Zinc proteomes, phylogenetics and evolution†
Leonardo
Decaria
a,
Ivano
Bertini
ab and
Robert J. P.
Williams
*c
aMagnetic Resonance Center (CERM), University of Florence, Via L. Sacconi 6, 50019 Sesto Fiorentino, Italy
bDepartment of Chemistry, University of Florence, Via della Lastruccia 3, 50019 Sesto Fiorentino, Italy
cDepartment of Inorganic Chemistry, University of Oxford, South Parks Road, Oxford OX1 3QR, United Kingdom. E-mail: bob.williams@chem.ox.ac.uk
First published on 25th August 2010
Abstract
Evolution has not been studied in detail with reference to the changing environment. This requires a study of the inorganic chemistry of organisms, especially metalloproteins. The evolution of organisms has been analysed many times previously using comparative studies, fossils, and molecular sequences of proteins, DNA and 16s rRNA (Zhang and Gladyshev, Chem. Rev., 2009, 109, 4828). These methods have led to the confirmation of Darwin's original proposal that evolution followed from natural selection in a changing environment often pictured as a tree. In all cases, the main tree in its upper later reaches has been well studied but its lower earlier parts are not so well defined. To approach this topic we have treated evolution as due to the intimate combination of the effect of chemical changes in the environment and in the organisms (Williams and da Silva, The Chemistry of Evolution, 2006, Elsevier). The best chemicals to examine are inorganic ions as they are common to both. As a more detailed example of the chemical study of organisms we report in this paper a bioinformatic approach to the characterization of the zinc proteomes. We deduce them from the 821 totally sequenced DNA of organisms available on NCBI, exploiting a published method developed by one of us (Andreini, Bertini and Rosato, Acc. Chem. Res., 2009, 42, 1471). Comparing the derived zinc-finger-containing proteins and zinc hydrolytic enzymes in organisms of different complexity there is a correlation in their changes during evolution related to environmental change.
Introduction
Evolution has a well-mapped outline especially from the Cambrian period to today. Many authors have described it in diagrams with a tree-like structure using substantially two methods: comparison of organisms while exploiting also fossil evidence as Darwin did, or, more recently, using sequence studies of DNA, RNA and proteins.1 In all cases the recent upper reaches of the tree have been well studied but the earlier trunk and lower branches have not been so easy to study. In an effort to overcome some of the difficulties, we have begun a study of the environmental organisms and changes from the earliest possible dates since it is changes in the environment, which have had the greatest effect on early evolution.2 The environment can be examined and dated in sediments and organisms are considered to have arisen in large groups from bacteria to higher animals relative to these datings. The dating of organisms is aided by knowledge of the ages of fossils. The best chemicals for a comparative study during evolution are the inorganic elements as they are common to both. This paper is the first example of a detailed examination based on bioinformatic analysis of one element, zinc, from organism DNA sequences, devised by one of us.3Methods
We obtained 821 complete proteomes, 52 from archaea, 723 from bacteria and 46 from eukarya available on NCBI. We selected zinc proteins from them as an example of inorganic element chemistry involvement in organisms. We were able to do so from our ability to recognize zinc-binding domains in sequences of proteins. We recognized separately two major groups of zinc proteins, those of the hydrolytic enzymes and those of the zinc fingers as there are clear identifying structural data on both.4,5 In the case of the zinc fingers the metal sites are recognized by the coordination of the zinc by a combination of four residues of His or Cys in a particular sequence arrangement, giving rise to a tetrahedral geometry. The hydrolytic enzymes have zinc coordinated by a particular sequence of three residues selected from His, Glu, Asp and Arg with one or two water molecules. The knowledge of the site structures allows us to recognise the zinc binding domains in a protein or an enzyme as published.3 As reported in that paper, we used the HMMER program to search the NCBI refseq proteins database for matches to the hidden Markov models (HMMs) representing the selected domains. The HMMs were taken from the Pfam database without modification. We selected 10−3 as an Evalue cut-off. We thereby obtained a set of 271 zinc-binding proteins and divided them by their functions as mentioned above. The specific activities of the enzymes are known and we divided the hydrolases further as proposed by the Enzyme Commission, once their sequences were recognized. Further information about the methods is included in the ESI.†Results
In order to give a comparative account of the zinc protein data and their analyses, we have considered organisms in the following ways. The prokaryotes were divided into archaea and eubacteria, and the eubacteria were further divided into those with a small proteome of less than 1500 domains and those with a larger domain, see the Table 1. The smaller proteins are mostly of invasive bacteria found in animal hosts while the larger are of bacteria which in general are free-living. The two have considerable differences in the numbers of their zinc proteins. We shall take the archaea and the larger eubacteria as representing possible early forms of life or at least life of low complexity. There is little difference in the zinc protein content of these eubacteria and archaea and between anaerobic and aerobic species of both. Amongst eukaryotes we have placed multicellular metazoans in order of complexity, and quite probably in the order of their evolution, as is conventional; C. elegans, D. melanogaster, and Homo sapiens. We do not have data on a single-cell free-living eukaryote organisms and have used the small parasites, P. falsiparsium and T. brucei as examples. We have looked separately at the zinc proteins of one plant, an Arabidopsis and the yeast, Saccharomyces cerevisiae as an example of a fungus. In order that the reader can appreciate our approach to the common features of changes in the environment and this description of the evolution of organisms, we note that the prokaryotes are always considered as arising even before there was oxygen in the atmosphere and that the single cell eukaryotes arose about two billion years ago with the first step rise in oxygen, followed by the multicellular organisms with the second rise in oxygen after about one billion years ago.1,2 Accompanying the steps in oxygen were steps in trace metals in the environment including zinc.2 The data on the times of evolution of organisms can therefore be tied in time to those of the environment which enables us to compare zinc analyses of both. We turn to the examination of the zinc data on organisms in a comparative format.| Total proteome | %Zn-finger | %EC:3.4 | |
|---|---|---|---|
| a Average Value: Numbers in the first column refer to groupings of proteome sizes and the numbers in brackets refer to the numbers of proteomes examined. The average of which is given in the second column. | |||
| Archea (52) | 2176a | 0.180 | 0.923 |
| Bact. under 1500 (93) | 940a | 0.383 | 1.611 |
| Bact. over 1500 (630) | 3671a | 0.177 | 1.227 |
| P. falciparum | 6265 | 1.538 | 0.675 |
| T. brucei | 9279 | 1.369 | 0.787 |
| C. elegans | 22![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 844 844 |
2.889 | 1.064 |
| D. melanogaster | 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 513 513 |
3.734 | 2.613 |
| H. sapiens | 37![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 742 742 |
4.849 | 1.200 |
| A. thaliana | 32![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 615 615 |
2.370 | 0.584 |
Discussion
We shall refer to the total content of the zinc proteins in the organisms, see the Table 1, but to make a better comparison, we shall also give the zinc proteins as a percentage in their proteomes, Fig. 1. The increases in zinc finger proteins in both numbers and percentages there are seen to follow the order of complexity of organisms and probably the order of their evolution, Fig. 2. While the average number in all prokaryotes is less than six the number in Homo sapiens has increased to 1800. Both in numbers and percentages there are to be two step changes in the increase of these proteins between prokaryotes and unicellular metazoan organisms and between these unicellular organisms and the three multicellular organisms.6,7 There is a larger increase in the percentage in Homo sapiens, Fig. 3. Of great interest is to note that as mentioned above the zinc in the environment increased with oxygen increase in two steps which are close to these steps of organism evolution at 2.5 to 2.0 billion years ago and 1.0 to 0.5 billion years ago. The greater increase is in the second period when multicellular organisms arose. Increases in complexity of organisms is paralleled by the need for increases of message systems and we note that the zinc fingers are very important transcription factors for hormonal messengers. The smaller percentage and number of transcription factors in plants, Fig. 3 can be correlated with the much lower complexity of them compared with Homo sapiens. The value of percentage zinc fingers in unicellular yeast of 1.9% is similar to that in the unicellular metazoans.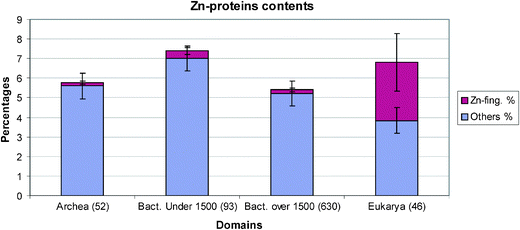 | ||
| Fig. 1 The average zinc protein contents for archaea, small and large bacteria and eukarya. Archaea and large bacteria have averages near to 0.2% of Zn-finger proteins in their proteomes, while small bacteria have about 0.4%. Eukarya Zn-finger content rises up to 3%. | ||
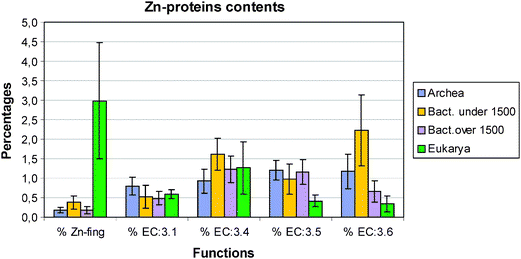 | ||
| Fig. 2 Zn–protein distribution in the four groups archaea, small and large bacteria and eukarya. EC:3.4 = protease/peptidase; EC:3.5 = hydrolases of C–N bonds other than in peptides; EC:3.6 = acid anhydride hydrolases. | ||
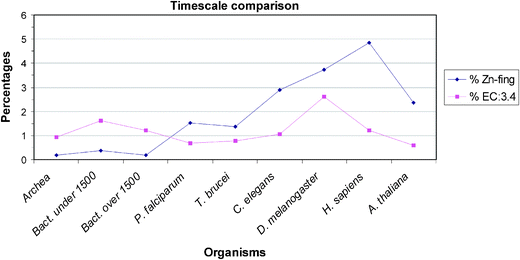 | ||
| Fig. 3 A timescale comparison of small genomes in small and large prokaryotes, then in unicellular and finally in multicellular eukaryotes. It is notable that the percentages in Zn-finger content rise within this evolutionary series. The percentage value of EC:3.4 in small bacteria is higher than in the large bacteria. Small bacteria are usually parasites, they need a bigger pool of proteases/peptidases to break down extracellular proteins for food. | ||
Turning to the other enzymes and proteins we have studied there are no metallothioneins in prokaryotes (not shown), plants and yeast but small numbers in metazoans. Greatest interest centres on the large numbers of hydrolytic zinc enzymes. We noted above the greater percentage of them in small eubacteria and their low level in eukarya, Fig. 2. In particular we see in Fig. 3 that the percentage of EC:3.4 enzymes, that is the peptidases and proteases, has a very different pattern from that of zinc fingers. Except for the fly, D. melanogaster, the percentage varies little being slightly lower in all the other eukaryotes than in prokaryotes. The high value in the fly could be related to its need to metamorphose. This will be examined in a wider range of organisms. The content of the zinc hydrolytic enzymes is high in all the organisms and we consider that this is a reflection of the need to hydrolyse proteins for food in all organisms and to hydrolyse connective tissue for growth in eukaryotes. It is noticeably lower in the multicellular plant and in yeast (0.8%) than in multicellular animals.
The changes in the EC:6 enzymes are of considerable interest as they include those for the hydrolytic reactions of phosphates. In particular zinc is associated in the earliest forms of life with the activities of RNA enzymes. In Fig. 2 we observe that the average values for all eukaryotes is much lower, less than 0.5%, than for all prokaryotes, greater than 0.6%. The parasytic eubacteria have the high value of above 2.0% on average. Notice however that they have very small genomes indicative of a loss of many genes but not of EC:4 and EC:6 enzymes. Unlike most of the other zinc proteins it appears that these enzymes have not evolved greatly from their initial functions.
The conclusion of this paper is that much light is thrown on the development of complexity and probably of evolution of organisms from a comparison of the zinc in the environment with that in organisms. It will be possible to examine our conclusions more closely as we acquire more data. It is certainly necessary to repeat the analysis with data on other metal ions. We stress the great advantage of such examinations of the inorganic content of organisms with their co-existing environment particularly in the study of early evolution.2 In this paper we have not attempted to trace special recent features of the evolution of zinc use such as the loss of cobalt enzymes, using vitamin B12, and their replacement by zinc enzymes in higher plants. Again zinc is required in the synthesis of shikimic acid, an essential part of the pathway to all amino acids carrying aromatic side-chains but the zinc enzyme is absent in higher animals. When did these gains or losses of zinc genes occur? The bioinformatic approach to metallomics of one of us3 as used here should be able to provide such information.
References
- Y. Zhang and V. N. Gladyshev, Chem. Rev., 2009, 109, 4828–4861 CrossRef CAS.
- R. J. P. Williams and J. J. R. Fraùsto da Silva, The Chemistry of Evolution, Elsevier, Chichester, 2006 Search PubMed.
- C. Andreini, I. Bertini and A. Rosato, Acc. Chem. Res., 2009, 42, 1471–1479 CrossRef CAS.
- W. Maret and Y. Li, Chem. Rev., 2009, 109, 4682–4707 CrossRef CAS.
- A. Messerschmidt, W. Bode and M. Cygler, Handbook of Metalloproteins, Elsevier, Chichester, 2004 Search PubMed.
- A. D. Anbar and A. H. Knoll, Science, 2002, 297, 1137–1142 CrossRef CAS.
- M. A. Saito, D. M. Sigman and F. M. M. Morel, Inorg. Chim. Acta, 2003, 356, 308–318 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available: Additional data. See DOI: 10.1039/c0mt00024h |
| This journal is © The Royal Society of Chemistry 2010 |
