied in these simulations. In ref. 143, 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/16
0/16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0 PC bilayer systems were investigated, and the convergence of structural and dynamical properties with system size and with time in MD simulations were studied.
0 PC bilayer systems were investigated, and the convergence of structural and dynamical properties with system size and with time in MD simulations were studied.
A MD simulation of a hydrated 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0-SM lipid bilayer was carried out in ref. 144. Properties of the bilayer calculated from the simulation are compared with those of 16
0-SM lipid bilayer was carried out in ref. 144. Properties of the bilayer calculated from the simulation are compared with those of 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/16
0/16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0 PC bilayers. The 18
0 PC bilayers. The 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0-SM bilayers are significantly more ordered and compact than 16
0-SM bilayers are significantly more ordered and compact than 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/16
0/16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0 PC bilayers at the same temperature.144
0 PC bilayers at the same temperature.144
In ref. 145, MD simulations of 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/14
0/14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0 PC model system in the fluid phase was combined with several experimental methods. The combination of experiment and simulation offers a powerful set of tools to investigate the lipid structure and dynamics. Whereas experiments are essential for force-field validation and developments, simulations help to interpret and complement experiments and can, in turn, initiate further experimental studies. Similarly, combined MD simulations and experiments of fluid phase 16
0 PC model system in the fluid phase was combined with several experimental methods. The combination of experiment and simulation offers a powerful set of tools to investigate the lipid structure and dynamics. Whereas experiments are essential for force-field validation and developments, simulations help to interpret and complement experiments and can, in turn, initiate further experimental studies. Similarly, combined MD simulations and experiments of fluid phase 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/16
0/16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0 PC bilayers were performed in ref. 146.
0 PC bilayers were performed in ref. 146.
For a literature on MD simulation studies of several fully hydrated bilayers (e.g., 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/16
0/16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0 PC, 16
0 PC, 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/16
0/16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0 PE) in gel phase, see ref. 129, 147 and 148 Studies of the gel to liquid-crystal phase transitions of fully hydrated bilayers were also performed.147,149,150 It should be mentioned also a series of MD simulations of the specific branch-chained lipid bilayers151–155 and a ceramide bilayer.156–158
0 PE) in gel phase, see ref. 129, 147 and 148 Studies of the gel to liquid-crystal phase transitions of fully hydrated bilayers were also performed.147,149,150 It should be mentioned also a series of MD simulations of the specific branch-chained lipid bilayers151–155 and a ceramide bilayer.156–158
4 Sterols, anesthetics and other inclusions in lipid bilayers
4.1 Cholesterol
Sterols are essential constituents of mammalian cell membranes, one of them is CHOL. The significance of CHOL in biological membranes has been known for a long time. A large number of experimental and theoretical studies has been devoted to unravel the modes of action of this molecule (for reviews see, e.g.,ref. 29, 35, 38, 98, 159 and 170).CHOL′s preference for specific fatty acid chains was investigated in MD computer simulations109 of a lipid bilayer membrane consisting of CHOL and 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/22
0/22![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6(n-3)cis PC in a 1
6(n-3)cis PC in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 ratio. Three bilayer systems were studied by MD in work:111 18
3 ratio. Three bilayer systems were studied by MD in work:111 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/22
0/22![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6(n-3)cis PC, 18
6(n-3)cis PC, 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/22
0/22![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5(n-6)cis PC, and 18
5(n-6)cis PC, and 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/22
0/22![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6(n-3)cis PC with 25 mol % CHOL. It was found that the distribution of lateral stress within the hydrophobic core of the membrane is sensitively dependent on the degree of chain unsaturation and on the presence of CHOL. Replacing (n-3) fatty acids with (n-6) chains, or incorporating CHOL into the membrane, shifts the repulsive lateral chain pressure away from the lipid/water interface toward the bilayer interior.111
6(n-3)cis PC with 25 mol % CHOL. It was found that the distribution of lateral stress within the hydrophobic core of the membrane is sensitively dependent on the degree of chain unsaturation and on the presence of CHOL. Replacing (n-3) fatty acids with (n-6) chains, or incorporating CHOL into the membrane, shifts the repulsive lateral chain pressure away from the lipid/water interface toward the bilayer interior.111
An all-atom MD simulation of lipid bilayers with different CHOL/SM molar ratios was reported in ref. 171 Five hydrated systems were built with molar CHOL/18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0-SM ratios of 0/100 (pure SM), 20/80, 30/70, 35/65, and 40/60. The results revealed structural and dynamic changes suggesting the random distribution of lipids along the bilayer planes is supplanted at CHOL concentrations above 30 mol % by the formation of a liquid-ordered phase, which is thought to be the precursor to lipid raft formation. The packing of molecules in the bilayer is shown to be associated with the hydrogen bonding between CHOL and SM. The molecules tend to migrate toward distributions in which the SM molecule forms on average one hydrogen bond with a CHOL molecule.171
0-SM ratios of 0/100 (pure SM), 20/80, 30/70, 35/65, and 40/60. The results revealed structural and dynamic changes suggesting the random distribution of lipids along the bilayer planes is supplanted at CHOL concentrations above 30 mol % by the formation of a liquid-ordered phase, which is thought to be the precursor to lipid raft formation. The packing of molecules in the bilayer is shown to be associated with the hydrogen bonding between CHOL and SM. The molecules tend to migrate toward distributions in which the SM molecule forms on average one hydrogen bond with a CHOL molecule.171
MD simulations to consider 1,6-diphenyl-1,3,5-hexatriene fluorescent probes in a fluid hydrated 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/16
0/16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0 PC bilayers with 5 and 20 mol % CHOL were used in ref. 172. It was shown that while the fluorescent probe affects a number of membrane properties, the perturbations induced by the probe depend on the concentration of CHOL in a membrane. The fluorescent probe was found to influence the mass density distribution of lipids across the membrane and to promote the ordering of acyl chains around the probe. Yet, these perturbations get relatively weaker for increasing CHOL concentration.
0 PC bilayers with 5 and 20 mol % CHOL were used in ref. 172. It was shown that while the fluorescent probe affects a number of membrane properties, the perturbations induced by the probe depend on the concentration of CHOL in a membrane. The fluorescent probe was found to influence the mass density distribution of lipids across the membrane and to promote the ordering of acyl chains around the probe. Yet, these perturbations get relatively weaker for increasing CHOL concentration.
4.2 Anesthetics
Another important membrane inclusions can be anesthetics (e.g., lidocaine, benzocaine, halothane, hexafluoroethane, short chain alcohols like methanol, ethanol, 1-alkanols).105,173–180 The specific molecular mechanism of action of anesthetics and details of their interactions with biological membranes are, to a large extent, unknown or poorly understood. For instance, lidocaine-family drugs are widely used as local anesthetics in medical treatment to prevent or relieve pain. Clearly, the lidocaine - membrane (and other anesthetics) interaction perturbs the bilayer structure. It is speculated175 that this change in the local order will also affect the lipid protein (ion channel) interaction which is claimed to be essential for the anesthetic activity. In ref. 179 it was demonstrated that addition of local anesthetic benzocaine increases disorder in the membrane. Since the action of general anesthetics was known to be pressure dependent, MD simulations of such a molecule, halothane, embedded in a 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/14
0/14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0 PC membrane, performed under physiological conditions and also at elevated pressures were carried out in ref. 181. The results clearly show that at high pressures the halothane molecules tend to cluster together. A possible mechanism for pressure reversal of general anesthetics from computer simulations is discussed in ref. 182. Recent paper, ref. 180, contains an account of a series of simulations of PC bilayers discussing a possible effect of halotane general anesthetic on K+ ion channel.
0 PC membrane, performed under physiological conditions and also at elevated pressures were carried out in ref. 181. The results clearly show that at high pressures the halothane molecules tend to cluster together. A possible mechanism for pressure reversal of general anesthetics from computer simulations is discussed in ref. 182. Recent paper, ref. 180, contains an account of a series of simulations of PC bilayers discussing a possible effect of halotane general anesthetic on K+ ion channel.
In ref. 183, long-time MD simulations were performed to investigate the effect of 1-alkanols of various carbon chain lengths onto the structure and dynamics of 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/14
0/14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0 PC bilayers. All investigated 1-alkanols assembled inside the lipid bilayer within tens of nanoseconds. Their hydroxyl groups bound preferentially to the lipid carbonyl group and the hydrocarbon chains stretched into the hydrophobic core of the bilayer. The studies showed that all 1-alkanols drastically affected the bilayer properties. Insertion of long-chain 1-alkanols decreased the area per lipid while increasing the thickness of the bilayer and the order of the lipids. The bilayer elasticity was reduced and the diffusive motion of the lipids within the bilayer plane was suppressed. On the other hand, integration of ethanol into the bilayer enlarged the area per lipid. The bilayer became softer and lipid diffusion was enhanced.183
0 PC bilayers. All investigated 1-alkanols assembled inside the lipid bilayer within tens of nanoseconds. Their hydroxyl groups bound preferentially to the lipid carbonyl group and the hydrocarbon chains stretched into the hydrophobic core of the bilayer. The studies showed that all 1-alkanols drastically affected the bilayer properties. Insertion of long-chain 1-alkanols decreased the area per lipid while increasing the thickness of the bilayer and the order of the lipids. The bilayer elasticity was reduced and the diffusive motion of the lipids within the bilayer plane was suppressed. On the other hand, integration of ethanol into the bilayer enlarged the area per lipid. The bilayer became softer and lipid diffusion was enhanced.183
Another action mechanism discussed in the literature is the change of electrostatic potential inside membrane (called also dipole potential). It was demonstrated recently by MD simulations that addition of lidocaine to 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/14
0/14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0 PC membrane causes noticeable increase, by up to 200 mV, of the dipole potential176 which may affect the work of ion channels and result in anesthetic action.
0 PC membrane causes noticeable increase, by up to 200 mV, of the dipole potential176 which may affect the work of ion channels and result in anesthetic action.
4.3 Small molecules
One of the key membrane functions is the regulation of the transport of small molecules across the membrane. While the membrane transport, as a rule, involves special channel forming peptides and proteins, various small, uncharged molecules, such as O2, CO2, water, NO, CO, etc., can permeate in small amounts the cell membrane without the aid of any transmembrane protein. In ref. 184 the effects of the hydrocarbon chain length of lipid molecules on the permeation process of small molecules (O2, CO, NO, and water) through lipid bilayers were investigated. MD simulations of three saturated lipid bilayer systems were performed: 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/12
0/12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0 PC, 14
0 PC, 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/14
0/14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0 PC, 16
0 PC, 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/16
0/16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0 PC.184
0 PC.184
Cell membranes need to be hydrated by water for their proper functioning. As a matter of fact, water is an essential constituent of biomembranes. Therefore, it is important to understand the structural and dynamical properties of water molecules located at the interface with lipids and other biomolecules. MD computer simulations to study the orientational dynamics of water next to bilayers containing 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/12
0/12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0 PC with different hydration levels were performed in ref. 185, next to bilayers of 18
0 PC with different hydration levels were performed in ref. 185, next to bilayers of 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1(n-9)cis/18
1(n-9)cis/18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1(n-9)cis PC and 18
1(n-9)cis PC and 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1(n-9)cis/18
1(n-9)cis/18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1(n-9)cis PS - in ref. 124, next to 18
1(n-9)cis PS - in ref. 124, next to 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1(n-9)cis/18
1(n-9)cis/18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1(n-9)cis PC bilayer - in ref. 186; see also the review in ref. 187 concerning simulations of aqueous solutions next to phospholipid membrane surfaces. Water permeability for a bilayer composed of a 2
1(n-9)cis PC bilayer - in ref. 186; see also the review in ref. 187 concerning simulations of aqueous solutions next to phospholipid membrane surfaces. Water permeability for a bilayer composed of a 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 molar ratio of ceramide NS 24
1 molar ratio of ceramide NS 24![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0 (ceramide 2) – CHOL – free FA 24
0 (ceramide 2) – CHOL – free FA 24![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0 was estimated with extended ensemble MD in ref. 158.
0 was estimated with extended ensemble MD in ref. 158.
The interaction of dimethylsulfoxide molecule ((CH3)2SO) with gel-phase bilayers of ceramide 2 was investigated in ref. 157. The liquid-crystalline phase of ceramides is expected to be markedly more permeable to solutes than the gel-phase structure.
Another long-standing problem in membrane biophysics is related to the ion permeation across protein-free lipid membranes. Pore formation in lipid membranes and subsequent pore-mediated ion transport, salt-induced effects in the plasma membrane, the electrostatic properties of membranes are also traditionally attractive topics for computer simulation studies (see, e.g.,ref. 188–200 and the literature lists). MD simulations of biologically realistic transmembrane potential gradients across a 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/14
0/14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0 PC bilayer are presented in ref. 201. These simulations are the first to model this gradient in all-atom detail, with the field generated solely by explicit ion dynamics.
0 PC bilayer are presented in ref. 201. These simulations are the first to model this gradient in all-atom detail, with the field generated solely by explicit ion dynamics.
MD simulations to consider how mono- (NaCl) and divalent (CaCl2) salt affects properties of inner and outer membranes of mitochondria were employed in ref. 202. This work seems to be the first computational study concerning mitochondrial membrane properties in the presence of salt. Six different membrane systems mimicking mitochondrial membranes with and without salt were studied. Three of them corresponded to the inner membrane and three to the outer one. The membranes were composed of PC, PE, and cardiolipin. Linoleic (18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2(n-6)cis) chain was used as the acyl chains of the lipids, i.e., two chains in PC and PE and four chains in each cardiolipin. The main focus of the study was on the basic modifications induced by ions to the bilayer. It was discovered that the influence of salt on structural properties is rather minute, only weakly affecting lipid packing, conformational ordering, and membrane electrostatic potential. The changes induced by salt were found202 to be more prominent in dynamical properties related to ion binding and formation of ion-lipid complexes and lipid aggregates, as rotational diffusion of lipids is slowed down by ions, especially in the case of CaCl2. In the same spirit, lateral diffusion of lipids is slowed down rather considerably for increasing concentration of CaCl2.
2(n-6)cis) chain was used as the acyl chains of the lipids, i.e., two chains in PC and PE and four chains in each cardiolipin. The main focus of the study was on the basic modifications induced by ions to the bilayer. It was discovered that the influence of salt on structural properties is rather minute, only weakly affecting lipid packing, conformational ordering, and membrane electrostatic potential. The changes induced by salt were found202 to be more prominent in dynamical properties related to ion binding and formation of ion-lipid complexes and lipid aggregates, as rotational diffusion of lipids is slowed down by ions, especially in the case of CaCl2. In the same spirit, lateral diffusion of lipids is slowed down rather considerably for increasing concentration of CaCl2.
4.4 Large penetrants
Some of the penetrants (e.g., drug molecules) are comparatively large. To reach their biological target, drugs have to cross cell membranes, and understanding passive membrane permeation is therefore crucial for rational drug design. Behavior of small solutes203–205 and large drugs205–207 in a lipid bilayer from computer simulations is also widely discussed. MD simulations of bilayers consisted of the single tail cationic surfactant behenyl trimethyl ammonium chloride with stearyl alcohol as the cosurfactant were reported in ref. 208.MD simulations and X-ray diffraction analysis study of several 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1(n-9)trans/18
1(n-9)trans/18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1(n-9)trans PE membranes containing free FAs was performed in ref. 209. The study was aimed at understanding the interactions of several structurally related FAs with biomembranes, which is necessary for further rational lipid drug design in membrane-lipid therapy. FAs able to affect biophysical properties of cell membranes, in turn will also alter localization and/or function of membrane protein involved in the regulation of cellular processes. For the above reasons, FAs or other lipids could be a tool to modulate pathophysiological conditions via cell membrane properties.209
1(n-9)trans PE membranes containing free FAs was performed in ref. 209. The study was aimed at understanding the interactions of several structurally related FAs with biomembranes, which is necessary for further rational lipid drug design in membrane-lipid therapy. FAs able to affect biophysical properties of cell membranes, in turn will also alter localization and/or function of membrane protein involved in the regulation of cellular processes. For the above reasons, FAs or other lipids could be a tool to modulate pathophysiological conditions via cell membrane properties.209
A coarse-grained MD method (this approach is discussed in more detail in the last section of our review) was used in ref. 210 to model the permeation properties of flat 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/16
0/16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0 PC bilayers with various amounts of incorporated monopalmitoylphosphatidylcholine lysolipid (i.e., PC molecule with one 16
0 PC bilayers with various amounts of incorporated monopalmitoylphosphatidylcholine lysolipid (i.e., PC molecule with one 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0 chain). The enhanced permeability of the membranes at their gel to liquid-crystalline phase transition was explored. A peak in the permeability was shown to coincide with the phase transition temperature from the gel to liquid-crystalline state when lysolipid is present. This peak in permeability correlates with a jump in the area per lipid near the same temperature as well as increased fluctuations in the lipid bilayer free volume.
0 chain). The enhanced permeability of the membranes at their gel to liquid-crystalline phase transition was explored. A peak in the permeability was shown to coincide with the phase transition temperature from the gel to liquid-crystalline state when lysolipid is present. This peak in permeability correlates with a jump in the area per lipid near the same temperature as well as increased fluctuations in the lipid bilayer free volume.
Transmembrane lipid translocation (flip-flop) processes can also be discussed in this context. These processes are involved in a variety of properties and functions of cell membranes; flip-flops are one of the least understood dynamical processes in membranes. In ref. 211, the atomic-scale MD simulations were performed on 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/14
0/14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0 PC bilayer. It was shown that the computational approach can actually provide a great deal of insight into the mechanism (or one of the mechanisms) associated with lipid flip-flops.
0 PC bilayer. It was shown that the computational approach can actually provide a great deal of insight into the mechanism (or one of the mechanisms) associated with lipid flip-flops.
One of the methods to introduce foreign molecules into cells is electroporation. Electric fields can induce pore formation and other structural defects in lipid membranes. The mechanism of pore formation by direct MD simulations of 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1(n-9)cis/18
1(n-9)cis/18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1(n-9)cis PC bilayers with applied electric fields of different strengths is investigated in ref. 123, that of asymmetric membranes 18
1(n-9)cis PC bilayers with applied electric fields of different strengths is investigated in ref. 123, that of asymmetric membranes 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1(n-9)cis/18
1(n-9)cis/18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1(n-9)cis PC – 18
1(n-9)cis PC – 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1(n-9)cis/18
1(n-9)cis/18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1(n-9)cis PS is studied in ref. 125,212.
1(n-9)cis PS is studied in ref. 125,212.
An important contributor to the thermodynamic driving force is the available free volume across a membrane. Thus, the diffusion properties of the penetrants are obviously related to the properties of the free volume clusters (e.g., their size, shape, orientation, etc.) present in the membrane, and therefore a detailed analysis of the voids can also provide some information on the permeability properties of the membrane. Such a study was performed for bilayers composed of 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/18
0/18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1(n-9)cis PC, 18
1(n-9)cis PC, 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/18
0/18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2(n-6)cis PC, 18
2(n-6)cis PC, 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/18
0/18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3(n-3)cis PC, 18
3(n-3)cis PC, 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/20
0/20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4(n-6)cis PC and 18
4(n-6)cis PC and 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/22
0/22![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6(n-3)cis PC molecules in ref. 112. It was found that there are no broad enough preformed channels in the bilayers through small penetrants, such as water molecules, can readily go through; however, the existing channels can largely facilitate the permeation of these molecules.
6(n-3)cis PC molecules in ref. 112. It was found that there are no broad enough preformed channels in the bilayers through small penetrants, such as water molecules, can readily go through; however, the existing channels can largely facilitate the permeation of these molecules.
Below we will focus mainly on the papers in which two subjects are touched upon, namely the effect of cholesterol on (i) the bond order parameter profiles of the lipid hydrocarbon chains with respect to the normal in various lipid/CHOL bilayers and (ii) the lateral diffusion coefficients of lipids in such bilayers.
5 Effect of cholesterol on bond order parameters of phospholipids
Bond order parameters of the hydrocarbon chains of lipids with respect to the membrane normal are important characteristics of a membrane system. They are assessable experimentally by the NMR technique. The bond ordering in various phospholipid and phospholipid/CHOL bilayers have been studied repeatedly within recent years by computer simulation methods.56,57,61,81,93,109,115,121,122,126–132,134,136,137,139,141,143,213–221The presence of CHOL has been found to increase the degree of orientational order of C–H bonds of saturated hydrocarbon chains of various phospholipids, e.g., PC, PS, SM, PE, in the liquid-crystalline phase. This is known as the condensing effect of CHOL.159
Such an effect was observed for 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/12
0/12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0 PC,217 14
0 PC,217 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/14
0/14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0 PC,214,218 16
0 PC,214,218 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/16
0/16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0 PC,213,215,217 16
0 PC,213,215,217 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/18
0/18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1(n-9)cis PE,108 16
1(n-9)cis PE,108 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/22
0/22![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6(n-3)cis PE,108 16
6(n-3)cis PE,108 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/18
0/18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1(n-9)cis PC,130 18
1(n-9)cis PC,130 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/22
0/22![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6(n-3)cis PC,109 18
6(n-3)cis PC,109 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/18
0/18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1(n-9)cis PS,127 for five different systems containing 16
1(n-9)cis PS,127 for five different systems containing 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/16
0/16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0 PC molecules and modified CHOL molecules (sterol concentration of 20 mol %),221 for eight different PC bilayer systems with 20 mol % CHOL comprising two 18-carbon long monounsaturated acyl chains, where the position of the cis double bond is varied systematically along the acyl chains, namely 18
0 PC molecules and modified CHOL molecules (sterol concentration of 20 mol %),221 for eight different PC bilayer systems with 20 mol % CHOL comprising two 18-carbon long monounsaturated acyl chains, where the position of the cis double bond is varied systematically along the acyl chains, namely 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/18
0/18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0 PC, 18
0 PC, 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1(n-3)cis/18
1(n-3)cis/18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1(n-3)cis PC, 18
1(n-3)cis PC, 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1(n-5)cis/18
1(n-5)cis/18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1(n-5)cis PC, 18
1(n-5)cis PC, 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1(n-7)cis/18
1(n-7)cis/18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1(n-7)cis PC, 18
1(n-7)cis PC, 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1(n-9)cis/18
1(n-9)cis/18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1(n-9)cis PC, 18
1(n-9)cis PC, 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1(n-11)cis/18
1(n-11)cis/18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1(n-11)cis PC, 18
1(n-11)cis PC, 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1(n-13)cis/18
1(n-13)cis/18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1(n-13)cis PC, and 18
1(n-13)cis PC, and 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1(n-15)cis/18
1(n-15)cis/18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1(n-15)cis PC.134,135
1(n-15)cis PC.134,135
The condensing effect of CHOL was investigated for two monounsaturated phospholipid systems with 40 mol % CHOL and without CHOL composed of short-chain 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1(n-5)cis/14
1(n-5)cis/14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1(n-5)cis PC and long-chain 22
1(n-5)cis PC and long-chain 22![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1(n-9)cis/22
1(n-9)cis/22![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1(n-9)cis PC,220 for 18
1(n-9)cis PC,220 for 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1(n-9)cis/18
1(n-9)cis/18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1(n-9)cis PC bilayer studied by MD using different force fields,61 for 16
1(n-9)cis PC bilayer studied by MD using different force fields,61 for 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/16
0/16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0 PC, 16
0 PC, 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/18
0/18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1(n-9)cis PC, and 18
1(n-9)cis PC, and 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1(n-9)cis/18
1(n-9)cis/18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1(n-9)cis PC bilayers with varying concentrations of CHOL (10,20, 25, and 33 mol %),137 for a system of 16
1(n-9)cis PC bilayers with varying concentrations of CHOL (10,20, 25, and 33 mol %),137 for a system of 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/18
0/18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1(n-9)cis PC – CHOL (40 mol %),132 for a mixture of 18
1(n-9)cis PC – CHOL (40 mol %),132 for a mixture of 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0-SM (∼ 15 mol %), 18
0-SM (∼ 15 mol %), 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1(n-9)cis/18
1(n-9)cis/18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1(n-9)cis PC (∼ 78 mol %), and CHOL (∼ 7 mol %),121 for a ternary mixture of 18
1(n-9)cis PC (∼ 78 mol %), and CHOL (∼ 7 mol %),121 for a ternary mixture of 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0-SM – 18
0-SM – 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1(n-9)cis/18
1(n-9)cis/18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1(n-9)cis PC – CHOL with a 1
1(n-9)cis PC – CHOL with a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 composition,122 and a binary mixture of 18
1 composition,122 and a binary mixture of 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0-SM – 18
0-SM – 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1(n-9)cis/18
1(n-9)cis/18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1(n-9)cis PC with a 1
1(n-9)cis PC with a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 composition,122 for 18
1 composition,122 for 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0-SM -– CHOL (∼ 46 mol %),216 16
0-SM -– CHOL (∼ 46 mol %),216 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/18
0/18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1(n-9)cis PC – 16
1(n-9)cis PC – 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/SM,126 for 18
0/SM,126 for 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0-SM – CHOL (0, 22, and 50 mol %) bilayer systems and 15
0-SM – CHOL (0, 22, and 50 mol %) bilayer systems and 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/18
0/18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0 PC – CHOL bilayer (22 mol %),219 for pure bilayers of 18
0 PC – CHOL bilayer (22 mol %),219 for pure bilayers of 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0-SM, 18
0-SM, 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1(n-9)cis-SM, and 16
1(n-9)cis-SM, and 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/18
0/18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1(n-9)cis PC and those with 34 mol % CHOL.131 for bilayers of 18
1(n-9)cis PC and those with 34 mol % CHOL.131 for bilayers of 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0-SM and 16
0-SM and 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/18
0/18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1(n-9)cis PC with low content of CHOL.139
1(n-9)cis PC with low content of CHOL.139
This effect was also observed for a ternary system of 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/18
0/18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1(n-9)cis PC, ganglioside-monosialated, and CHOL, for a binary system of 16
1(n-9)cis PC, ganglioside-monosialated, and CHOL, for a binary system of 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/18
0/18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1(n-9)cis PC and CHOL, and for pure bilayer 16
1(n-9)cis PC and CHOL, and for pure bilayer 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/18
0/18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1(n-9)cis PC,136 for a ternary mixture of 16
1(n-9)cis PC,136 for a ternary mixture of 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/18
0/18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1(n-9)cis PC, 16
1(n-9)cis PC, 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/18
0/18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1(n-9)cis phosphatidic acid (PA), and CHOL with a 3
1(n-9)cis phosphatidic acid (PA), and CHOL with a 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 composition,128 for several unsaturated bilayers of 18
1 composition,128 for several unsaturated bilayers of 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/18
0/18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1(n-9)cis PC, 18
1(n-9)cis PC, 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/18
0/18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2(n-6)cis PC, 18
2(n-6)cis PC, 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/18
0/18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3(n-3)cis PC, 18
3(n-3)cis PC, 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/20
0/20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4(n-6)cis PC, and 18
4(n-6)cis PC, and 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/22
0/22![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6(n-3)cis PC with 40 mol % CHOL115 in comparison with the pure bilayers.93,115
6(n-3)cis PC with 40 mol % CHOL115 in comparison with the pure bilayers.93,115
It should be mentioned that the ability of CHOL to condense fluid phospholipid membranes has been known for more than 80 years. The bond order parameters in the phospholipid tails were found to increase with addition of CHOL or increasing CHOL content.109,115,213,215,218,220 With saturated lipids, CHOL promotes the formation of highly ordered raft-like membrane domains, whereas domains rich in unsaturated lipids with a double bond in the middle remain highly fluid despite the presence of CHOL.134
In 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/18
0/18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1(n-9)cis PC – CHOL bilayers126,130 interactions with CHOL were found to increase the order of not only the saturated 16
1(n-9)cis PC – CHOL bilayers126,130 interactions with CHOL were found to increase the order of not only the saturated 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0 chain, but also the monounsaturated 18
0 chain, but also the monounsaturated 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1(n-9)cis acyl (the same is observed in other monounsaturated PC bilayers134). Additionally, it was found that the effect of CHOL on the order parameters occurs primarily in the upper half of the chains, consistent with the preferred location of the CHOL molecule (the corresponding methylene groups are located at the same depth along the bilayer normal as the CHOL rings). The order parameters were found to decrease214,217 upon approaching the center of the membrane.
1(n-9)cis acyl (the same is observed in other monounsaturated PC bilayers134). Additionally, it was found that the effect of CHOL on the order parameters occurs primarily in the upper half of the chains, consistent with the preferred location of the CHOL molecule (the corresponding methylene groups are located at the same depth along the bilayer normal as the CHOL rings). The order parameters were found to decrease214,217 upon approaching the center of the membrane.
The presence of CHOL in a bilayer was shown115 to promote the extension of both saturated and PU lipid chains. The condensing effect of CHOL on the order parameters was more pronounced for the double C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C bonds of PU chains than for single C–C bonds of saturated chains.115 On the other hand, CHOL was shown to exhibit a preferential affinity for saturated over PU lipid chains108,109,159 It was shown recently by MD simulations117 of bilayers 16
C bonds of PU chains than for single C–C bonds of saturated chains.115 On the other hand, CHOL was shown to exhibit a preferential affinity for saturated over PU lipid chains108,109,159 It was shown recently by MD simulations117 of bilayers 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/18
0/18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1(n-9)cis PC, 18
1(n-9)cis PC, 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/20
0/20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4(n-6)cis PC, 20
4(n-6)cis PC, 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4(n-6)cis/20
4(n-6)cis/20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4(n-6)cis PC, each containing 10 mol % of CHOL, that in dipolyunsaturated bilayer 20
4(n-6)cis PC, each containing 10 mol % of CHOL, that in dipolyunsaturated bilayer 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4(n-6)cis/20
4(n-6)cis/20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4(n-6)cis PC the CHOL head groups show preference for the membrane interior.
4(n-6)cis PC the CHOL head groups show preference for the membrane interior.
MD simulations with saturated 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/16
0/16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0 PC bilayers containing 20 and 40 mol % CHOL222 show that CHOL increases the order and rigidity of the bilayers, which restricts bilayer deformations and prevents pore formation. Increasing the CHOL content slows the rate of 16
0 PC bilayers containing 20 and 40 mol % CHOL222 show that CHOL increases the order and rigidity of the bilayers, which restricts bilayer deformations and prevents pore formation. Increasing the CHOL content slows the rate of 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/16
0/16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0 PC molecule flip - flop by orders of magnitude. Other simulations118 with saturated 16
0 PC molecule flip - flop by orders of magnitude. Other simulations118 with saturated 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/16
0/16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0 PC bilayers containing 0, 20 and 40 mol % CHOL and PU bilayer 20
0 PC bilayers containing 0, 20 and 40 mol % CHOL and PU bilayer 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4(n-6)cis/20
4(n-6)cis/20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4(n-6)cis PC with CHOL molecules demonstrate that CHOL prefers more ordered and rigid bilayers and has the lowest affinity for bilayers with two PU chains.118
4(n-6)cis PC with CHOL molecules demonstrate that CHOL prefers more ordered and rigid bilayers and has the lowest affinity for bilayers with two PU chains.118
The heterogeneous interactions among three different constituents of ref. 128, namely 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/18
0/18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1(n-9)cis PC, 16
1(n-9)cis PC, 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/18
0/18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1(n-9)cis PA, and CHOL, resulted in a broad spectrum of lipid properties, including varied lipid ordering as a function of lipid closeness to CHOL. The results presented in ref. 221 demonstrate that the “side” methyl groups of CHOL are important structural elements. Their role seems to be somehow related to the maintenance of optimal tilt of the sterol ring system, thus promoting the nonspecific mechanism by which CHOL increases order in various saturated membranes, such as 16
1(n-9)cis PA, and CHOL, resulted in a broad spectrum of lipid properties, including varied lipid ordering as a function of lipid closeness to CHOL. The results presented in ref. 221 demonstrate that the “side” methyl groups of CHOL are important structural elements. Their role seems to be somehow related to the maintenance of optimal tilt of the sterol ring system, thus promoting the nonspecific mechanism by which CHOL increases order in various saturated membranes, such as 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/16
0/16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0 PC.221
0 PC.221
Differences between double-bond parametrizations by varying the position of the double bond systematically along the lipid monounsaturated hydrocarbon chains were compared in ref. 135, where 7 monoenoic one-component bilayers 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1(n-j)cis/18
1(n-j)cis/18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1(n-j)cis PC were considered (j = 3, 5, 7, 9, 11, 13 and 15), and 7 mixed PC bilayers with CHOL. The results give rise for concern: they indicate that the double-bond description may change not only the quantitative but also the qualitative nature of membrane behavior.
1(n-j)cis PC were considered (j = 3, 5, 7, 9, 11, 13 and 15), and 7 mixed PC bilayers with CHOL. The results give rise for concern: they indicate that the double-bond description may change not only the quantitative but also the qualitative nature of membrane behavior.
6 Lateral self-diffusion
The lateral diffusion coefficients of phospholipids have been studied using MD computer simulations in various model bilayer systems with or without CHOL,13,81,83,109,114,115,213,215,218,223 such as pure 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/14
0/14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0 PC bilayer;13 pure bilayers composed of 16
0 PC bilayer;13 pure bilayers composed of 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/16
0/16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0 PC molecules under different simulation conditions;83 pure bilayers composed of 16
0 PC molecules under different simulation conditions;83 pure bilayers composed of 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/16
0/16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0 PC, 16
0 PC, 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/18
0/18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1(n-9)cis PC, 16
1(n-9)cis PC, 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/18
0/18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2(n-6)cis PC, 16
2(n-6)cis PC, 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/20
0/20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4(n-6)cis PC and 16
4(n-6)cis PC and 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/22
0/22![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6(n-3)cis PC molecules;114 pure bilayer of 16
6(n-3)cis PC molecules;114 pure bilayer of 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/18
0/18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1(n-9)cis PC under different simulation conditions,56 pure 14
1(n-9)cis PC under different simulation conditions,56 pure 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/14
0/14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0 PC and 18
0 PC and 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1(n-9)cis/18
1(n-9)cis/18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1(n-9)cis PC bilayers.81 14
1(n-9)cis PC bilayers.81 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/14
0/14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0 PC bilayer containing ∼25 mol % CHOL or ergosterol;218 16
0 PC bilayer containing ∼25 mol % CHOL or ergosterol;218 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/16
0/16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0 PC bilayer with 0–40 mol % CHOL;213 16
0 PC bilayer with 0–40 mol % CHOL;213 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/16
0/16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0 PC bilayer system with ∼5–50 mol % CHOL;215 18
0 PC bilayer system with ∼5–50 mol % CHOL;215 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/22
0/22![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6(n-3)cis PC bilayer system with 25 mol % CHOL;109 bilayers containing 40 mol % CHOL composed of 18
6(n-3)cis PC bilayer system with 25 mol % CHOL;109 bilayers containing 40 mol % CHOL composed of 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/18
0/18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1(n-9)cis PC, 18
1(n-9)cis PC, 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/18
0/18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2(n-6)cis PC, 18
2(n-6)cis PC, 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/18
0/18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3(n-3)cis PC, 18
3(n-3)cis PC, 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/20
0/20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4(n-6)cis PC, and 18
4(n-6)cis PC, and 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/22
0/22![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6(n-3)cis PC molecules, and corresponding pure PC bilayers, by MD simulations on short time scales,115 pure 14
6(n-3)cis PC molecules, and corresponding pure PC bilayers, by MD simulations on short time scales,115 pure 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/14
0/14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0 PC bilayer and 14
0 PC bilayer and 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/14
0/14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0 PC bilayer with 50 mol % CHOL by an atomistic-level MD simulation with a ghost probe (a two-dimensional undulating bloblike entity) and by a coarse-grained model.223
0 PC bilayer with 50 mol % CHOL by an atomistic-level MD simulation with a ghost probe (a two-dimensional undulating bloblike entity) and by a coarse-grained model.223
In spite of extensive literature on the dynamics of lipid bilayers, the determination of the lateral self-diffusion coefficient of lipid molecules in the membrane remains to a great extent a controversial problem. The calculation results of this coefficient appear to depend significantly on the time and length scales probed by the employed computational method.
It was found by various computer simulations109,213,215,218 that for temperatures slightly above the main transition temperature for the pure lipid bilayer, the lateral diffusion coefficient (mainly of saturated and monounsaturated lipids) decreases significantly while passing from the liquid-disordered phase to the liquid ordered phase by increasing the CHOL concentration. The lateral diffusion was analysed on short and medium time scales;13 in particular, at short times, the center of mass of a lipid was shown to move mainly due to conformational changes of the hydrocarbon chains.13
The presence of CHOL was shown to change the membrane properties significantly, in particular the fluidity and the molecular packing. Obviously, the situation is not unambiguous: the value of the lipid self-diffusion coefficient depends on the temperature, the CHOL content, and the lipid type i.e., lipid headgroups and both tails.
No clear trend in the lateral diffusion was found in ref. 114 that could be linked to lipid unsaturation in the set of pure (single – component) 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/18
0/18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1(n-9)cis PC, 16
1(n-9)cis PC, 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/18
0/18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2(n-6)cis PC, 16
2(n-6)cis PC, 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/20
0/20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4(n-6)cis PC and 16
4(n-6)cis PC and 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/22
0/22![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6(n-3)cis PC bilayers: these all had similar lateral diffusion coefficients.114 At the same time, for an increasing level of unsaturation, considerable effects on dynamical properties, such as an accelerated dynamics of the PC head groups and glycerol backbones and a speeded up rotational dynamics of the lipid molecules were found.114 It was shown in ref. 115 that on short time scales the lateral self-diffusion coefficient of PC molecules 18
6(n-3)cis PC bilayers: these all had similar lateral diffusion coefficients.114 At the same time, for an increasing level of unsaturation, considerable effects on dynamical properties, such as an accelerated dynamics of the PC head groups and glycerol backbones and a speeded up rotational dynamics of the lipid molecules were found.114 It was shown in ref. 115 that on short time scales the lateral self-diffusion coefficient of PC molecules 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/18
0/18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1(n-9)cis PC, 18
1(n-9)cis PC, 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/18
0/18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2(n-6)cis PC, 18
2(n-6)cis PC, 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/18
0/18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3(n-3)cis PC, 18
3(n-3)cis PC, 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/20
0/20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4(n-6)cis PC, and 18
4(n-6)cis PC, and 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/22
0/22![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6(n-3)cis PC of the lipid bilayer containing 40 mol % CHOL is smaller than that for the corresponding pure PC bilayer. The diffusion coefficients increase with increasing the degree of unsaturation of one of the PC chains in bilayers of both types (i.e., in pure bilayers or in bilayers with CHOL).115 These “trends” were verified by pulsed field gradient (pfg) NMR experiments for unsaturated bilayers with 40 mol % and without CHOL composed of 18
6(n-3)cis PC of the lipid bilayer containing 40 mol % CHOL is smaller than that for the corresponding pure PC bilayer. The diffusion coefficients increase with increasing the degree of unsaturation of one of the PC chains in bilayers of both types (i.e., in pure bilayers or in bilayers with CHOL).115 These “trends” were verified by pulsed field gradient (pfg) NMR experiments for unsaturated bilayers with 40 mol % and without CHOL composed of 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/18
0/18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1(n-9)cis PC, 18
1(n-9)cis PC, 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/18
0/18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2(n-6)cis PC, 18
2(n-6)cis PC, 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/20
0/20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4(n-6)cis PC, and 18
4(n-6)cis PC, and 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/22
0/22![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6(n-3)cis PC;115 a correspondence between MD and pfg-NMR “trends” has been observed in spite of the difference in the diffusion mechanisms on various time and length scales.
6(n-3)cis PC;115 a correspondence between MD and pfg-NMR “trends” has been observed in spite of the difference in the diffusion mechanisms on various time and length scales.
A phenomenological memory function approach was developed in ref. 224, which can be used to describe the behavior of the mean square displacement of lipid atoms and molecules over the whole time interval, and the dynamics of selected lipid atoms and lipid molecules in a hydrated 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/14
0/14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0 PC lipid bilayer was investigated.224
0 PC lipid bilayer was investigated.224
7 Coarse grained simulations
The properties of lipid membranes can be studied at different time and length scales. For some properties it is important to take into account all the details of a system, all atoms including hydrogens. Other properties require much larger length and time scale than that where atomistic simulations are possible. Examples are undulations of membrane surface, formation of different aggregates as micelles, vesicles, lammelar or hexagonal phase transformations, problem related to domain formations at the membrane surfaces, etc. In such cases, one need to simplify description of individual lipids, so that groups of atoms are lumped into pseudo-particles to arrive at a coarse-grained, or mesoscopic, description of a bilayer. Coarse-grained models are emerging as a practical alternative to all-atom simulations for the characterization of membrane phenomena over long time scales. Recently several reviews and discussions appeared describing different aspects of coarse-grained, or multiscale modelling of bilayers.37,38,225–235There exists a large variety of coarse-grained models of lipids differing by the level of details, account for the solvent and the way how interaction potentials are defined. It is rather common to unite groups consisting of 2–5 heavy atoms into a single coarse-grained site, as it is shown in Fig. 3 for a 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/14
0/14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0 PC lipid. Further, while some of coarse-grained models use explicit “coarse-grained” water, other models are formulated to use in implicit solvent, where the effect of solvent (water) is described by effective potentials. One of the most widely used explicit solvent coarse grained models is based on the Martini force field.236,237 The Martini force field is based essentially on a four-to-one mapping, i.e., on average four heavy atoms are represented by a single interaction center. The interaction potential consists of a Lennard-Jones and eventually electrostatic terms, which are tuned to reproduce experimental partitioning data. Recently the Martini force field was generalized to include protein models238 and carbohydrates.239 During a few latest years the Martini force field have been used in a large variety of studies: formation of lipid domains and rafts by spontaneous separation of the mixture into a liquid-ordered and a liquid-disordered phase,240 monolayer collapse,241 lipid self-assembly and vesicle formation,242 flip-flop motions of CHOL and lipids in membrane,222 nanoparticle transport and accumulation in lipid bilayers,243–246 spontaneous curvature and stability of asymmetric bilayers,247 membrane curvature and lipid packing,248 freezing of small lipid vesicles,249 voltage-sensitive dyes with a lipid membrane,250 and surfactant micellization.251 It is still worth to note, that Martine force field has certain limitations, for example approximate description of the electrostatic interactions via shifted electrostatic potential which is cut at 12 Å, and dielectric permittivity set to 15. Also, coarse-grained water in the Martini force field freezes at normal temperature, which is typically counterweighted by introduction of artificial “antifreeze” particles.237 In order to improve properties of water in the Martini force field, as well as to provide better description of electrostatic and polarization interactions, a polarizable coarse-grained model for water has been introduced in a recent paper.252
0 PC lipid. Further, while some of coarse-grained models use explicit “coarse-grained” water, other models are formulated to use in implicit solvent, where the effect of solvent (water) is described by effective potentials. One of the most widely used explicit solvent coarse grained models is based on the Martini force field.236,237 The Martini force field is based essentially on a four-to-one mapping, i.e., on average four heavy atoms are represented by a single interaction center. The interaction potential consists of a Lennard-Jones and eventually electrostatic terms, which are tuned to reproduce experimental partitioning data. Recently the Martini force field was generalized to include protein models238 and carbohydrates.239 During a few latest years the Martini force field have been used in a large variety of studies: formation of lipid domains and rafts by spontaneous separation of the mixture into a liquid-ordered and a liquid-disordered phase,240 monolayer collapse,241 lipid self-assembly and vesicle formation,242 flip-flop motions of CHOL and lipids in membrane,222 nanoparticle transport and accumulation in lipid bilayers,243–246 spontaneous curvature and stability of asymmetric bilayers,247 membrane curvature and lipid packing,248 freezing of small lipid vesicles,249 voltage-sensitive dyes with a lipid membrane,250 and surfactant micellization.251 It is still worth to note, that Martine force field has certain limitations, for example approximate description of the electrostatic interactions via shifted electrostatic potential which is cut at 12 Å, and dielectric permittivity set to 15. Also, coarse-grained water in the Martini force field freezes at normal temperature, which is typically counterweighted by introduction of artificial “antifreeze” particles.237 In order to improve properties of water in the Martini force field, as well as to provide better description of electrostatic and polarization interactions, a polarizable coarse-grained model for water has been introduced in a recent paper.252
 | ||
Fig. 3 Atomistic (left) and coarse grained 10-site (right) models of a 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/14 0/14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0 PC lipid. 0 PC lipid. | ||
A coarse-grained model for lipids and proteins in water solution, build initially on similar principles as Martini force field with 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 mapping (counting all atoms), and refined by atomistic simulations, is described in a series of works by Schulten group.253,254 This model was used to study selfassembly of lipoprotein systems255,256 and the effect of proteins on membrane curvature.254,257,258
1 mapping (counting all atoms), and refined by atomistic simulations, is described in a series of works by Schulten group.253,254 This model was used to study selfassembly of lipoprotein systems255,256 and the effect of proteins on membrane curvature.254,257,258
In the papers ref. 259 and 260, another coarse-grained lipid model with explicit water was considered, in which coarse-grained sites of lipids were presented as ellipsoidal particles interacting by Gay-Berne potential with embeded charges on the head group and dipoles at the ester groups. Water was presented as a one-site spherical particle with a dipole. This model provides a coarse-grainde description of hydrated bilayer in more details than Martini force field. In ref. 260 this model was used to study membrane electrostatics, pressure distribution, spontaneous curvature, water permeation and some other properties of 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/14
0/14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0 PC and 18
0 PC and 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1/18
1/18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 PC bilayers.
1 PC bilayers.
Another class of coarse-grained lipid models do not treat water explicitly. Instead, effective solvent-mediated potentials between lipid sites are used.261–264 Potentially, such models can be used for simulations of very large lipid systems since computer time is not spent for simulation of water. Parametrization of such models is however more difficult. In some cases, a Lennard-Jones potential or its modifications are used to describe coarse-grained sites.261,265,266 In a more consequitive approach, effective potentials for coarse grained simulations are derived from fully atomistic simulations of lipids in water. There exist two main approaches to derive coarse-grained potentials from atomistic simulation. One is based of the force-matching procedure in which expression for the pairwise coarse grained force field is fitted to reproduce the n-body potential of mean force for the coarse-grained sites in the atomistic system.267,268 This approach was used for multiscale simulations of 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/14
0/14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0 bilayers,264 14
0 bilayers,264 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/14
0/14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0 PC - CHOL lipid mixtures in plain bilayers as well as liposomes,264,269 and 18
0 PC - CHOL lipid mixtures in plain bilayers as well as liposomes,264,269 and 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1/18
1/18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 PC – 18
1 PC – 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1/18
1/18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 PE lipid mixtures.270 A hybrid algorithm with a part of coarse-grained interaction potential presented by the Gay-Berne potential was also considered.271
1 PE lipid mixtures.270 A hybrid algorithm with a part of coarse-grained interaction potential presented by the Gay-Berne potential was also considered.271
In another approach the effective potentials between lipid coarse-grained sites are derived from the structural properties of lipids simulated in atomistic details. Radial distribution functions between sites of different lipids, as well as distributions of intramolecular distances are used for parametrization of inter- and intramolecular potentials. Computations of the effective potentials can in this case be carried out using the inverse Monte Carlo272 or inverse Boltzmann273 methods. Effective potentials, computed in ref. 263 from atomistic simulations of DMPC lipids, were used to describe processes of spontaneous formation of bicells, micelles and multilammelar structures.274,275 In ref. 276 the iterative inverse Boltzmann approach was used to derive effective potentials for coarse-grained 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/16
0/16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0 PC lipid model which are suitable for description of both liquid crystalline and amorphopus states of this lipid. In ref. 277, the inverse Monte Carlo method was used to derive effective potentials for two-dimensional model of lipid - CHOL domain forming mixtures.
0 PC lipid model which are suitable for description of both liquid crystalline and amorphopus states of this lipid. In ref. 277, the inverse Monte Carlo method was used to derive effective potentials for two-dimensional model of lipid - CHOL domain forming mixtures.
We mention also a number of other coarse-grained lipid models built on different principles and which were used to study pore formation under tension,278 protein interactions in membranes,279,280 behavior of CHOL in membrane,281 phase transitions in lipid monolayers and bilayers,282 process of self assembly and gel-phase transition,283 simulations of membranes under tension.284
8 Conclusions
Computer simulations of various lipid membrane systems allow to elucidate at the molecular level the detailed relations between the chemical structure and physical properties of various lipid molecules and membrane inclusions, to explain individual peculiarities of natural objects, to make forecasts concerning their behavior, etc. An understanding of the molecular basis of various physical properties of lipids and other membrane constituents allows one to narrow down the list of hypotheses under consideration about the possible functions of various components (such as acyl chains) in lipid membranes, e.g., the maintenance of proper bilayer fluidity and permeability, of the activity of membrane-bound enzymes, etc. Thus, together with the continued improvements of force fields and significant development of simulation approaches (methodologies, algorithms and mutually complementary methods), rapid advances in computing power, the long-term prospects of computer simulations in membrane studies seem to be highly promising.Acknowledgements
This work has been supported by the Swedish Institute (Visby programme grant 00961/2008), Swedish Research Council, grant No. 3731.2010.4 for leading research schools of Russian Federation, and grant No. 10-03-00201 of Russian Foundation for Basic Research.References
- P. S. Niemelä, S. Ollila, M. T. Hyvönen, M. Karttunen and I. Vattulainen, PLoS Comput. Biol., 2007, 3, 304–312 Search PubMed.
- A. J. Kox, J. P. J. Michels and F. W. Wiegel, Nature, 1980, 287, 317–319 CrossRef CAS.
- P. van der Ploeg and H. J. C. Berendsen, J. Chem. Phys., 1982, 76, 3271–3276 CrossRef CAS.
- P. van der Ploeg and H. J. C. Berendsen, Mol. Phys., 1983, 49, 233–248 CrossRef CAS.
- H. Heller, M. Schaefer and K. Schulten, J. Phys. Chem., 1993, 97, 8343–8360 CrossRef CAS.
- S. J. Marrink and H. J. C. Berendsen, J. Phys. Chem., 1994, 98, 4155–4168 CrossRef CAS.
- D. P. Tieleman and H. J. C. Berendsen, J. Chem. Phys., 1996, 105, 4871–4880 CrossRef CAS.
- R. Pastor, R. Venable and M. Karplus, J. Chem. Phys., 1988, 89, 1112–1127 CrossRef CAS.
- R. Pastor, R. Venable, M. Karplus and A. Szabo, J. Chem. Phys., 1988, 89, 1128–1140 CrossRef CAS.
- O. Mouritsen, in Molecular Description of Biological Membrane Components by Computer Aided Conformational Analysis, vol. 1, ed. R. Brasseur, CRC Press, Roca Raton, FL, 1990, pp. 3–83 Search PubMed.
- T.-X. Xiang, Biophys. J., 1993, 65, 1108–1120 CrossRef CAS.
- J. Essex, M. Hann and W. Richards, Philosophical Transactions of the Royal Society of London, Ser. B - Biological Sciences, 1994, 344, 239–260 Search PubMed.
- J. Wohlert and O. Edholm, J. Chem. Phys., 2006, 125, 204703 CrossRef.
- P. Bjelkmar, P. S. Niemelå, I. Vattulainen and E. Lindahl, PLoS Comput. Biol., 2009, 5, e1000289 Search PubMed.
- K. V. Damodaran and K. M. Merz, in Reviews in Computational Chemistry, ed. K. B. Lipkowitz and D. B. Boyd, VCH Publishers, Inc.N.Y., 1994, vol. 5, pp. 269–298 Search PubMed.
- O. Mouritsen and K. Jorgensen, Chem. Phys. Lipids, 1994, 73, 3–25 CrossRef CAS.
- R. W. Pastor, Curr. Opin. Struct. Biol., 1994, 4, 486–492 CAS.
- O. Mouritsen, M. Sperotto, J. Risbo, Z. Zhang and M. Zuckermann, in Advances in Computational Biology, vol. 2, JAI Press, 1996, pp. 15–64 Search PubMed.
- E. Jacobsson, Trends Biochem. Sci., 1997, 22, 339–344 CrossRef.
- D. P. Tieleman, S. J. Marrink and H. J. C. Berendsen, Biochim. Biophys. Acta, Biomembr., 1997, 1331, 235–270 CrossRef CAS.
- D. Tobias, K. Tu and M. Klein, Curr. Opin. Colloid Interface Sci., 1997, 2, 15–26 CAS.
- M. Merz, Curr. Opin. Struct. Biol., 1997, 7, 511–517 CrossRef CAS.
- H. J. C. Berendsen and D. P. Tieleman, in Encyclopedia of computational chemistry, ed. R. Schleyer, Chichester, N.Y.et al., J. Wiley & Sons, 1998, pp. 1639–1650 Search PubMed.
- S. E. Feller and A. D. MacKerell, J. Phys. Chem. B, 2000, 104, 7510–7515 CrossRef CAS.
- S. E. Feller, Curr. Opin. Colloid Interface Sci., 2000, 5, 217–223 CrossRef CAS.
- L. R. Forrest and M. S. P. Sansom, Curr. Opin. Struct. Biol., 2000, 10, 174–181 CrossRef CAS.
- S. E. Feller, in Lipid Bilayers. Structure and Interactions, John Katsaras and Thomas Gutberlet, ed. Berlin, Heidelberg, N.Y. Springer-Verlag, 2001, pp. 89–107 Search PubMed.
- D. J. Tobias, in Computational Biochemistry and Biophysics, ed. Becker O. H.MacKerell A. D., Jr., Roux B., Watanabe M.N.Y.: Dekker, 2001, pp. 465–496 Search PubMed.
- H. L. Scott, Curr. Opin. Struct. Biol., 2002, 12, 495–502 CrossRef CAS.
- T. Hansson, C. Oostenbrink and W. F. van Gunsteren, Curr. Opin. Struct. Biol., 2002, 12, 190–196 CrossRef CAS.
- L. Saiz and M. L. Klein, Acc. Chem. Res., 2002, 35, 482–489 CrossRef CAS.
- L. Saiz, S. Bandyopadhyay and M. L. Klein, Biosci. Rep., 2002, 22, 151–173 CrossRef CAS.
- L. Vigh, P. Escribá, A. Sonnleitner, M. Sonnleitner, S. Piotto, B. Maresca, I. Horváth and J. L. Harwood, Prog. Lipid Res., 2005, 44, 303–344 CrossRef CAS.
- Y. H. M. Chan and S. G. Boxer, Curr. Opin. Chem. Biol., 2007, 11, 581–587 CrossRef CAS.
- L. S. Vermeer, B. L. de Groot, V. Reat, A. Milon and J. Czaplicki, Eur. Biophys. J., 2007, 36, 919–931 CrossRef CAS.
- Computational Modeling of Membrane Bilayers. (Current Topics in Membranes), ed. S. Feller, Elsevier Inc., San Diego, 2008, vol. 60, p. 448 Search PubMed.
- S. J. Marrink, A. H. de Vries and D. P. Tieleman, Biochim. Biophys. Acta, Biomembr., 2009, 1788, 149–168 CrossRef CAS.
- S. A. Pandit and H. L. Scott, Biochim. Biophys. Acta, Biomembr., 2009, 1788, 136–148 CrossRef CAS.
- J. A. D. MacKerell and L. Nilsson, Curr. Opin. Struct. Biol., 2008, 18, 194–199 CAS.
- E. Lindahl and M. Sansom, Curr. Opin. Struct. Biol., 2008, 18, 425–431 CrossRef CAS.
- B. Grant, A. Gorfe and J. McCammon, Curr. Opin. Struct. Biol., 2010, 20, 142–147 CrossRef CAS.
- A. Amadei, I. Daidone, A. Nola and M. Aschi, Curr. Opin. Struct. Biol., 2010, 20, 155–161 CrossRef CAS.
- B. Villoutreix and O. Sperandio, Curr. Opin. Struct. Biol., 2010, 20, 168–179 CrossRef CAS.
- J. Hermans, H. J. C. Berendsen, W. F. van Gunsteren and J. P. M. Postma, Biopolymers, 1984, 23, 1513–1518 CAS.
- L. D. Schuler, X. Daura and W. F. van Gunsteren, J. Comput. Chem., 2001, 22, 1205–1218 CrossRef CAS.
- I. Chandrasekhar, M. Kastenholz, R. D. Lins, C. Oostenbrink, L. D. Schuler, D. P. Tieleman and W. F. van Gunsteren, Eur. Biophys. J., 2003, 32, 67–77 CAS.
- A. D. Mackerell, J. Wiórkiewicz-Kuczera and M. Karplus, J. Am. Chem. Soc., 1995, 117, 11946–11975 CrossRef.
- A. D. MacKerell, D. Bashford, M. Bellott, R. L. Dunbrack, J. D. Evanseck, M. J. Field, S. Fisher, J. Gao, S. Ha, D. Joseph-McCarthy, L. Kuchnir, K. Kuczera, F. T. K. Lau, C. Mattos, S. Michnick, T. Ngo, D. T. Nguyen, B. Prodhom, W. E. Reiher III, B. Roux, M. Schlenkrich, J. C. Smith, R. Stote, J. Straub, M. Watanabe, J. Wiórkiewicz-Kuczera, D. Yin and M. Karplus, J. Phys. Chem. B, 1998, 102, 3586–3616 CrossRef CAS.
- O. Berger, O. Edholm and F. Jähnig, Biophys. J., 1997, 72, 2002–2013 CrossRef CAS.
- E. Lindahl, B. Hess and D. van der Spoel, J. Mol. Model., 2001, 7, 306–317 CAS.
- S. E. Feller, D. Yin, R. W. Pastor and A. D. MacKerell, Biophys. J., 1997, 73, 2269–2279 CrossRef CAS.
- J. B. Klauda, B. R. Brooks, A. D. MacKerell, R. M. Venable and R. W. Pastor, J. Phys. Chem. B, 2005, 109, 5300–5311 CrossRef CAS.
- J. B. Klauda, R. M. Venable, J. A. Freites, J. W. O'Connor, D. J. Tobias, C. Mondragon-Ramires, I. Vorobyov, A. D. MacKerell and R. W. Pastor, J. Phys. Chem., 2010, 114, 7830–7843 Search PubMed.
- L. Kale, R. Skeel, M. Bhandarkar, R. Brunner, A. Gursoy, N. Krawetz, J. Phillips, A. Shinozaki, K. Varadarajan and K. Schulten, J. Comput. Phys., 1999, 151, 283–312 CrossRef.
- J. Wang, R. M. Wolf, J. W. Caldwell, P. A. Kollman and D. A. Case, J. Comput. Chem., 2004, 25, 1157–1174 CrossRef.
- B. Jójárt and T. A. Martinek, J. Comput. Chem., 2007, 28, 2051–2026 CrossRef CAS.
- D. Poger and A. E. Mark, J. Chem. Theory Comput., 2010, 6, 325–336 CrossRef CAS.
- R. W. Benz, F. Castro-Roman, D. J. Tobias and S. H. White, Biophys. J., 2005, 88, 805–817 CrossRef CAS.
- C.-J. Högberg and A. P. Lyubartsev, J. Phys. Chem. B, 2006, 110, 14326–14336 CrossRef.
- C. Anezo, A. H. de Vries, H.-D. Höltje, D. P. Tieleman and S.-J. Marrink, J. Phys. Chem. B, 2003, 107, 9424–9433 CrossRef CAS.
- S. W. I. Siu, R. Vácha, P. Jungwirth and R. A. Böckmann, J. Chem. Phys., 2008, 128, 125103 CrossRef.
- P. Prakash and R. Sankararamakrishnan, J. Comp. Chem., 2010, 31, 266–277 CAS.
- S. W. Chiu, S. A. Pandit, H. L. Scott and E. Jakobsson, J. Phys. Chem. B, 2009, 113, 2748–2763 CrossRef CAS.
- D. Poger, W. F. van Gunsteren and A. E. Mark, J. Comput. Chem., 2010, 31, 1117–1125 CAS.
- M. O. Jensen, O. G. Mouritsen and G. H. Peters, Biophys. J., 2004, 86, 3556–3575 CrossRef CAS.
- M. T. Hyvönen and P. T. Kovanen, Eur. Biophys. J., 2005, 34, 294–305 CrossRef.
- J. Sonne, M.Ø. Jensen, F. Y. Hansen, L. Hemmingsen and G. H. Peters, Biophys. J., 2007, 92, 4157–4167 CrossRef CAS.
- C.-J. Högberg, A. M. Nikitin and A. P. Lyubartsev, J. Comput. Chem., 2008, 29, 2359–2369 CrossRef.
- J. Gullingsrud and K. Schulten, Biophys. J., 2004, 86, 3496–3509 CrossRef CAS.
- J. B. Klauda, N. Kučerka, B. R. Brooks, R. W. Pastor and J. F. Nagle, Biophys. J., 2006, 90, 2796–2807 CrossRef CAS.
- P. Prakash and R. Sankararamakrishnan, J. Comp. Chem., 2009, 31, 266–277.
- M. Roark and S. E. Feller, J. Phys. Chem. B, 2009, 113, 13229–13234 CrossRef CAS.
- J. Sonne, F. Y. Hansen and G. H. Peters, J. Chem. Phys., 2005, 122, 124903 CrossRef.
- U. R. Pedersen, G. H. Peters and P. Westh, Biophys. Chem., 2007, 125, 104–111 CrossRef CAS.
- J. Taylor, N. E. Whiteford, G. Bradley and G. W. Watson, Biochim. Biophys. Acta, Biomembr., 2009, 1788, 638–649 CrossRef CAS.
- J. Henin, W. Shinoda and M. L. Klein, J. Phys. Chem. B, 2008, 112, 7008–7015 CrossRef CAS.
- K. V. Damodaran, K. M. Merz and B. P. Gaber, Biochemistry, 1992, 31, 7656–7664 CrossRef CAS.
- U. Essmann, L. Perera and M. L. Berkowitz, Langmuir, 1995, 11, 4519–4531 CrossRef CAS.
- P. B. Moore, C. F. Lopez and M. L. Klein, Biophys. J., 2001, 81, 2484–2494 CAS.
- C. F. Lopez, S. O. Nielsen, M. L. Klein and P. B. Moore, J. Phys. Chem. B, 2004, 108, 6603–6610 CrossRef CAS.
- L. Rosso and I. R. Gould, J. Comput. Chem., 2008, 29, 24–37 CrossRef CAS.
- F. Castro-Román, R. W. Benz, S. H. White and D. J. Tobias, J. Phys. Chem. B, 2006, 110, 24157–24164 CrossRef CAS.
- J. B. Klauda, B. R. Brooks and R. W. Pastor, J. Chem. Phys., 2006, 125, 144710 CrossRef.
- M. Patra, M. Karttunen, Hyvönen, E. Falck and I. Vattulainen, J. Phys. Chem. B, 2004, 108, 4485–4494 CrossRef CAS.
- R. M. Venable, L. E. Chen and R. W. Pastor, J. Phys. Chem. B, 2009, 113, 5855–5862 CrossRef CAS.
- M. P. Allen and D. J. Tildesley, Computer simulations of liquids, Clarendon, Oxford, 2nd edn, 1987 Search PubMed.
- X. W. Wu and B. R. Brooks, J. Chem. Phys., 2005, 122, 044107 CrossRef.
- J. B. Klauda, X. Wu, R. W. Pastor and B. R. Brooks, J. Phys. Chem. B, 2007, 111, 4393–4400 CrossRef CAS.
- E. A. Dratz and A. J. Deese, in The health effects of polyunsaturated fatty acids in seafood, ed. A. P. Simopoulous, R. R. Kifer, R. E. Martin, N.Y., Acad. Press, 1986, pp. 319–351 Search PubMed.
- A. L. Rabinovich and P. O. Ripatti, Uspekhi sovremennoi biologii (Progress in modern biology), 1994, 114, 581–594 Search PubMed.
- K. Gawrisch, N. V. Eldho and L. L. Holte, Lipids, 2003, 38, 445–452 CrossRef CAS.
- W. Stillwell and S. R. Wassall, Chem. Phys. Lipids, 2003, 126, 1–27 CrossRef CAS.
- A. L. Rabinovich, P. O. Ripatti, N. K. Balabaev and F. A. M. Leermakers, Phys. Rev. E: Stat., Nonlinear, Soft Matter Phys., 2003, 67, 011909 CrossRef CAS.
- R. C. Valentine and D. L. Valentine, Prog. Lipid Res., 2004, 43, 383–402 CrossRef CAS.
- S. E. Feller and K. Gawrisch, Curr. Opin. Struct. Biol., 2005, 15, 416–422 CrossRef CAS.
- K. Gawrisch, O. Soubias and M. Mihailescu, Prostaglandins, Leukotrienes Essent. Fatty Acids, 2008, 79, 131–134 CrossRef CAS.
- W. Stillwell, Chem. Phys. Lipids, 2008, 153, 1–2 CrossRef CAS.
- S. R. Wassall and W. Stillwell, Biochim. Biophys. Acta, Biomembr., 2009, 1788, 24–32 CrossRef CAS.
- D. B. Jump, D. Botolin, Y. Wang, J. Xu, O. Demeure and B. Christian, Chem. Phys. Lipids, 2008, 153, 3–13 CrossRef CAS.
- R. S. Chapkin, J. Seo, D. N. McMurray and J. R. Lupton, Chem. Phys. Lipids, 2008, 153, 14–23 CrossRef CAS.
- S. R. Shaikh and M. Edidin, Chem. Phys. Lipids, 2008, 153, 24–33 CrossRef CAS.
- H. Y. Kim, Chem. Phys. Lipids, 2008, 153, 34–46 CrossRef CAS.
- R. A. Siddiqui, K. Harvey and W. Stillwell, Chem. Phys. Lipids, 2008, 153, 47–56 CrossRef CAS.
- N. V. Eldho, S. E. Feller, S. Tristram-Nagle, I. V. Polozov and K. Gawrisch, J. Am. Chem. Soc., 2003, 125, 6409–6421 CrossRef CAS.
- L. Koubi, L. Saiz, M. Tarek, D. Scharf and M. Klein, J. Phys. Chem. B, 2003, 107, 14500–14508 CrossRef CAS.
- F. A. M. Leermakers, A. L. Rabinovich and N. K. Balabaev, Phys. Rev. E: Stat., Nonlinear, Soft Matter Phys., 2003, 67, 011910 CrossRef CAS.
- A. L. Rabinovich, P. O. Ripatti and N. K. Balabaev, Proc. SPIE., 2003, 5127, 54–58 CAS.
- S. R. Shaikh, V. Cherezov, M. Caffrey, W. Stillwell and S. R. Wassall, Biochemistry, 2003, 42, 12028–12037 CrossRef CAS.
- M. C. Pitman, F. Suits, A. D. MacKerell and S. E. Feller, Biochemistry, 2004, 43, 15318–15328 CrossRef CAS.
- A. L. Rabinovich, P. O. Ripatti and N. K. Balabaev, Russian J. Phys. Chem., 2004, 78, 1004–1009 Search PubMed.
- M. Carrillo-Tripp and S. E. Feller, Biochemistry, 2005, 44, 10164–10169 CrossRef CAS.
- A. L. Rabinovich, N. K. Balabaev, M. G. Alinchenko, V. P. Voloshin, N. N. Medvedev and P. Jedlovszky, J. Chem. Phys., 2005, 122, 084906 CrossRef CAS.
- A. L. Rabinovich, P. O. Ripatti and N. K. Balabaev, in Molecular simulation studies in material and biological sciences, ed. Kh. Kholmurodov. Nova Science Publ., Inc., N.Y., 2006, pp. 15–29 Search PubMed.
- S. Ollila, M. T. Hyvönen and I. Vattulainen, J. Phys. Chem. B, 2007, 111, 3139–3150 CrossRef CAS.
- A. L. Rabinovich, V. V. Kornilov, N. K. Balabaev, F. A. M. Leermakers and A. V. Filippov, Biochemistry (Moscow), Ser. A: Membr. and Cell Biol., 2007, 1, 343–357 Search PubMed.
- S. E. Feller, Chem. Phys. Lipids, 2008, 153, 76–80 CrossRef CAS.
- S. J. Marrink, A. H. de Vries, T. A. Harroun, J. Katsaras and S. R. Wassall, J. Am. Chem. Soc., 2008, 130, 10–11 CrossRef CAS.
- W. F. D. Bennett, J. L. MacCallum, M. J. Hinner, S. J. Marrink and D. P. Tieleman, J. Am. Chem. Soc., 2009, 131, 12714–12720 CrossRef CAS.
- M. Bachar, P. Brunelle, D. P. Tieleman and A. Rauk, J. Phys. Chem. B, 2004, 108, 7170–7179 CrossRef CAS.
- A. H. de Vries, A. E. Mark and S. J. Marrink, J. Phys. Chem. B, 2004, 108, 2454–2463 CrossRef CAS.
- S. A. Pandit, S. Vasudevan, S. W. Chiu, R. J. Mashl, E. Jakobsson and H. L. Scott, Biophys. J., 2004, 87, 1092–1100 CrossRef CAS.
- S. A. Pandit, E. Jakobsson and H. L. Scott, Biophys. J., 2004, 87, 3312–3322 CrossRef CAS.
- D. P. Tieleman, BMC Biochem., 2004, 5, 10 CrossRef.
- S. Y. Bhide and M. L. Berkowitz, J. Chem. Phys., 2005, 123, 224702 CrossRef.
- P. T. Vernier, M. J. Ziegler, Y. Sun, W. V. Chang, M. A. Gundersen and D. P. Tieleman, J. Am. Chem. Soc., 2006, 128, 6288–6289 CrossRef CAS.
- J. Aittoniemi, P. S. Niemelä, M. T. Hyvönen, M. Karttunen and I. Vattulainen, Biophys. J., 2007, 92, 1125–1137 CrossRef CAS.
- S. Y. Bhide, Z. Zhang and M. L. Berkowitz, Biophys. J., 2007, 92, 1284–1295 CrossRef CAS.
- M. H. Cheng, L. T. Liu, A. C. Saladino, Y. Xu and P. Tang, J. Phys. Chem. B, 2007, 111, 14186–14192 CrossRef CAS.
- S. Leekumjorn and A. K. Sum, J. Phys. Chem. B, 2007, 111, 6026–6033 CrossRef CAS.
- S. A. Pandit, S. W. Chiu, E. Jakobsson, A. Grama and H. L. Scott, Biophys. J., 2007, 92, 920–927 CrossRef CAS.
- Z. Zhang, S. Y. Bhide and M. L. Berkowitz, J. Phys. Chem. B, 2007, 111, 12888–12897 CrossRef CAS.
- Q. Zhu, K. H. Cheng and M. W. Vaughn, J. Phys. Chem. B, 2007, 111, 11021–11031 CrossRef CAS.
- M. Dahlberg and A. Maliniak, J. Phys. Chem. B, 2008, 112, 11655–11663 CrossRef CAS.
- H. Martinez-Seara, T. Róg, M. Pasenkiewicz-Gierula, I. Vattulainen, M. Karttunen and R. Reigada, Biophys. J., 2008, 95, 3295–3305 CrossRef CAS.
- H. Martinez-Seara, T. Róg, M. Karttunen, R. Reigada and I. Vattulainen, J. Chem. Phys., 2008, 129, 105103 CrossRef.
- S. Mondal and C. Mukhopadhyay, Langmuir, 2008, 24, 10298–10305 CrossRef CAS.
- S. A. Pandit, S. W. Chiu, E. Jakobsson, A. Grama and H. L. Scott, Langmuir, 2008, 24, 6858–6865 CrossRef CAS.
- M. Roark and S. E. Feller, Langmuir, 2008, 24, 12469–12473 CrossRef CAS.
- Z. Zhang, L. Lu and M. L. Berkowitz, J. Phys. Chem. B, 2008, 112, 3807–3811 CrossRef CAS.
- S. P. Soni, J. A. Ward, S. E. Sen, S. E. Feller and S. R. Wassall, Biochemistry, 2009, 48, 11097–11107 CrossRef CAS.
- K. Lähdesmäki, O. H. S. Ollila, A. Koivuniemi, P. T. Kovanen and M. T. Hyvönen, Biochim. Biophys. Acta, Biomembr., 2010, 1798, 938–946 CrossRef.
- J. Wohlert and O. Edholm, Biophys. J., 2004, 87, 2433–2445 CrossRef CAS.
- A. H. de Vries, I. Chandrasekhar, W. F. van Gunsteren and P. H. Hünenberger, J. Phys. Chem. B, 2005, 109, 11643–11652 CrossRef CAS.
- S. W. Chiu, S. Vasudevan, E. Jakobsson, R. J. Mashl and H. L. Scott, Biophys. J., 2003, 85, 3624–3635 CrossRef CAS.
- J. S. Hub, T. Salditt, M. C. Rheinstädter and B. L. de Groot, Biophys. J., 2007, 93, 3156–3168 CrossRef CAS.
- J. B. Klauda, M. F. Roberts, A. G. Redfield, B. R. Brooks and R. W. Pastor, Biophys. J., 2008, 94, 3074–3083 CrossRef CAS.
- S. Leekumjorn and A. K. Sum, Biochim. Biophys. Acta, Biomembr., 2007, 1768, 354–365 CrossRef CAS.
- J. Pimthon, R. Willumeit, A. Lendlein and D. Hofmann, Biochim. Biophys. Acta, 2009, 921, 136–148.
- S.-S. Qin, Z. W. Yu and Y.-X. Yu, J. Phys. Chem. B, 2009, 113, 8114–8123 CrossRef CAS.
- P. S. Coppock and J. T. Kindt, Langmuir, 2009, 25, 352–359 CrossRef CAS.
- W. Shinoda, M. Mikami, T. Baba and M. Hato, J. Phys. Chem. B, 2003, 107, 14030–14035 CrossRef CAS.
- W. Shinoda, M. Mikami, T. Baba and M. Hato, J. Phys. Chem. B, 2004, 108, 9346–9356 CrossRef CAS.
- W. Shinoda, M. Mikami, T. Baba and M. Hato, Chem. Phys. Lett., 2004, 390, 35–40 CrossRef CAS.
- K. Shinoda, W. Shinoda, T. Baba and M. Hato, J. Chem. Phys., 2004, 121, 9648–9654 CrossRef CAS.
- W. Shinoda, T. Shinoda, T. Baba and M. Mikami, Biophys. J., 2005, 89, 3195–3202 CrossRef CAS.
- S. A. Pandit and H. L. Scott, J. Chem. Phys., 2006, 124, 014708 CrossRef.
- R. Notman, W. K. den Otter, M. G. Noro, W. J. Briels and J. Anwar, Biophys. J., 2007, 93, 2056–2068 CrossRef CAS.
- C. Das, P. D. Olmsted and M. G. Noro, Soft Matter, 2009, 5, 4549–4555 RSC.
- H. Ohvo-Rekilä, B. Ramstedt, P. Leppimäki and J. P. Slotte, Prog. Lipid Res., 2002, 41, 66–97 CrossRef CAS.
- D. Bach and E. Wachtel, Biochim. Biophys. Acta, Biomembr., 2003, 1610, 187–197 CrossRef CAS.
- H. M. McConnell and A. Radhakrishnan, Biochim. Biophys. Acta, Biomembr., 2003, 1610, 159–173 CrossRef CAS.
- J. R. Silvius, Biochim. Biophys. Acta, Biomembr., 2003, 1610, 174–183 CrossRef CAS.
- Z. Cournia, A. C. Vaiana, G. M. Ullmann and J. C. Smith, Pure Appl. Chem., 2004, 76, 189–196 CrossRef CAS.
- I. P. W. McMullen, R. N. A. H. Lewis and R. N. McElhaney, Curr. Opin. Colloid Interface Sci., 2004, 8, 459–468 CrossRef CAS.
- Z. Cournia, J. C. Smith and G. M. Ullmann, J. Comput. Chem., 2005, 26, 1383–1399 CrossRef CAS.
- F. A. M. Leermakers and A. L. Rabinovich, Phys. Rev. E: Stat., Nonlinear, Soft Matter Phys., 2007, 76, 031904 CrossRef CAS.
- R. Zidovetzki and I. Levitan, Biochim. Biophys. Acta, Biomembr., 2007, 1768, 1311–1324 CrossRef CAS.
- M. L. Berkowitz, Biochim. Biophys. Acta, Biomembr., 2009, 1788, 86–96 CrossRef CAS.
- J. Pan, S. Tristram-Nagle and J. F. Nagle, Phys. Rev. E: Stat., Nonlinear, Soft Matter Phys., 2009, 80, 021931 CrossRef.
- S. R. Wassall and W. Stillwell, Chem. Phys. Lipids, 2008, 153, 57–63 CrossRef CAS.
- J. Zidar, F. Merzel, M. Hodošček, K. Rebolj, K. Sepčić, P. Maček and D. Janežič, J. Phys. Chem. B, 2009, 113, 15795–15802 CrossRef CAS.
- M. Fraňová, J. Repáková, P. Čapková, J. M. Holopainen and I. Vattulainen, J. Phys. Chem. B, 2010, 114, 2704–2711 CrossRef CAS.
- M. Patra, E. Salonen, E. Terama, I. Vattulainen, R. Faller, B. W. Lee, J. Holopainen and M. Karttunen, Biophys. J., 2006, 90, 1121–1135 CrossRef CAS.
- C.-J. Högberg, A. Maliniak and A. P. Lyubartsev, Biophys. Chem., 2007, 125, 416–424 CrossRef.
- V. Castro, B. Stevensson, S. V. Dvinskikh, C.-J. Högberg, A. P. Lyubartsev, H. Zimmermann, D. Sandström and A. Maliniak, Biochim. Biophys. Acta, Biomembr., 2008, 1778, 2604–2611 CrossRef CAS.
- C.-J. Högberg and A. P. Lyubartsev, Biophys. J., 2008, 94, 525–531 CrossRef.
- E. Terama, O. H. S. Ollila, E. Salonen, A. C. Rowat, C. Trandum, P. Westh, M. Patra, M. Karttunen and I. Vattulainen, J. Phys. Chem. B, 2008, 112, 4131–4139 CrossRef CAS.
- R. C. Bernardi, D. E. Gomes, R. Gobato, C. A. Taft, A. T. Ota and P. G. Pascutti, Mol. Phys., 2009, 107, 1437–1443 CrossRef CAS.
- R. D. Porasso, W. F. D. Bennett, S. D. Oliveira-Costa and J. J. L. Cascales, J. Phys. Chem. B, 2009, 113, 9988–9994 CrossRef CAS.
- S. Vemparala, C. Domene and M. L. Klein, Acc. Chem. Res., 2010, 43, 103–110 CrossRef CAS.
- P. L. Chau, P. Jedlovszky, P. N. M. Hoang and S. Picaud, J. Mol. Liq., 2009, 147, 128–134 CrossRef CAS.
- L. Chau, P. N. M. Hoang, S. Picaud and P. Jedlovszky, Chem. Phys. Lett., 2007, 438, 294–297 CrossRef.
- B. Griepernau, S. Leis, M. F. Schneider, M. Sikor, D. Steppich and R. A. Böckmann, Biochim. Biophys. Acta, Biomembr., 2007, 1768, 2899–2913 CrossRef CAS.
- T. Sugii, S. Takagi and Y. Matsumoto, J. Chem. Phys., 2005, 123, 184714 CrossRef.
- Z. Zhang and M. L. Berkowitz, J. Phys. Chem. B, 2009, 113, 7676–7680 CrossRef CAS.
- S. Y. Bhide and M. L. Berkowitz, J. Chem. Phys., 2006, 125, 094713 CrossRef.
- M. L. Berkowitz, D. L. Bostick and S. Pandit, Chem. Rev., 2006, 106, 1527–1539 CrossRef CAS.
- J. N. Sachs, H. Nanda, H. I. Petrache and T. B. Woolf, Biophys. J., 2004, 86, 3772–3782 CrossRef CAS.
- A. A. Gurtovenko and I. Vattulainen, J. Am. Chem. Soc., 2005, 127, 17570–17571 CrossRef CAS.
- A. A. Gurtovenko, J. Chem. Phys., 2005, 122, 244902 CrossRef.
- A. A. Gurtovenko and I. Vattulainen, Biophys. J., 2007, 92, 1878–1890 CrossRef CAS.
- R. A. Böckmann, B. L. de Groot, S. Kakorin, E. Neumann and H. Grubmüller, Biophys. J., 2008, 95, 1837–1850 CrossRef.
- A. Cordomi, O. Edholm and J. J. Perez, J. Phys. Chem. B, 2008, 112, 1397–1408 CrossRef CAS.
- A. Dickey and R. Faller, Biophys. J., 2008, 95, 2636–2646 CrossRef CAS.
- A. A. Gurtovenko and I. Vattulainen, J. Phys. Chem. B, 2008, 112, 1953–1962 CrossRef CAS.
- A. A. Gurtovenko and I. Vattulainen, J. Phys. Chem. B, 2008, 112, 4629–4634 CrossRef CAS.
- A. Cordomi, O. Edholm and J. J. Perez, J. Chem. Theory Comput., 2009, 5, 2125–2134 CrossRef CAS.
- A. A. Gurtovenko and I. Vattulainen, J. Chem. Phys., 2009, 130, 215107 CrossRef.
- A. A. Gurtovenko and I. Vattulainen, J. Phys. Chem. B, 2009, 113, 7194–7198 CrossRef CAS.
- R. Vácha, S. W. I. Siu, M. Petrov, R. A. Böckmann, J. Barucha-Kraszewska, P. Jurkiewicz, M. Hof, M. L. Berkowitz and P. Jungwirth, J. Phys. Chem. A, 2009, 113, 7235–7243 CrossRef CAS.
- J. N. Sachs, P. S. Crozier and T. B. Woolf, J. Chem. Phys., 2004, 121, 10847–10851 CrossRef CAS.
- S. Pöyry, T. Róg, M. Karttunen and I. Vattulainen, J. Phys. Chem. B, 2009, 113, 15513–15521 CrossRef CAS.
- D. Bemporad, J. W. Essex and C. Luttmann, J. Phys. Chem. B, 2004, 108, 4875–4884 CrossRef CAS.
- D. Bemporad, J. W. Essex and C. Luttmann, Biophys. J., 2004, 87, 1–13 CrossRef CAS.
- D. Bemporad, J. W. Essex and C. Luttmann, Biochim. Biophys. Acta, Biomembr., 2005, 1718, 1–21 CrossRef CAS.
- J. Ulander and A. D. J. Haymet, Biophys. J., 2003, 85, 3475–3484 CrossRef CAS.
- P. Mukhopadhyay, H. J. Vogel and D. P. Tieleman, Biophys. J., 2004, 86, 337–345 CrossRef CAS.
- A. Debnath, K. G. Ayappa, V. Kumaran and P. K. Maiti, J. Phys. Chem. B, 2009, 113, 10660–10668 CrossRef CAS.
- A. Cordomí, J. Prades, J. Frau, O. Vögler, S. S. Funari, J. J. Perez, P. V. Escribá and F. Barceló, J. Lipid Res., 2010, 51, 1113–1124 CAS.
- N. D. Winter and G. C. Schatz, J. Phys. Chem. B, 2010, 114, 5053–5060 CrossRef CAS.
- A. A. Gurtovenko and I. Vattulainen, J. Phys. Chem. B, 2007, 111, 13554–13559 CrossRef CAS.
- P. T. Vernier, M. J. Ziegler, Y. Sun, M. A. Gundersen and D. P. Tieleman, Phys. Biol., 2006, 3, 233–247 Search PubMed.
- C. Hofsäss, E. Lindahl and O. Edholm, Biophys. J., 2003, 84, 2192–2206 CrossRef CAS.
- P. Jedlovszky and M. Mezei, J. Phys. Chem. B, 2003, 107, 5311–5321 CrossRef CAS.
- E. Falck, M. Patra, M. Karttunen, M. T. Hyvönen and I. Vattulainen, Biophys. J., 2004, 87, 1076–1091 CrossRef CAS.
- G. A. Khelashvili and H. L. Scott, J. Chem. Phys., 2004, 120, 9841–9847 CrossRef CAS.
- S. A. Pandit, D. Bostick and M. L. Berkowitz, Biophys. J., 2004, 86, 1345–1356 CrossRef CAS.
- J. Czub and M. Baginski, Biophys. J., 2006, 90, 2368–2382 CrossRef CAS.
- I. Róg and M. Pasenkiewicz-Gierula, Biophys. J., 2006, 91, 3756–3767 CrossRef.
- N. Kučerka, J. D. Perlmutter, J. Pan, S. Tristram-Nagle, J. Katsaras and J. N. Sachs, Biophys. J., 2008, 95, 2792–2805 CrossRef CAS.
- S. Pöyry, T. Róg, M. Karttunen and I. Vattulainen, 112, 2008, 112, pp. 2922–2929.
- W. F. D. Bennett, J. L. MacCallum and D. P. Tieleman, J. Am. Chem. Soc., 2009, 131, 1972–1978 CrossRef CAS.
- G. S. Ayton and G. A. Voth, Biophys. J., 2004, 87, 3299–3311 CrossRef CAS.
- E. Flenner, J. Das, M. C. Rheinstädter and I. Kosztin, Phys. Rev. E: Stat., Nonlinear, Soft Matter Phys., 2009, 79, 011907 CrossRef.
- S. O. Nielsen, C. F. Lopez, G. Srinivas and M. L. Klein, J. Phys.: Condens. Matter, 2004, 16, R481–R512 CrossRef CAS.
- G. Brannigan, L. C. L. Lin and F. L. H. Brown, Biophys. J., 2006, 35, 104–124 CAS.
- M. Müller, K. Katsov and M. Schick, Phys. Rep., 2006, 434, 113–176 CrossRef CAS.
- M. Venturoli, M. M. Sperotto, M. Kranenburg and B. Smit, Phys. Rep., 2006, 437, 1–54 CrossRef CAS.
- G. S. Ayton, W. G. Noid and G. A. Voth, Curr. Opin. Struct. Biol., 2007, 17, 192–198 CrossRef CAS.
- J. Shillcock and R. Lipowsky, Biophys. Rev. Lett., 2007, 2, 33–55 CrossRef CAS.
- M. L. Klein and W. Shinoda, Science, 2008, 321, 798–800 CrossRef CAS.
- J. Southern, J. Pitt-Francis, J. Whiteley, D. Stokeley, H. Kobashi, R. Nobes, Y. Kadooka and D. Gavaghan, Prog. Biophys. Mol. Biol., 2008, 96, 60–89 CrossRef CAS.
- G. S. Ayton and G. A. Voth, Curr. Opin. Struct. Biol., 2009, 19, 138–144 CrossRef CAS.
- S. V. Bennun, M. I. Hoopes, C. Xing and R. Faller, Chem. Phys. Lipids, 2009, 159, 59–66 CrossRef CAS.
- G. S. Ayton, E. Lyman and G. A. Voth, Faraday Discuss., 2010, 144, 347–358 RSC.
- S. J. Marrink, A. H. de Vries and A. E. Mark, J. Phys. Chem. B, 2004, 108, 750–760 CrossRef CAS.
- S. J. Marrink, H. J. Risselada, S. Yefimov, D. P. Tieleman and A. H. de Vries, J. Phys. Chem. B, 2007, 111, 7812–7824 CrossRef CAS.
- L. Monticelli, S. K. Kandasamy, X. Periole, R. G. Larson, D. P. Tieleman and S. J. Marrink, J. Chem. Theory Comput., 2008, 4, 819–834 CrossRef CAS.
- C. A. Lopez, A. J. Rzepiela, A. H. de Vries, L. Dijkhuizen, P. H. Hunenberger and S. J. Marrink, J. Chem. Theory Comput., 2009, 5, 3195–3210 CrossRef CAS.
- H. J. Risselada and S. J. Marrink, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 17367–17372 CrossRef CAS.
- S. Baoukina, L. Monticelli, H. J. Risselada, S. J. Marrink and D. P. Tieleman, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 12462–12467.
- Z. L. Wang and X. H. He, J. Chem. Phys., 2009, 130, 094905 CrossRef.
- Y. Li, X. Chen and N. Gu, J. Phys. Chem. B, 2008, 112, 16647–16653 CrossRef CAS.
- L. Monticelli, E. Salonen, P. C. Ke and I. Vattulainen, Soft Matter, 2009, 5, 4433–4445 RSC.
- Y. Li and N. Gu, J. Phys. Chem. B, 2010, 114, 2749–2754 CrossRef CAS.
- X. B. Lin, Y. Li and N. Gu, J. Comput. Theor. Nanosci., 2010, 7, 269–276 CrossRef CAS.
- S. Esteban-Martin, H. J. Risselada, J. Salgado and S. J. Marrink, J. Am. Chem. Soc., 2009, 131, 15194–15202 CrossRef CAS.
- H. J. Risselada and S. J. Marrink, Phys. Chem. Chem. Phys., 2009, 11, 2056–2067 RSC.
- H. J. Risselada and S. J. Marrink, Soft Matter, 2009, 5, 4531–4541 RSC.
- M. J. Hinner, S. J. Marrink and A. H. de Vries, J. Phys. Chem. B, 2009, 113, 15807–15819 CrossRef CAS.
- S. A. Sanders and A. Z. Panagiotopoulos, J. Chem. Phys., 2010, 132, 114902 CrossRef.
- S. O. Yesylevskyy, L. V. Schåfer, D. Sengupta and S. J. Marrink, PLoS Comput. Biol., 2010, 6, e1000810 Search PubMed.
- A. Y. Shih, A. Arkhipov, P. L. Freddolino and K. Schulten, J. Phys. Chem. B, 2006, 110, 3674–3684 CrossRef CAS.
- A. Arkhipov, Y. Yin and K. Schulten, Biophys. J., 2008, 95, 2806–2821 CrossRef CAS.
- A. Y. Shih, P. L. Freddolino, A. Arkhipov and K. Schulten, J. Struct. Biol., 2007, 157, 579–592 CrossRef CAS.
- A. Y. Shih, A. Arkhipov, P. L. Freddolino, S. G. Sligar and K. Schulten, J. Phys. Chem. B, 2007, 111, 11095–11104 CrossRef CAS.
- Y. Yin, A. Arkhipov and K. Schulten, Structure, 2009, 17, 882–892 CrossRef CAS.
- A. Arkhipov, Y. Yin and K. Schulten, Biophys. J., 2009, 97, 2727–2735 CrossRef CAS.
- M. Orsi, D. Y. Haubertin, W. E. Sanderson and J. W. Essex, J. Phys. Chem. B, 2008, 112, 802–815 CrossRef CAS.
- M. Orsi, J. Michel and J. W. Essex, J. Phys.: Condens. Matter, 2010, 22, 155106 CrossRef.
- J. C. Shelley, M. Y. Shelley, R. C. Reeder, S. Bandyopadhyay and M. L. Klein, J. Phys. Chem. B, 2001, 105, 4464–4470 CrossRef CAS.
- I. R. Cooke and M. Deserno, J. Chem. Phys., 2005, 123, 224710 CrossRef.
- A. P. Lyubartsev, Eur. Biophys. J., 2005, 35, 53–61 CrossRef CAS.
- S. Izvekov and G. A. Voth, J. Chem. Theory Comput., 2006, 2, 637–648 CrossRef CAS.
- J. C. Shelley, M. Y. Shelley, R. C. Reeder, S. Bandyopadhyay, P. B. Moore and M. L. Klein, J. Phys. Chem. B, 2001, 105, 9785–9792 CrossRef CAS.
- M. J. Stevens, J. Chem. Phys., 2004, 121, 11942–11948 CrossRef CAS.
- S. Izvekov and G. A. Voth, J. Phys. Chem. B, 2005, 109, 2469–2473 CrossRef CAS.
- W. G. Noid, J.-W. Chu, G. S. Ayton, V. Krishna, S. Izvekov, G. A. Voth, A. Das and H. C. Andersen, J. Chem. Phys., 2008, 128, 244114 CrossRef CAS.
- S. Izvekov and G. A. Voth, J. Phys. Chem. B, 2009, 113, 4443–4455 CrossRef CAS.
- L. Lu and G. A. Voth, J. Phys. Chem. B, 2009, 113, 1501–1510 CrossRef CAS.
- G. S. Ayton and G. A. Voth, J. Phys. Chem. B, 2009, 113, 4413–4424 CrossRef CAS.
- A. P. Lyubartsev and A. Laaksonen, Phys. Rev. E: Stat. Phys., Plasmas, Fluids, Relat. Interdiscip. Top., 1995, 52, 3730–3737 CrossRef CAS.
- D. Reith, M. Pütz and F. Müller-Plathe, J. Comput. Chem., 2003, 24, 1624–1636 CrossRef CAS.
- A. Lyubartsev, Y. Tu and A. Laaksonen, J. Comput. Theor. Nanosci., 2009, 6, 951–959 CrossRef CAS.
- A. Lyubartsev, A. Mirzoev, L. J. Chen and A. Laaksonen, Faraday Discuss., 2010, 144, 43–56 RSC.
- K. R. Hadley and C. McCabe, J. Chem. Phys., 2010, 132, 134505 CrossRef CAS.
- T. Murtola, E. Falck, M. Karttunen and I. Vattulainen, J. Chem. Phys., 2007, 126, 075101 CrossRef.
- W. K. den Otter, J. Chem. Phys., 2009, 131, 205101 CrossRef.
- B. West, F. L. H. Brown and F. Schmid, Biophys. J., 2009, 96, 101–115 CrossRef CAS.
- B. W. Reynwar and M. Deserno, Biointerfaces, 2008, 3, FA117–FA124 Search PubMed.
- F. de Meyer and B. Smit, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 3654–3658 CrossRef CAS.
- F. Schmid, D. Duchs, O. Lenz and B. West, Comput. Phys. Commun., 2007, 177, 168–171 CrossRef CAS.
- M. Hömberg and M. Müller, J. Chem. Phys., 2010, 132, 155104 CrossRef.
- J. Neder, B. West, P. Nielaba and F. Schmid, J. Chem. Phys., 2010, 132, 115101 CrossRef.
| This journal is © The Royal Society of Chemistry 2011 |
