Microwave-assisted synthesis of highly water-soluble graphene towards electrical DNA sensor†
Bong Gill
Choi
a,
HoSeok
Park
b,
Min Ho
Yang
a,
Young Mee
Jung
c,
Sang Yup
Lee
a,
Won Hi
Hong
*a and
Tae Jung
Park
*d
aDepartment of Chemical & Biomolecular Engineering (BK21), KAIST, 335 Gwahangno, Yuseong-gu, Daejeon 305-701, Republic of Korea. E-mail: whhong@kaist.ac.kr
bDepartment of Chemical Engineering, Kyung Hee University, 1 Seocheon-dong, Giheung-gu, Yongin-si, Gyeonggi-do 446-701, Republic of Korea
cDepartment of Chemistry, Kangwon National University, Chunchon, 200-701, Republic of Korea
dBioProcess Engineering Research Center, Center for Systems & Synthetic Biotechnology, Institute for the BioCentury, KAIST, 335 Gwahangno, Yuseong-gu, Daejeon 305-701, Republic of Korea. E-mail: tjpark@kaist.ac.kr
First published on 25th October 2010
Abstract
Graphene sheets have the potential for practical applications in electrochemical devices, but their development has been impeded by critical problems with aggregation of graphene sheets. Here, we demonstrated a facile and bottom-up approach for fabrication of DNA sensor device using water-soluble sulfonated reduced graphene oxide (SRGO) sheets via microwave-assisted sulfonation (MAS), showing enhanced sensitivity, reliability, and low detection limit. Key to achieving these performances is the fabrication of the SRGOs, where the MAS method enabled SRGOs to be highly dispersed in water (10 mg mL−1) due to the acidic sulfonated groups generated within 3 min of the functionalization reaction. The water-soluble SRGO-DNA (SRGOD) hybrids prepared by electrostatic interactions between a flat single layer of graphene sheets and DNAs are suitable for fabrication of electrical DNA sensor devices because of the unique electrical characteristics of SRGODs. The high sensing performance of SRGOD sensors was demonstrated with detection of DNA hybridization using complementary DNAs, single base mismatched DNAs, and noncomplementary DNAs, with results showing higher sensitivity and lower detection limit than those of reduced graphene oxide-based DNA sensors. Simple and easy fabrication of DNA sensor devices using SRGODs is expected to provide an effective way for electrical detection of DNA hybridization using miniature sensors without the labor-intensive labeling of the sensor and complex measurement equipment.
Introduction
Graphene is a leading building block for various electrochemical devices due to its remarkable electrical, optical, and mechanical properties.1–6 A key challenge for the practical applications of graphene is to improve the processability in stabilizing exfoliated graphene in suspension while maintaining its intrinsic physical and electric properties close to ideal values.1–6 Recently, many efforts have been devoted to overcome this challenge, particularly through the functionalization and dispersion of graphene sheets based on solution chemistry.7–9 Among the various functionalization methods of graphene, introduction of dangling sulfonic groups (–SO3−) into the basal and/or edge planes of graphene allows for the high concentration, full exfoliation, and long-term stability of the graphene sheets in water because of the high acidicity of sulfonic groups (pKa = −6).10,11 T. Samulski et al. reported the chemically covalent functionalization of graphene by p-phenyl-SO3H groups.10 G. Shi et al. suggested a different method of non-covalent functionalization of graphene using sulfonated polyaniline.11 These methodologies, however, require polymeric or surfactant stabilizers, long reaction times, and severe experimental conditions. In this regard, there is a need for the development of a facile and efficient approach for processing graphene sheets for large-scale production.To date, microwave-assisted processes have been regarded as a suitable technique for the functionalization of carbon allotropes, such as carbon nanotubes (CNTs) and graphene, in terms of the simplicity, time-saving, versatility, high yields, and high product purity.12,13 In particular, the introduction of functional groups, such as carboxylated and sulfonated groups via the microwave method, alters the carbon materials to possess easily dispersable characteristics in water. The hydrophilic functional groups can modulate the architecture of nanoscale–hybrid complexes by means of covalent or noncovalent interactions with biomolecules, such as peptides, RNA and DNA, for applications in molecular electronics, optoelectronics, and sensors.14–17 Among various practical applications, DNA hybridization sensors based on carbon nanomaterials have received much attention in diagnosing pathogenic or genetic diseases.18–20
In this research, we report the facile, rapid, and scalable method of synthesizing highly water-soluble graphene sheets through microwave-assisted sulfonation (MAS). The graphene sheets functionalized by sulfonic groups can be readily dispersed in various organics and water solvents. Additionally, the complex interactions of the functionalized graphenes with DNA provided an advanced resistive sensing platform for the electrical detection of label-free DNA.
Experimental
Chemicals
Graphite powder (<20 μM) and hydrazine solution (65 wt% in water) were purchased from Aldrich. Sulfuric acid (97%) and nitric acid (70%) aqueous solutions were obtained from Junsei.Preparation of graphene oxide (GO)
GO was synthesized by the modified Hummers method.21 As-prepared graphite oxide powders were purified thoroughly by dialysis with de-ionized (DI) water for one week to remove remaining impurities until the filtrated solution was neutral. The powder was obtained by filtration with DI water several times, and then dried at 60 °C under vacuum. The fully exfoliated GOs were prepared by the following procedure. The graphite oxide powders (1 mg) were dispersed in water (10 mL), then the dispersion was fully exfoliated by ultrasonic treatment for 30 min (Fisher Scientific Model 500 ultrasonic, 350 W) and centrifuged for 30 min. The high purified GOs were collected by filtrating the resulting brown dispersion from the centrifuge. Prior to use the GOs, the GO powders were dried at 60 °C under vacuum.Synthesis of sulfonated reduced graphene oxide (SRGO)
20 mg of GOs were added into 20 mL of a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (vol%) mixture of nitric acid (70%) and sulfuric acid (97%) aqueous solutions in the reaction chamber lined with Teflon PFA and controlled with a pressure (0–200 psi). This mixture was then treated with microwave radiation (CEM MDS-2100 microwave digestion system) at 50% of a total of 900 W and 20 psi of pressure for 3 min. The resulting mixture was carefully added to DI water (500 mL) (the acid mixture is highly corrosive). The dispersion of functionalized sulfonated GOs (SGOs) was purified by dialysis with DI water until the filtrated solution was neutral. The functionalized GOs were filtrated, washed with DI water several times, and dried at 60 °C under vacuums. In order to prepare the SRGOs, the obtained functionalized GOs (10 mg) by microwave-assisted method were dispersed in DI water (10 mL), and the highly homogeneous black solution of SRGO was obtained by reduction with an addition of 100 μL hydrazine solutions at 90 °C for 2 h. The resulting mixture was purified by dialysis for one week to remove the remaining hydrazine and impurities. The final SRGO products were filtered and dried under vacuum at 60 °C and stored.
1 (vol%) mixture of nitric acid (70%) and sulfuric acid (97%) aqueous solutions in the reaction chamber lined with Teflon PFA and controlled with a pressure (0–200 psi). This mixture was then treated with microwave radiation (CEM MDS-2100 microwave digestion system) at 50% of a total of 900 W and 20 psi of pressure for 3 min. The resulting mixture was carefully added to DI water (500 mL) (the acid mixture is highly corrosive). The dispersion of functionalized sulfonated GOs (SGOs) was purified by dialysis with DI water until the filtrated solution was neutral. The functionalized GOs were filtrated, washed with DI water several times, and dried at 60 °C under vacuums. In order to prepare the SRGOs, the obtained functionalized GOs (10 mg) by microwave-assisted method were dispersed in DI water (10 mL), and the highly homogeneous black solution of SRGO was obtained by reduction with an addition of 100 μL hydrazine solutions at 90 °C for 2 h. The resulting mixture was purified by dialysis for one week to remove the remaining hydrazine and impurities. The final SRGO products were filtered and dried under vacuum at 60 °C and stored.
Preparation of SRGO/DNA (SRGOD) hybrids and resistive biosensor
SRGOD hybrids and resistive biosensors were prepared as follows. As-prepared 1 mg of SRGO was added in tris-ethylenediaminetetraacetic acid (Tris–EDTA, pH 8.0) buffer solution (10 mM Tris-HCl, 1 mM EDTA, and 100 mM NaCl) containing 1 μM of probe DNA (5 mL) for DNA immobilization and stirred for 1 h. The SRGOD hybrids were obtained after dialysis for 10 h to remove unbound probe DNAs. To fabricate the SRGOD resistive biosensors, 2 μL of SRGOD solutions was drop-casted onto the interdigitated gold electrodes and dried under ambient conditions. RGO-DNA (RGOD) resistive biosensors as a control sample were prepared by following the same protocol as the SRGOD resistive biosensor using RGO instead of SRGO. The probe-modified electrodes were blocked with a poly(ethylene glycol) (PEG, molecular weight 380–420) aqueous solution for 12 h in order to block the target DNAs from directly stacking onto SRGOs. After rinsing with water several times, the blocked biosensors were treated with blow drying. Label-free DNA hybridization on the surface of resistive sensor was performed by dispersion in phosphate-buffered saline (PBS, pH 7.4) solution with 2 μL of target DNAs for 1 h.Characterization
Transmission electron microscopy (TEM) images were collected on an E.M. 912 Ω energy-filtering TEM (EF TEM 120kV) and a JEM-3010 HR-TEM (300 kV). The scanning electron microscopy (SEM) micrographs were obtained using a field emission scanning electron microscope (FEI Sirion model) equipped with an in-house Schottky emitter in high stability. Atomic force microscopy (AFM) images were recorded in non-contact mode using a NanoMan Digital Instruments 3100 AFM (Veeco) with etched silicon aluminium coated tip. FT-IR spectra were collected on a JASCO Fourier transform infrared spectroscopy (FT-IR) 470 and by attenuated total reflection. Each spectrum was recorded from 4000 to 650 cm−1 using 12 scans at a resolution of 4 cm−1. The pressure was equal in all samples to avoid differences caused by the pressure and penetrating depth. Solid-state Magic Angle Spinning Nuclear Magnetic Resonance (NMR) spectra were collected on a Bruker Avance 400 spectrometer operating at 10 kHz with using a 4 mm Magic Angle Spinning. Spectra based on free induction decays with moderate decoupling power were averaged over 4000 scans with a recycle delay of 4 s. The Raman spectra were recorded using a Jobin Yvon/HORIBA LabRam ARAMIS Raman spectrometer equipped with an integral BX 41 confocal microscope. The radiation from an air cooled HeNe laser (632.8 nm) was used as the excitation source. The zeta potentials of SRGO solutions were measured by a Malvern Zetasizer Nano-ZS particle analyzer. DNA hybridization through electrical sensing was carried out by measuring current versus voltage (I–V) characteristics of SRGOD and RGOD sensor devices through manual probe station integrated with Keithley measurement (Cascade SUMMIT, 11862B). The current values from all devices were taken with a voltage sweep condition from −2 V to 2 V at room temperature under ambient conditions.Results and discussion
Scheme 1 illustrates a schematic representation of the steps to synthesize water-soluble SRGO sheets and SRGOD hybrids for label-free DNA detection. 20 mg of graphene oxide (GO), which was prepared by a modified Hummers method,20 was added to a mixture of nitric acid (50 vol%) and sulfuric acid (50 vol%) aqueous solutions. This mixture was then treated with microwave radiation for 3 min under controlled pressure and power causing extensive carboxylation and sulfonation on the GOs to obtain SGOs. Subsequently, after the reduction of SGOs, SRGOs were obtained, possessing reduced carboxylic groups and residual sulfonic acid groups, and dispersed in water at a concentration of 10 mg mL−1. The quality of single layer GO, SGO and SRGO was analyzed as below. In order to obtain the SRGOD complexes, the SRGOs were complexed with single-stranded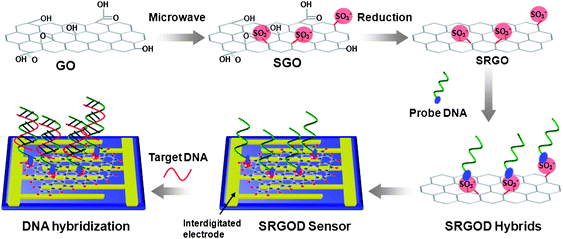 | ||
| Scheme 1 Illustration for the preparation of SRGO and procedure for the electrical detection of label-free DNA hybridization using SRGOD hybrids. | ||
DNA (ssDNA) probes via electrostatic interactions between the sulfonic acid groups of SRGOs and the amine groups of probe DNAs. The solution of SRGOD hybrids was dropped onto the surface of an interdigitated gold electrode for the assembly of electrochemical DNA sensing platforms. This resistive type sensors based on the SRGOD hybrids was used to detect the DNA hybridization between the target and probe ssDNAs.
Fig. 1a presents the photo images of SRGO suspensions with respect to different concentrations in aqueous and organic solvents. The highly stable homogeneous solutions of SRGOs were immediately obtained in water, alcohol, N,N-dimethylformamide (DMF), dimethyl sulfoxide (DMSO), tetrahydrofuran (THF), and acetone. In particular, the high concentration of 10 mg mL−1 of SRGOs was homogeneously dispersed in water without the aggregation of sheets even after long term dispersion for one month. The zeta potential values of SRGOs showed −51.5 mV (at 2 mg mL−1 in water), which is a reasonable value for stable colloids based on a general colloidal science.22 In contrast, reduced graphene oxides (RGOs) were reaggregated under identical experimental conditions. Consequently, the high acidic sulfonic moieties tethered to the RGO sheets played an important role in accomplishing the long term dispersion at a high concentration. Fig. 1b, 1c, and 1d show TEM and AFM images of SGO and SRGO. SRGOs in an aqueous solution were fully exfoliated into a single layer of graphene sheets analogous to the RGOs (Fig. 1b). After drop casting onto the surface of mica, SGO and SRGO revealed a thickness of 1.6 nm and 0.85 nm, respectively, as shown in AFM analysis (Fig. 1c and 1d). Similar to other functionalized RGOs,10 the water-soluble SRGOs were slightly thicker than the single graphene sheets of ∼0.34 nm, due to the existence of dangling sulfonic groups prepared by MAS.
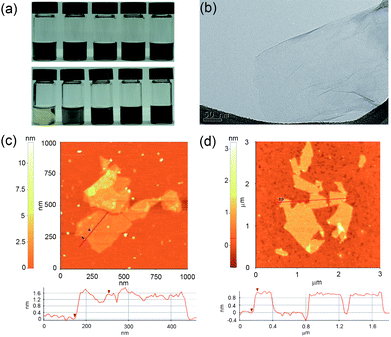 | ||
| Fig. 1 (a) Photographs of SRGO dispersions of various organic solvents (top) from left: ethanol, DMF, DMSO, THF, and acetone (1 mg mL−1) and various concentrations in DI water (bottom) from left: 0.05, 0.5, 1, 2, and 10 mg mL−1, (b) TEM image of SRGO, and AFM images of (c) SGO and (d) SRGO. | ||
FT-IR and X-ray photoelectron spectroscopy (XPS) were used to investigate the chemical properties of SGOs and SRGOs after MAS and reduction, as shown in Fig. 2 and 3. The characteristic bands of the FT-IR spectroscopy of GO were assigned as follows: 3300 cm−1 for νO–H, 1720 cm−1 for νC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, 1620 cm−1 for νadsorbed H2O, 1410 cm−1 for νO–H deform, and 1050 cm−1 for νC–O stretch.23 Compared to the bands of GOs, the bands of sulfonated groups, as well as the carboxylic acid of SGOs, were more prominent after microwave radiation. For SGOs carboxylate moieties, a broad band at 3041 cm−1 for ν–OH stretch and the strong band at 1710 cm−1 for νC
O, 1620 cm−1 for νadsorbed H2O, 1410 cm−1 for νO–H deform, and 1050 cm−1 for νC–O stretch.23 Compared to the bands of GOs, the bands of sulfonated groups, as well as the carboxylic acid of SGOs, were more prominent after microwave radiation. For SGOs carboxylate moieties, a broad band at 3041 cm−1 for ν–OH stretch and the strong band at 1710 cm−1 for νC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretch were more prominent after microwave radiation. In addition, SGOs revealed the emergence of strong bands at 1200 cm−1 for νSO2 symmetric stretch and 1046 cm−1 for νSO3– stretch of sulfonic acid groups.12 After reduction of SGOs with hydrazine, the band intensities for the carboxylate groups at 3041 cm−1, 1710 cm−1, and 1600 cm−1 significantly decreased. The characteristic bands for the sulfonic groups at 1200 cm−1 and 1048 cm−1 were still apparent, though slightly decreased in intensity. The results of FT-IR spectra were consistent with those of C1s spectra of GOs and SRGOs from XPS as shown in Fig. 3. The C1s spectrum of GOs could be deconvoluted into four peaks: C–C (284.5 eV), C–O (286.5 eV), C
O stretch were more prominent after microwave radiation. In addition, SGOs revealed the emergence of strong bands at 1200 cm−1 for νSO2 symmetric stretch and 1046 cm−1 for νSO3– stretch of sulfonic acid groups.12 After reduction of SGOs with hydrazine, the band intensities for the carboxylate groups at 3041 cm−1, 1710 cm−1, and 1600 cm−1 significantly decreased. The characteristic bands for the sulfonic groups at 1200 cm−1 and 1048 cm−1 were still apparent, though slightly decreased in intensity. The results of FT-IR spectra were consistent with those of C1s spectra of GOs and SRGOs from XPS as shown in Fig. 3. The C1s spectrum of GOs could be deconvoluted into four peaks: C–C (284.5 eV), C–O (286.5 eV), C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O (288 eV), and O–C
O (288 eV), and O–C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O (289.1 eV).24,25 After SGOs were reduced to SRGO, most of the C–O, C
O (289.1 eV).24,25 After SGOs were reduced to SRGO, most of the C–O, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, and O–C
O, and O–C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O peaks were weakened while an additional peak assigned to C–S (285.4 eV) appeared.11,26 Microwave radiation extensively functionalized the chemical groups of GOs in the presence of a strong oxidant mixture, proved by FT-IR results. The hydroxyl, epoxy, and carboxyl functionalities were more abundantly introduced onto the GOs by strong oxidation of GOs under microwave systems. In addition, the acid sulfonate functionalization were present on GOs in the form of C–S covalent functionalization, verified by XPS results. Importantly, the reaction of the MAS treated-GOs with strong reductant reduced the hydroxyl, epoxy, and carboxyl functionalities, but maintained the sulfonate functionalities. The reduction of SGOs to SRGO was further confirmed by Raman and NMR (ESI, Fig. S1† for Raman and Fig. S2† for NMR).
O peaks were weakened while an additional peak assigned to C–S (285.4 eV) appeared.11,26 Microwave radiation extensively functionalized the chemical groups of GOs in the presence of a strong oxidant mixture, proved by FT-IR results. The hydroxyl, epoxy, and carboxyl functionalities were more abundantly introduced onto the GOs by strong oxidation of GOs under microwave systems. In addition, the acid sulfonate functionalization were present on GOs in the form of C–S covalent functionalization, verified by XPS results. Importantly, the reaction of the MAS treated-GOs with strong reductant reduced the hydroxyl, epoxy, and carboxyl functionalities, but maintained the sulfonate functionalities. The reduction of SGOs to SRGO was further confirmed by Raman and NMR (ESI, Fig. S1† for Raman and Fig. S2† for NMR).
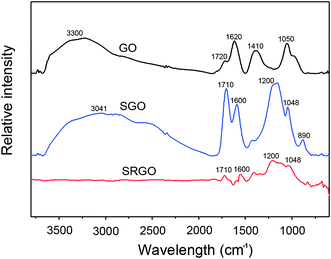 | ||
| Fig. 2 FT-IR spectra of GO, SGO and SRGO, respectively. | ||
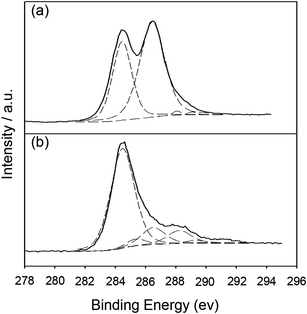 | ||
| Fig. 3 The C1s XPS spectra of (a) GO and (b) SRGO. | ||
To demonstrate the usefulness of SRGOs as a sensing platform for detecting DNA hybridizations, a sensor using the SRGOD complex was fabricated and tested. The sequences of the DNA strands used in this experiment were designed as follows: probe DNA, 5′-TGATGGAACTTGCTT-NH2-3′; complementary target DNA (ctDNA), 5′-AAGCAAGTTCCATCA-3′; single-base mismatched DNAs (mmDNA), 5′-AAGCAAGCTCCATCA-3′ (mmDNA1; single-base mismatched target DNA type 1); 5′-AAGCAAGATCCATCA-3′ (mmDNA2; single-base mismatched target DNA type 2); 5′-AAGCAAGGTCCATCA-3′ (mmDNA3; single-base mismatched target DNA type 3); non-complementary DNA (ncDNA), 5′-TGCGTTCACGGTAGG-3′. The probe DNA strands can easily interact with the water-soluble single sheets of SRGOs to form a monolayer on the SRGOD complex. The formation of the monolayer is driven by the non-covalent interaction between the sulfonic groups on the SRGOs and the terminal amine groups of the probe DNA molecules (ESI, Fig. S3† of FT-IR result). The SRGOD hybrid solution of 2 μL was drop-deposited onto the interdigitated area of the electrodes as shown in Fig. 4. The SRGODs deposited on electrodes were then dried under vacuum to remove residual solvents. In order to prevent the target DNAs from directly stacking onto SRGOs through non-specific interactions, the blocking method was used with PEG. The density of SRGODs across the electrode fingers can be easily controlled by varying the concentration of the SRGOD solution. The dropped solution was sufficient to connect the 3 μm gaps between electrodes. The reproducibility of drop-casting SRGOD onto the electrodes for sensor preparation was checked by analysis of SEM and AMF images, which showed very flat thin layers of SRGODs connecting the electrodes (see Fig. 4b and ESI, Fig. S4† for AFM images). The resulting interdigitated sensor assembly provided reproducible performance. The TEM and AFM results were used to characterize the SRGOD complexes deposited on the electrode, as shown in Fig. 4c and 4d. The SRGOD hybrid formed a flat thin layer, which was shown from AFM results with an apparent height of 3 nm for the SRGOD, consistent with the monolayer of DNA molecules adsorbed onto SRGOs.19 The favorable interactions of water-soluble single sheets of SRGOs with the probe DNA strands and the electrostatic interactions between the sulfonic groups on the SRGOs and the amine groups of probe DNAs, induced the smooth monolayer of SRGOD complex. Tarlov et al. demonstrated that creating monolayers of DNA probes influenced the improvement of DNA hybridization due to the good accessibility of target DNAs to the well-oriented probe DNAs.27 It is expected that the monolayer morphology of SRGOD complex influences the effective specific hybridization with target DNAs. Therefore, the strategy based on SRGO through MAS was suitable for the fabrication of an electrical DNA sensor due to the efficient immobilization of nucleic acids.
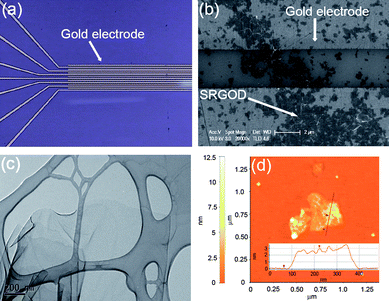 | ||
| Fig. 4 (a) Photograph of interdigitated electrodes, (b) SEM image of SRGODs across two gold electrodes, (c) TEM image of SRGOD, and (d) AFM image of SRGOD. Inset represents the cross-sectional contour plot between the mica surface and the SRGOD of the scan line in the area. | ||
In resistance-based sensors, the signal transduction is the detection of the change in current (I) versus voltage (V) after the immobilization of target DNAs on the blocked SRGOD sensor (Fig. 5a). The significant attenuation of the electrical conductance of the device (∼ 66%) is clearly observed when 700 nM of ctDNAs was introduced to the sensor. In contrast, only a small decrease in current was selectively and competitively observed for ncDNA (∼3%) and for mmDNA1 (∼26%). These observations indicate that the electrochemical sensing platform based on SRGOD can capture the hybridization efficiency of label-free DNAs because of the excellent electrical properties of the graphene sheets, the large surface area for the accessibility of target DNAs, and the sequence-specific binding between probe and target DNA molecules.
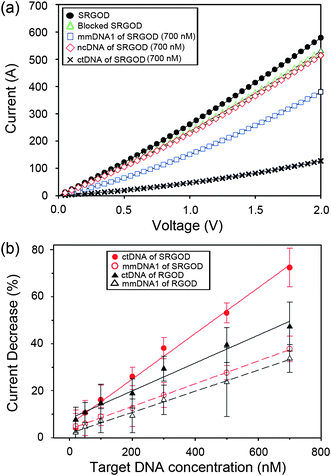 | ||
| Fig. 5 (a) I–V plots of SRGOD sensor immobilized with probe DNA, blocked with PEG, and hybridized with noncomplementary (700 nM) and target (700 nM) DNA; and (b) comparison of electrical sensing characteristics for hybridizing ctDNA and mmDNA1 on the SRGOD and RGOD sensors, respectively. | ||
To verify the performance of the DNA sensor, the changes in the current responses of SRGOD were further measured with respect to various concentrations of the ctDNA and mmDNA molecules, and then compared to the RGO-DNA (RGOD) resistive sensor under the same experimental conditions. The change in conductance response by the immobilization of DNAs was calculated by the following equation:
In comparison of RGOs, the sulfonate functionality of RGOs delivered many advantages for application of DNA sensors in terms of easy processing of graphene, strong immobilization of probe DNAs through electrostatic interactions, and sensitive electrical change by the scattering strength on defective sites. The fully exfoliated SRGOs could easily interact with probe DNAs in an aqueous solution, enabling an array of probe DNAs on a flat layer of SRGOs. The smooth monolayer of the SRGOD complex on the sensor device provided a large accessible surface area and the favorable morphology for hybridization of target DNAs. In contrast to SRGODs, RGODs without MAS method displayed aggression and non-uniform RGODs on sensor devices (see ESI, Fig. S4† of AFM image). The introduction of oxygen groups onto the graphene sheets induced the increase of defect sites on the electrical transport of graphene. When DNAs with negative phosphate groups adsorbed onto the defect points of graphene sheets, the scattering strength of the defect points increased, and consequently decreased the electrical conductance of graphene sheets.28,29 Subsequently, it is expected that the hybridization of target DNAs with probe DNAs on defective sites changes the electrical conductance due to the increase of anionic phosphate groups on the defective carbon nanomaterials.28,29 The well-defined architectures of the SRGOD complexes provided the good accessibility of the target DNAs to the probe DNAs and served as an excellent electrical signal transduction represented by the change in electrical conductance.
Conclusions
In summary, we have demonstrated a fast, facile, and scalable functionalization synthesis of water-soluble graphene sheets through MAS for the implementation of high performance label-free DNA biosensors. The SRGO sheets fabricated by MAS method could be exfoliated into flat single layers and dispersed in water without any assistance from polymeric or surfactant stabilizers. The sulfonic acidic groups of SRGOs interacted with amine groups of DNA molecules by electrostatic interactions, thereby forming flat single layers. The SRGOD hybrids with conformational rearrangement allowed the favorable morphology and accessible surface area for specific interactions with target DNA molecules. The sensor assembly based on SRGOD complexes showed a more clear discrimination in electrical resistance upon exposure to ctDNAs and mmDNAs compared to that of RGOD sensor. In addition, the SRGOD sensor showed a well-defined linear electrical response in various concentrations of target DNAs in the range from 20 nM to 700 nM, with a low detection limit of 42 nM. The simple fabrication process, reproducibility, and fast readout of the label-free DNA sensor for the detection of DNA hybridization suggested here make the SRGOD-based sensor a promising candidate for highly efficient nanoscale diagnostic devices.Acknowledgements
This work was supported by the IT Leading R&D Support Project from the Ministry of Knowledge Economy (Korea) through KEIT and by a grant from the National Fisheries Research and Development Institute (NFRDI), Korea.Notes and references
- X. Li, X. Wang, L. Zhang, S. Lee and H. Dai, Science, 2008, 319, 1229–1232 CrossRef CAS.
- F. Schedin, A. K. Geim, S. V. Morozov, E. W. Hill, P. Blake, M. I. Katsnelson and K. S. Novoselov, Nat. Mater., 2007, 6, 652–655 CrossRef CAS.
- N. Mohanty and V. Berry, Nano Lett., 2008, 8, 4469–4476 CrossRef CAS.
- M. D. Stoller, S. Park, Y. Zhu, J. An and R. S. Ruoff, Nano Lett., 2008, 8, 3498–3502 CrossRef CAS.
- J. L. Vickery, A. J. Patil and S. Mann, Adv. Mater., 2009, 21, 2180–2184 CrossRef CAS.
- M. Zhou, Y. Zhai and S. Dong, Anal. Chem., 2009, 81, 5603–5613 CrossRef CAS.
- S. Park and R. S. Ruoff, Nat. Nanotechnol., 2009, 4, 217–224 CrossRef CAS.
- H. Yang, C. Shan, F. Li, D. Han, Q. Zhang and L. Niu, Chem. Commun., 2009, 3880–3382 RSC.
- J. R. Lomeda, C. D. Doyle, D. V. Kosynkin, W.-F. Hwang and J. M. Tour, J. Am. Chem. Soc., 2008, 130, 16201–16206 CrossRef CAS.
- Y. Si and E. T. Samulski, Nano Lett., 2008, 8, 1679–1682 CrossRef CAS.
- H. Bai, Y. Xu, L. Zhao, C. Li and G. Shi, Chem. Commun., 2009, 1667–1669 RSC.
- Y. Wang, Z. Lqbal and S. Mitra, J. Am. Chem. Soc., 2006, 128, 95–99 CrossRef CAS.
- A. V. Murugan, T. Muraliganth and A. Manthiram, Chem. Mater., 2009, 21, 5004–5006 CrossRef CAS.
- B. L. Allen, P. D. Kichambare and A. Star, Adv. Mater., 2007, 19, 1439–1451 CrossRef CAS.
- R. J. Chen, H. C. Choi, S. Bangsaruntip, E. Yenilmez, X. Tang, Q. Wang, Y.-L. Chang and H. Dai, J. Am. Chem. Soc., 2004, 126, 1563–1568 CrossRef CAS.
- J. Chen, J. Zhang, K. Wang, X. Lin, L. Huang and G. Chen, Anal. Chem., 2008, 80, 8028–8034 CrossRef CAS.
- H.-S. Zhuang, J.-L. Huang and G.-N. Chen, Anal. Chim. Acta, 2004, 512, 347–353 CrossRef CAS.
- X. Tang, S. Bansaruntip, N. Nakayama, E. Yenilmez, Y.-l. Chang and Q. Wang, Nano Lett., 2006, 6, 1632–1636 CrossRef CAS.
- C.-H. Lu, H.-H. Yang, C.-L. Zhu, X. Chen and G.-N. Chen, Angew. Chem., Int. Ed., 2009, 48, 4785–4787 CrossRef CAS.
- X. Dong, Y. Shi, W. Huang, P. Chen and L.-J. Li, Adv. Mater., 2010, 22, 1649–1653 CrossRef CAS.
- W. S. Hummers and R. E. Offeman, J. Am. Chem. Soc., 1958, 80, 1339 CrossRef CAS.
- D. H. Everett, Basic Principles of Colloid Science, The Royal Society of Chemistry, London, 1988 Search PubMed.
- Y. Xu, H. Bai, G. Lu, C. Li and G. Shi, J. Am. Chem. Soc., 2008, 130, 5856–5857 CrossRef CAS.
- S. Stankovich, D. A. Dikin, R. D. Piner, K. A. Kohlhaas, A. Kleinhammes, Y. Jia, Y. Wu, S. T. Nguyen and R. S. Ruoff, Carbon, 2007, 45, 1558–1565 CrossRef CAS.
- H. Ago, T. Kugler, F. Cacialli, W. R. Salaneck, M. S. P. Shaffer, A. H. Windle and R. H. Friend, J. Phys. Chem. B, 1999, 103, 8116–8121 CrossRef CAS.
- S. Stankovich, R. D. Piner, X. Chen, N. Wu, S. T. Nguyen and R. S. Ruoff, J. Mater. Chem., 2006, 16, 155–158 RSC.
- T. M. Herne and M. J. Tarlov, J. Am. Chem. Soc., 1997, 119, 8916–8920 CrossRef CAS.
- X. Wang, F. Liu, G. T. S. Andavan, X. Jing, K. Singh, V. R. Yazdanpanah, N. Bruque, R. R. Pandey, R. Lake, M. Ozkan, K. L. Wang and C. S. Ozkan, Small, 2006, 2, 1356–1365 CrossRef CAS.
- K. Kim, H. J. Park, B.-C. Woo, K. J. Kim, G. T. Kim and W. S. Yun, Nano Lett., 2008, 8, 3092–3096 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available: Raman, NMR and FT-IR spectroscopy; electrical detection data of DNA. See DOI: 10.1039/c0nr00562b |
| This journal is © The Royal Society of Chemistry 2010 |

