Nanotoxicology: no small matter
Neus
Feliu
and
Bengt
Fadeel
*
Division of Molecular Toxicology, Institute of Environmental Medicine, Karolinska Institutet, Nobels väg 13. 171 77, Stockholm, Sweden. E-mail: bengt.fadeel@ki.se; Fax: +46 8 34 38 49; Tel: +46 8 524 877 37
First published on 27th September 2010
Abstract
Engineered nanomaterials i.e. materials deliberately manufactured on a nanoscale offer exciting new opportunities in technology and medicine. However, the increasing use of nanomaterials in society also raises concerns as to their possible adverse effects on human health and the environment. This review considers the potential application of high-throughput screening approaches to assess hazards of engineered nanomaterials. The disciplinary identity of toxicology is also discussed as attention shifts towards nanoscale objects.
 Neus Feliu Neus Feliu | Neus Feliu is enrolled as a Ph.D. student at Karolinska Institutet. She received her undergraduate training in chemistry at the University of Barcelona and M.Sc. in Biomedical Materials at the Royal Institute of Technology in Stockholm. She is interested in the biomedical applications of nanomaterials including SPIONs and dendrimers. She also plays the cello.† |
 Bengt Fadeel Bengt Fadeel | Bengt Fadeel, M.D., Ph.D., is Head of the Division of Molecular Toxicology and Vice Chairman of the Institute of Environmental Medicine at Karolinska Institutet in Stockholm. He is the project coordinator of FP7-NANOMMUNE, an EU-US consortium focused on the assessment of hazardous effects of engineered nanomaterials on the immune system.† |
1. Introduction
The term ‘engineered nanomaterial’ is used to describe man-made materials that are produced on a nanoscale (Fig. 1). To what purpose are such materials made, apart from the obvious fact that it's fun to manipulate atoms1 or that perfect nanoscale objects such as fullerenes or dendrimers possess an inherent beauty,2 at least to some people? Nanomaterials are new materials i.e. fundamental physico-chemical properties of a material change as we shrink a material from bulk form to nano-size. Hence, the melting point of gold changes as a function of the particle diameter and nano-sized particles of iron oxide become superparamagnetic. Furthermore, taking a single sheet of carbon atoms and rolling it into a hollow tube produces a novel material that is lighter and stronger than steel and stiffer than a diamond. It is no small surprise that these novel properties have generated excitement in diverse areas of technology and medicine. However, these novel properties also raise concerns for human health and the environment.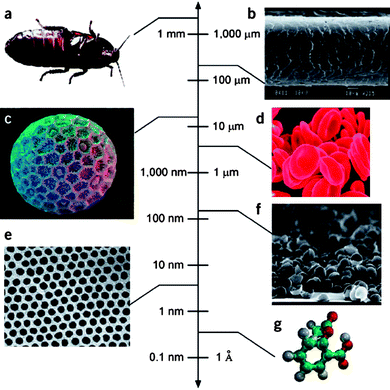 | ||
| Fig. 1 The scale of things. Engineered nanoparticles exist in the same size regime as biological structures. (a) Cockroach; (b) human hair; (c) Polygonum pollen grain; (d) red blood cells; (e) cobalt nanocrystal superlattice; (f) aggregate of half-shells of palladium; (g) aspirin molecule. Reprinted by permission from Macmillan Publishers Ltd: Whitesides GM. The ‘right’ size in nanobiotechnology. Nature Biotechnology, 2003, volume 21, number 10, pp. 1161–1165. | ||
One of the problems of nanotoxicological research today is that we are far too often looking for the keys under the lamppost. We purport to elucidate the general principles of the toxicological behavior of ‘nanoparticles’ when we are in reality only studying single examples of nanoparticles selected at random, or from convenience, from a vast universe of different man-made nanoparticles. What we need to do is to reverse the logic: we should study thousands of different nanomaterials in a coordinated and systematic manner in order to deduce whether common signatures or biological responses exist. Only then will our understanding of the toxicity of nanoscale materials transform into a predictive science.
Risk is commonly recognized as the product of hazard and exposure. However, while there has been an increasing emphasis placed on hazard assessment of nanomaterials in recent years, the extent of human or environmental exposure to engineered nanomaterials remains poorly understood. Moreover, it is important to consider the entire life cycle of nanomaterials and nanomaterial-based products, from production to disposal, and to understand the likelihood of worker or consumer exposure to nanomaterials at these different stages. Indeed, the exposure of nanoscientists/chemists to nanomaterials should not be neglected.
Have any advances been made in the field of nanotoxicology over the past 10 years? Yes, of course. For instance, as pointed out in a recent commentary,3 it is now well ingrained into researchers in the field that one must understand not only the model system that is being used but also the nature of the nanoparticle suspensions i.e. a detailed understanding of the physico-chemical properties of nanoparticles, and how these properties may change in a given biological system, is required for an adequate interpretation of the toxicological data. Nevertheless, we still lack an understanding of which nanoparticle properties that are driving the adverse effects, and the problem is aggravated by the sheer numbers of novel nanoscale objects that are emerging as chemists and physicists continue to rejoice in the manipulation of matter at the atomic scale.
High-throughput screening (HTS) approaches could offer a solution to some of these problems.
2. The scale of things
The cell is a collection of nanoscale machines.4 Indeed, consider the ribosome in the cytoplasm of the cell, a perfect example of a ‘nanoparticle’ (∼ 20 nm) designed by Mother Nature to perform the intricate task of synthesizing proteins. Furthermore, take a moment to contemplate the adenovirus, a common vector utilized in gene therapy. The adenovirus is a ‘nanoparticle’ of ∼ 30 nm in diameter and transports its cargo (nucleic acid) to specific target organs to combat disease, but its utility is limited by the fact that particles circulating in the blood stream are captured by macrophages in the liver. Nanomedicine grapples with similar issues when attempting to design nanoparticles for drug delivery. Understanding biology5 or “thinking nanobiologically”6 is a prerequisite for our prediction and understanding of the effects of engineered nanomaterials.George Whitesides7 remarked that “there already exists a highly developed science concerned with biologically relevant nanostructures: this science is called ‘chemistry’. One may add that there also exists a discipline that focuses on the elucidation of how the body handles foreign invasion by nanoscale and micron-sized objects (particles) and organisms: this science is called ‘immunology’. Indeed, as discussed in a recent review,8 a valid paradigm which could aid in our understanding of the biological and/or toxicological effects of nanoparticles is to consider the immune system and how it recognizes and responds to the multitude of nano- and microorganisms in our environment. The immune system is equipped with specialized, antigen-presenting cells that are responsible for integrating a myriad of external stimuli to produce an adaptive immune response.9 To interpret what to do with the antigen, antigen-presenting cells use so-called pattern recognition receptors to detect signature molecules (pathogen-associated molecular patterns) that herald infection. Deducing whether such general principles of interactions exist between man-made nanoparticles and biological systems remains a key challenge for toxicologists.8 The observation that nanoparticles are coated with proteins and lipids upon introduction into a biological system10 suggests that the immune system may “see” nanoparticles in much the same way as it senses microbes. There are likely many reciprocal lessons to be learned: immunology may inform nanotoxicology, as indicated above, and the use of engineered nanoparticles to probe the behavior of immune-competent cells could shed light on immunological mechanisms.
3. High-throughput screening
It is interesting to note how common it is for chemists to use the word ‘facile’ when describing a novel route of synthesis of a nanoscale material. In contrast, it is not easily accomplished or attained to perform toxicological testing on all these new materials. In fact, as pointed out by Maynard et al.,11 “the enormous diversity of engineered nanomaterials with different sizes, shapes, compositions and coatings matches, and possibly exceeds, that of conventional chemicals.” These authors proposed that benchmarked and validated high-throughput protocols are needed to screen for potential hazards. They also suggested that the selection of suitable in vitro screening assays and their validation should be accomplished within the next 5 years.11 The commentary was published in 2006 which means that we now have only 1 year left to reach this ambitious goal.High-throughput screening (HTS) is a method for scientific experimentation that comprises the screening of large chemical libraries for activity against biological targets via the use of automation, miniaturized assays, and large-scale data analysis.12 HTS (10.000–100.000 compounds tested per day) and ultra-HTS (excess of 100.000 data-points generated per day) are seen as key elements in the drug discovery pipeline in industry.
Miniaturization is key in HTS. The typical working volume for a 384-well plate, which accommodates 4 times more samples than a standard 96-well plate, is in the range of 30–100 μL with a standard volume of about 50 μL per well. The majority of assays, biochemical or cell-based, can be adapted to a 384-well format and this plate format has been established as the (current) format of choice among pharma and biotech companies.12 Some processes have been adapted toward 1536-well plate formats. The typical working volume is in the range of about 2.5–10 μL total volume, with a standard volume of ∼ 5 μL per well. In a very recent study, an ultrahigh-throughput screening platform using drop-based microfluidics was presented allowing for 1000 times faster screening (100 million reactions in 10 h) and a million-fold reduction in cost as compared to conventional techniques.13 In this study, the authors used aqueous drops dispersed in oil as picoliter-volume reaction vessels to identify new mutants of the enzyme horseradish peroxidase exhibiting catalytic rates more than 10 times faster than their parent (Fig. 2).
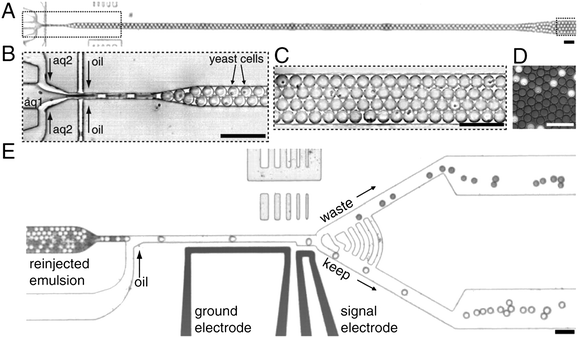 | ||
| Fig. 2 Ultrahigh-throughput screening platform. (A) A low-magnification image of the drop-making device. (B) A suspension of yeast cells displaying horseradish peroxidase (HRP) on their surface (aq1) is combined with a second aqueous stream containing the fluorogenic substrate, amplex ultrared (aq2). The yeast are at a concentration of 1 × 108 cells per mL, which gives an average of 0.3 cells per 6 pL drop after being diluted by half by the substrate stream. The aqueous drops are formed at a flow-focusing junction in a fluorocarbon oil, and the number of cells per drop follows a Poisson distribution: ∼ 22% contain a single cell. (C) The drops flow out of the device into a tube that acts as an incubation line where they incubate for 5 min. (D) A single layer of drops after incubation showing the fluorescence developing from the active HRP displayed on the surface of the cells. (E) From the delay line, the drops flow as a solid plug to a junction where oil is added to separate the drops. To visually demonstrate the sorting process, an emulsion containing light and dark drops was sorted; the light drops contain fluorescein, and the dark contain bromophenol blue. Scale bar, 80 μm. Reproduced from Agresti J.J. et al. Ultrahigh-throughput screening in drop-based microfluidics for directed evolution. Proc. Natl. Acad. Sci. USA, 2010;107(9):4004–9 (copyright 2010 by the National Academy of Sciences). | ||
In 2007, the U.S. Environmental Protection Agency (EPA) launched a project entitled ToxCast™ to predict (or forecast) toxicity of chemicals using computational chemistry, HTS, and various toxicogenomic technologies.14 The underlying hypothesis is that toxicological responses are driven by interactions between chemicals and biomolecular targets. The emphasis is on a multiple target matrix approach so that no single assay or endpoint will have a large impact on the interpretation of the “fingerprint” of the tested compound. In their introduction to the ToxCast™ program, Dix et al.14 noted one important difference between HTS for drug discovery and HTS as a tool to guide toxicology: the aim in drug discovery is to find a small number of active compounds amenable to subsequent optimization for drug development whereas in toxicology, HTS must determine the activity of all compounds tested, and false negatives are of greater concern. Several interesting studies have emanated from the ToxCast™ program since its inception.15–17 Judson et al.16 recently provided an overview of the entire ToxCast™ phase I assay results. In total, 309 chemicals were tested in 467 assays spanning nine technologies, including high-throughput cell-free assays and cell-based assays, in multiple human primary cells and cell lines plus rat primary hepatocytes. A total of 624 in vitro assay end-points (including multiple time-points) were measured for each chemical, generating > 200.000 concentration responses (see Fig. 3 for a heat map of the entire in vitro data set). Overall, chemicals were found to range in promiscuity across cellular and molecular pathways, from no activity to affecting dozens of pathways. The study also revealed associations between a small set of in vitro assays and rodent liver lesion formation.
![High-throughput screening of chemicals. Heat map of the entire ToxCast™ phase I assay results data set (624 assay measurements). Assays are arranged left to right, and chemicals are arranged top to bottom. The color bar at the top indicates the assay type: red (cell-free HTS), violet (multiplexed transcription reporter), yellow (biologically multiplexed activity profiling), green (high-content cell imaging), blue (multiplexed gene expression), pink (cell-based HTS), black (phase I and II XME cytotoxicity), white (real-time cell electronic sensing), and orange (HTS genotoxicity). Data values are −log10(AC50/LEC), where light pink is inactive and darker reds indicate increased activity (lower AC50/LEC). AC50, half-maximal activity concentration; HTS, high-throughput screening; LEC, lowest effective concentration; XME, xenobiotic metabolizing enzyme. Reproduced from Judson R.S. et al. In vitro screening of environmental chemicals for targeted testing prioritization: the ToxCast project. Environ. Health Perspect. 2010; 118(4):485–492 [doi: 10.1289/ehp.0901392] with permission from Environmental Health Perspectives.](/image/article/2010/NR/c0nr00535e/c0nr00535e-f3.gif) | ||
| Fig. 3 High-throughput screening of chemicals. Heat map of the entire ToxCast™ phase I assay results data set (624 assay measurements). Assays are arranged left to right, and chemicals are arranged top to bottom. The color bar at the top indicates the assay type: red (cell-free HTS), violet (multiplexed transcription reporter), yellow (biologically multiplexed activity profiling), green (high-content cell imaging), blue (multiplexed gene expression), pink (cell-based HTS), black (phase I and II XME cytotoxicity), white (real-time cell electronic sensing), and orange (HTS genotoxicity). Data values are −log10(AC50/LEC), where light pink is inactive and darker reds indicate increased activity (lower AC50/LEC). AC50, half-maximal activity concentration; HTS, high-throughput screening; LEC, lowest effective concentration; XME, xenobiotic metabolizing enzyme. Reproduced from Judson R.S. et al. In vitro screening of environmental chemicals for targeted testing prioritization: the ToxCast project. Environ. Health Perspect. 2010; 118(4):485–492 [doi: 10.1289/ehp.0901392] with permission from Environmental Health Perspectives. | ||
In recent years, high-content assays have been applied for toxicity assessment of engineered nanomaterials. In their seminal work, Shaw et al.18 evaluated 50 different nanomaterials at four doses in four cell types using four different assays. The goal was to analyze broad patterns of activity of the nanomaterials relative to one another, as opposed to extrapolating from results of a single in vitro assay. The cell types were selected to reflect a range of tissues relevant for evaluation of intravascularly administered agents: vascular cells (endothelial and smooth muscle cells), monocytes and hepatocytes. Hierarchical clustering of the data identified nanomaterials with similar patterns of biologic activity – or perturbation of activity – across several cellular contexts. Furthermore, a subset of nanoparticles was tested in mice, and nanoparticles with similar activity profiles in vitro exerted similar effects on monocyte number in vivo.18 High-content in vitro assays combined with genome-wide expression analysis of exposed cells has also been applied to assess the toxicity of poly(ethylene glycol)-coated versus non-coated quantum dots.19 Similarly, Jan et al.20 utilized high-content screening to assess the toxicity of quantum dots versus gold nanoparticles. George et al.21 demonstrated the use of a multiparameter cytotoxicity assay that evaluates oxidative stress to compare the effects of several metal oxide nanoparticles in bronchial epithelial and macrophage cell lines. Other investigators have proposed that zebrafish (Danio rerio) embryos may serve as an economically feasible, medium-throughput screening platform for assessment of nanoparticle toxicity.22 However, despite these interesting approaches, the full potential of automated HTS technologies for hazard assessment of nanomaterials has not yet been realized.
One of the causes of the high cost of drug development and of toxicological evaluation of chemicals is the lack of experimental model systems that can replace expensive and time-consuming animal studies. Three-dimensional in vitro assays that reconstitute tissue-tissue interfaces critical to organ function could serve to expand the capabilities of cell culture models. Legendre et al.23 developed an in vitro model that replicates the composition, organization, and barrier and spermatogenesis functions of the rat blood-testis barrier as a potential alternative to animal reproductive toxicity tests. Furthermore, Huh et al.24 reported very recently on a “lung-on-a-chip” microdevice that reproduces several structural, functional and mechanical properties of the human alveolar-capillary interface, the fundamental unit of the lung (Fig. 4). Applying this novel device to the study of nanoparticles, the authors showed that cyclic mechanical strain accentuates toxic and inflammatory responses to silica nanoparticles. Mechanical strain also enhanced epithelial and endothelial uptake of nanoparticles and stimulated their transport into the underlying microvasculature. Similar effects of physiological breathing on nanoparticle adsorption are observed in the intact mouse lung. There are limitations to the lung mimic device, such as the lack of alveolar macrophages, but this microengineering approach is promising and marks an important step in the in vitro modeling of critical tissue-tissue interfaces. The system is miniaturized and amenable to multiplexing24 and can potentially be adapted for automated HTS of drugs, chemicals or nanoparticles.
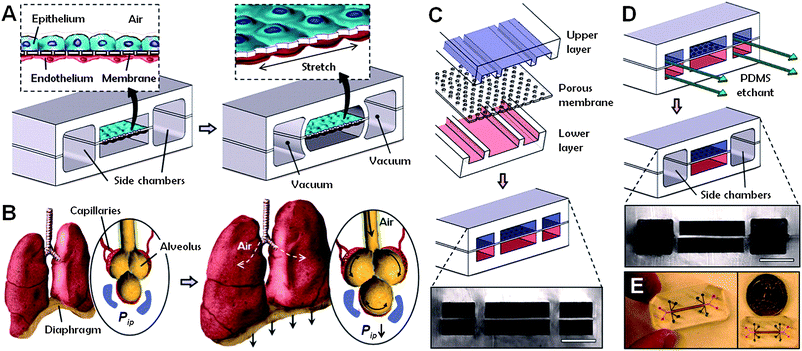 | ||
| Fig. 4 Lung-on-a-chip device. (A) The microfabricated device uses compartmentalized poly(dimethylsiloxane) (PDMS) microchannels to form an alveolar-capillary barrier on a thin, porous, flexible PDMS membrane coated with extracellular matrix (ECM) components. (B) During inhalation in the living lung, contraction of the diaphragm causes a reduction in intrapleural pressure (Pip), leading to distension of the alveoli and physical stretching of the alveolar-capillary interface. (C) Three PDMS layers are aligned and irreversibly bonded to form two sets of three parallel microchannels separated by a 10 μm thick PDMS membrane containing an array of through-holes with an effective diameter of 10 μm. (D) After permanent bonding, PDMS etchant is flowed through the side channels. Selective etching of the membrane layers in these channels produces two large side chambers to which vacuum is applied to cause mechanical stretching (i.e. ‘breathing’ lung mimic device). (E) Images of a lung-on-a-chip microfluidic device viewed from above. From: Huh D. et al. Reconstituting organ-level lung functions on a chip. Science, 2010, volume 328, pp. 1662–1668. Reprinted with permission from AAAS. | ||
In their recent review of the “21st century paradigm” for evaluating the hazards of nanoscale materials, Walker and Bucher25 cautioned that HTS approaches may only be applicable for a few classes of nanomaterials that are compatible with available test systems, due to the unpredictable and/or artifactual behavior of many of the current and future nanomaterials in (current) in vitro assays. The authors are absolutely right in highlighting the importance of a thorough characterization of the physico-chemical properties of nanomaterials, not only at synthesis but also in the experimental system used. However, the frequently noted interference of nanoparticles with existing toxicity assays, especially those in vitro assays that are based on the detection of light,26 should not prevent one from considering high-throughput approaches to screen for nanoparticle-induced toxicity. Indeed, there is an urgent need to establish standardized and validated cytotoxicity tests of nanomaterials, including assays based on new test principles (label-free detection) which cannot be influenced by nanoparticles themselves, and to adapt these to HTS. Chemical compounds can also produce artifacts in an aqueous environment due to the formation of colloidal chemical aggregates at certain pH, temperature and buffer conditions12 and this represents one of the challenges in modern drug discovery. However, with appropriate tools to characterize the test compound (chemical, nanomaterial) in situ, such artifacts may in principle be understood and avoided, or minimized.
The field of HTS of nanomaterials is in its infancy, but the goal is clear as well as bold: to utilize rapid, automated screening approaches to provide detailed and comparable toxicity data (‘signatures’) for thousands of different nanomaterials in order to promote the safe development of such materials. However, HTS will not replace conventional toxicology, but could aid in the prioritization of nanomaterials for further testing, including animal testing. HTS may also allow for the development of models that predict how nanoparticles react in biological systems. In fact, understanding the ‘behavior’ of nanoparticles in miniaturized test systems (384- or 1536-well plates) poses a critical challenge to toxicologists. However, the cell itself is a miniaturized system, a biological nano-cosmos. Therefore, understanding the behavior of engineered nanoparticles in a small reaction volume (for instance, in a single well of a miniaturized assay, or in a single human cell such as a macrophage or a red blood cell) is precisely the challenge that we need to address.
4. Is REACH out of reach?
Thomas Hartung suggested in his provocative commentary that the testing of substances for adverse effects on humans and the environment needs a radical overhaul if we are to meet the challenges of ensuring health and safety.27 He argued that three important technologies developed during the past decade may change the way in which we do toxicology: ‘omics’ technologies (such as genomics and proteomics), imaging techniques and robotized testing platforms. In fact, the combination of biochemical knowledge of cellular pathways with genomics, proteomics and metabonomics is rapidly emerging as systems biology, at the heart of which lies the integration rather than the reduction of information, and ‘systems toxicology’ can be considered as a new sub-branch of this field.27 Such a systems approach was put forward in a 2007 report by the US National Academy of Sciences on behalf of the US EPA.28 The overall aim, as explicated by Collins et al.,28 is to enable a shift from toxicity testing primarily in animal models to in vitro assays, in vivo assays using lower model organisms, and computational modeling, thus enabling the evolution of toxicology from a predominantly observational science to a predictive science.The European Union introduced the regulation known as Registration, Evaluation, Authorization and Restriction of Chemicals (REACH) by legislation in 2007. This regulation represents the largest safety assessment of chemicals that has ever been carried out, and calls for registration of ∼ 30.000 chemicals over a period of about one decade. Under REACH, the burden of proof in establishing the safety of a substance has been passed from the regulator to manufacturers, importers and producers, representing a significant financial cost to industry. Indeed, some have argued that the cost may be too high, and that uncertainties regarding the implementation of REACH for nanoscale substances or materials in particular would be detrimental to all parties involved in the commercialization of such materials.29 However, it will be far more counterproductive if unsafe nanomaterial-based products were to enter the market. The question, therefore, is not whether toxicity testing is needed, but how testing of the vast numbers of chemicals that are being manufactured should be conducted, and how to regulate nanoscale materials i.e. are nanoscale materials equivalent to their micro- or macroscale counterparts, or should they be considered as “new substances” and therefore be subjected to specific regulation? With regard to the first question, it would make a lot of sense to adopt systems toxicology approaches and HTS-based platforms, as outlined above. This could enable toxicologists to move away from the slow, traditional, chemical-by-chemical approach to a ‘category approach’ based on the grouping of chemicals with similar biologic profiles without the need for additional animal testing.30 In regard to the latter question, it appears, at present, that this has to be decided on a case-by-case basis. Not all nanomaterials are created equal. Nevertheless, if we accept that materials that are produced in the nanoscale acquire new physico-chemical properties not seen in the corresponding bulk form of the same material,31 then it is only logical that these materials should be viewed as “new substances”. For successful risk management of nanomaterials, it is important for the scientific community to understand what questions risk assessors and legislators need to ask, and what research will best answer them.32
There are scattered attempts to use gene expression profiling to address the toxicity of nanoparticles. Ding et al.33 reported on whole-genome expression analysis of human fibroblasts exposed to multi-walled carbon nano-onions versus multi-walled carbon nanotubes. The authors noted that multiple cellular pathways are perturbed after exposure to these nanomaterials, and could determine material-specific toxicogenomic profiles. Waters et al.34 conducted whole-genome microarray analyses of the RAW 264.7 murine macrophage cell line exposed to amorphous silica nanoparticles and found that cellular responses are highly conserved across particle sizes. Pan et al.,35 on the other hand, found that 1.4 nm gold nanoparticles capped with triphenylphosphine monosulfonate are much more cytotoxic to the HeLa human cervix carcinoma cell line than 15 nm nanoparticles of similar chemical composition. Genome-wide expression profiling indicated upregulation of stress-related genes after incubation with the smaller nanoparticles but not with the 15 nm nanoparticles. Overall, while transcriptomic approaches are gaining traction, what appears to be lacking is a systematic, side-by-side comparison of the impact of different classes of nanomaterials on gene expression profiles of exposed cells (in vitro) and tissues (in vivo). Such ‘genomic footprinting’ approaches,36 combined with proteomics-based safety evaluation of nanomaterials,37 could ultimately aid in the assessment – and prediction – of nanomaterial hazard.
Sydney Brenner famously complained that molecular biology research has become much more descriptive and much less experimental: he called this “low input, high throughput, no output science”.38 He argued that what one ought to do is not to collect more data but to organize it, so as to convert the vast amount of information that we are accumulating into knowledge. HTS and systems toxicology approaches will, by definition, lead to the accumulation of vast amounts of information. How, then, do we avoid drowning in a sea of data? Theoretical paradigms are helpful as platforms from which to view to data. In recent years, a number of paradigms have emerged in nanotoxicology: the bio-nano interface/protein corona paradigm,39 the oxidative stress paradigm40 and the pathogenic fibre paradigm,41 to name but a few. Ultimately, a paradigm is only a framework, a scaffold, and basic knowledge of biology will always be needed.
5. Closing remarks
In closing, it may be argued that there are no “nanomaterials”. There are, on the other hand, tens of thousands of different materials of different chemical composition produced on a nanoscale, and each new material should be studied on a case-by-case basis until common patterns or signatures can be assigned to these various nanomaterials or classes of nanomaterials. Single-walled carbon nanotubes are not spherical gold nanoparticles, and surface-modified/functionalized nanoparticles are different from the pristine nanomaterial.Our second sacrilegious conclusion is the following: There is no “nanotoxicology” (as in: “the toxicology of materials having one or more dimensions on the order of 100 nm or less”). Instead, one should consider the science of the interaction or interference of nanoscale objects with biological systems irrespective of any specific cut-off in size. Furthermore, we posit that in 10 years from today, there will no longer be a need for “nanotoxicology”. Instead, we will have only “toxicology” of chemicals or materials, including those materials that are produced and manipulated at the nanoscale. As pointed out in a recent commentary,42 we have no obvious use for a toxicological discipline that focuses exclusively on objects on a length scale of 0.1 to 1 m, and yet we try to put all things nano into one disciplinary bag. Instead, we should focus our attention on whether the nanoscale size per se endows materials/particles with specific and novel properties that require novel methods for assessment of the toxicological outcomes. We need to understand the nano-ness of nanoscale objects. The application of high-throughput screening as discussed herein could aid in the rapid assessment of large numbers of different nanoscale materials. This may allow not only for a better estimation of the potential risk of nanomaterial exposure but could also enhance our understanding of the beneficial and desirable biological properties of these materials. The future is small, and bright.
Acknowledgements
The authors are supported, in part, by the Seventh Framework Programme of the European Commission (EC-FP-7-NANOMMUNE-Grant Agreement No. 214281) and the Swedish Research Council.References
- C. Toumey, Nat. Nanotechnol., 2010, 5, 239–241 CrossRef CAS.
- C. Toumey, Nat. Nanotechnol., 2008, 3, 637–638 CrossRef CAS.
- M. J. Clift, P. Gehr and B. Rothen-Rutishauser, Arch. Toxicol., 2010 May 25 Search PubMed [Epub ahead of print].
- B. Alberts, Cell, 1998, 92, 291–294 CrossRef CAS.
- M. C. Roco, Curr. Opin. Biotechnol., 2003, 14, 337–346 CrossRef CAS.
- J. F. Nyland and E. K. Silbergeld, Hum. Exp. Toxicol., 2009, 28, 393–400 CrossRef CAS.
- G. M. Whitesides, Nat. Biotechnol., 2003, 21, 1161–1165 CrossRef CAS.
- A. A. Shvedova, V. E. Kagan and B. Fadeel, Annu. Rev. Pharmacol. Toxicol., 2010, 50, 63–88 CrossRef CAS.
- J. A. Hubbell, S. N. Thomas and M. A. Swartz, Nature, 2009, 462, 449–460 CrossRef CAS.
- A. E. Nel, L. Mädler, D. Velegol, T. Xia, E. M. Hoek, P. Somasundaran, F. Klaessig, V. Castranova and M. Thompson, Nat. Mater., 2009, 8, 543–557 CrossRef CAS.
- A. D. Maynard, R. J. Aitken, T. Butz, V. Colvin, K. Donaldson, G. Oberdörster, M. A. Philbert, J. Ryan, A. Seaton, V. Stone, S. S. Tinkle, L. Tran, N. J. Walker and D. B. Warheit, Nature, 2006, 444, 267–269 CrossRef CAS.
- L. M. Mayr and D. Bojanic, Curr. Opin. Pharmacol., 2009, 9, 580–588 CrossRef CAS.
- J. J. Agresti, E. Antipov, A. R. Abate, K. Ahn, A. C. Rowat, J. C. Baret, M. Marquez, A. M. Klibanov, A. D. Griffiths and D. A. Weitz, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 4004–4009 CrossRef CAS.
- D. J. Dix, K. A. Houck, M. T. Martin, A. M. Richard, R. W. Setzer and R. J. Kavlock, Toxicol. Sci., 2007, 95, 5–12 CAS.
- M. T. Martin, D. J. Dix, R. S. Judson, R. J. Kavlock, D. M. Reif, A. M. Richard, D. M. Rotroff, S. Romanov, A. Medvedev, N. Poltoratskaya, M. Gambarian, M. Moeser, S. S. Makarov and K. A. Houck, Chem. Res. Toxicol., 2010, 23, 578–590 CrossRef CAS.
- R. S. Judson, K. A. Houck, R. J. Kavlock, T. B. Knudsen, M. T. Martin, H. M. Mortensen, D. M. Reif, D. M. Rotroff, I. Shah, A. M. Richard and D. J. Dix, Environ. Health Perspect., 2010, 118, 485–492 CAS.
- D. M. Rotroff, B. A. Wetmore, D. J. Dix, S. S. Ferguson, H. J. Clewell, K. A. Houck, E. L. Lecluyse, M. E. Andersen, R. S. Judson, C. M. Smith, M. A. Sochaski, R. J. Kavlock, F. Boellmann, M. T. Martin, D. M. Reif, J. F. Wambaugh and R. S. Thomas, Toxicol. Sci., 2010 July 16 Search PubMed [Epub ahead of print].
- S. Y. Shaw, E. C. Westly, M. J. Pittet, A. Subramanian, S. L. Schreiber and R. Weissleder, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 7387–7392 CrossRef CAS.
- T. Zhang, J. L. Stilwell, D. Gerion, L. Ding, O. Elboudwarej, P. A. Cooke, J. W. Gray, A. P. Alivisatos and F. F. Chen, Nano Lett., 2006, 6, 800–808 CrossRef CAS.
- E. Jan, S. J. Byrne, M. Cuddihy, A. M. Davies, Y. Volkov, Y. K. Gunko and N. A. Kotov, ACS Nano, 2008, 2, 928–938 CrossRef CAS.
- S. George, S. Pokhrel, T. Xia, B. Gilbert, Z. Ji, M. Schowalter, A. Rosenauer, R. Damoiseaux, K. A. Bradley, L. Mädler and A. E. Nel, ACS Nano, 2010, 4, 15–29 CrossRef CAS.
- O. Bar-Ilan, R. M. Albrecht, V. E. Fako and D. Y. Furgeson, Small, 2009, 5, 1897–1910 CrossRef CAS.
- A. Legendre, P. Froment, S. Desmots, A. Lecomte, R. Habert and E. Lemazurier, Biomaterials, 2010, 31, 4492–4505 CrossRef CAS.
- D. Huh, B. D. Matthews, A. Mammoto, M. Montoya-Zavala, H. Y. Hsin and D. E. Ingber, Science, 2010, 328, 1662–1668 CrossRef CAS.
- N. J. Walker and J. R. Bucher, Toxicol. Sci., 2009, 110, 251–254 CrossRef CAS.
- N. Lewinski, V. Colvin and R. Drezek, Small, 2008, 4, 26–49 CrossRef CAS.
- T. Hartung, Nature, 2009, 460, 208–212 CrossRef CAS.
- F. S. Collins, G. M. Gray and J. R. Bucher, Science, 2008, 319, 906–907 CrossRef CAS.
- D. M. Bowman and G. van Calster, Nat. Nanotechnol., 2007, 2, 525–526 CrossRef CAS.
- K. van Leeuwen and G. Schaafsma, Nature, 2009, 462, 34 CrossRef CAS.
- M. Auffan, J. Rose, J. Y. Bottero, G. V. Lowry, J. P. Jolivet and M. R. Wiesner, Nat. Nanotechnol., 2009, 4, 634–641 CrossRef CAS.
- M. R. Gwinn and L. Tran, Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol., 2010, 2, 130–137 Search PubMed.
- L. Ding, J. Stilwell, T. Zhang, O. Elboudwarej, H. Jiang, J. P. Selegue, P. A. Cooke, J. W. Gray and F. F. Chen, Nano Lett., 2005, 5, 2448–2464 CrossRef CAS.
- K. M. Waters, L. M. Masiello, R. C. Zangar, B. J. Tarasevich, N. J. Karin, R. D. Quesenberry, S. Bandyopadhyay, J. G. Teeguarden, J. G. Pounds and B. D. Thrall, Toxicol. Sci., 2009, 107, 553–569 CAS.
- Y. Pan, A. Leifert, D. Ruau, S. Neuss, J. Bornemann, G. Schmid, W. Brandau, U. Simon and W. Jahnen-Dechent, Small, 2009, 5, 2067–2076 CrossRef CAS.
- T. L. Lee, W. Y. Chan and O. M. Rennert, ACS Nano, 2009, 3, 3830 CrossRef CAS author reply 3830–3831.
- H. Haniu, Y. Matsuda, K. Takeuchi, Y. A. Kim, T. Hayashi and M. Endo, Toxicol. Appl. Pharmacol., 2010, 242, 256–262 CrossRef CAS.
- E. C. Friedberg, Nat. Rev. Mol. Cell Biol., 2008, 9, 8–9 CrossRef CAS.
- I. Lynch, T. Cedervall, M. Lundqvist, C. Cabaleiro-Lago, S. Linse and K. A. Dawson, Adv. Colloid Interface Sci., 2007, 134–135, 167–174 CrossRef CAS.
- A. Nel, T. Xia, L. Mädler and N. Li, Science, 2006, 311, 622–627 CrossRef CAS.
- K. Donaldson, F. A. Murphy, R. Duffin and C. A. Poland, Part. Fibre Toxicol., 2010, 7, 5 CrossRef.
- R. A. Drezek and J. M. Tour, Nat. Nanotechnol., 2010, 5, 168–169 CrossRef CAS.
Footnote |
| † Photo: Ulf Sirborn, Karolinska Institutet |
| This journal is © The Royal Society of Chemistry 2010 |
