DOI:
10.1039/C0MD00116C
(Concise Article)
Med. Chem. Commun., 2010,
1, 355-360
Received
20th July 2010
, Accepted 14th August 2010
First published on
27th October 2010
Introduction
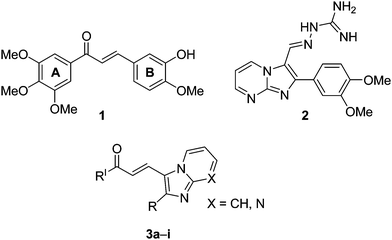
Chalcones (1,3-diaryl-2-propen-1-ones) represent an important group of natural products belonging to the flavonoids family.1,2 Chemically, these are open-chained molecules bearing two aromatic rings that are joined by a three-carbon enone fragment. These molecules possess interesting biological activities including cytotoxic3,4 antimalarial,5 antileishmanial,6 anti-inflammatory,7 anti-HIV,8 antifungal9 and as tyrosine kinase inhibitors.10 Natural and synthetic chalcones have been reported to possess strong antiproliferative effects in primary as well as established ovarian cancer cells11 and in gastric cancer (HGC-27) cells.12 Hydroxyl chalcones and isoliquiritigenin have been shown to be potent inhibitors of skin carcinogenesis.13 The remarkable biological potential of these chalcones is due to their possible interactions with various proteins related to cell apoptosis and proliferation. Recent studies have shown that these chalcones induce apoptosis in a variety of cell types, including breast cancers.14–16 The 3,4,5-trimethoxyphenyl ring is thought to be essential for retaining the anticancer activity of chalcones. Some of the recent advances in the development of anticancer agents involve structural modification of chalcones to improve their bioavailability and to study the role of various substituents on aryl or heteroaryl rings.17 In addition, chalcone derivatives wherein the B-ring is replaced by a heterocyclic ring have been systematically investigated.18 COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundImidazopyridine/pyrimidine are also well known compounds and many derivatives of this fused ring system have been evaluated for potential biological activity particularly for antitumor activity.19,20 Some representative COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundtrimethoxychalcone (1), imidazopyrimidine guanylhydrazone (COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound2) and COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundimidazopyridine/pyrimidine derivatives (3) are illustrated in Fig. 1.
Considering the potent bioactivities of compounds possessing an COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundimidazopyridine/pyrimidine core, we became interested to synthesize new chalcone derivatives incorporating an COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundimidazopyridine/pyrimidine skeleton and evaluated their anticancer activity. The promising activity obtained, prompted us to investigate their role in the cell proliferation and apoptosis of human breast cancer cell line (MCF-7). Further, it was considered of interest to investigate the effect of compound 3f on some of the proteins that regulate the cell cycle progression.
Results and discussion
Chemistry
The COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundimidazo[1,2-a]pyridine and COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundpyrimidine chalcone derivatives (3a–i) were prepared by the Claisen-Schmidt condensation of COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound3,4,5-trimethoxyacetophenone (9a) or COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound3,4-dimethoxyacetophenone (9b) with substituted imidazo[1,2-a]pyridine and pyrimidine aldehydes (8a–h) in the presence of KOH (50%). The imidazo[1,2-a]pyridine and pyrimidine aldehydes (8a–h) were obtained by means of the Vilsmeier reaction on the corresponding COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundimidazo[1,2-a] pyridine and pyrimidine (7a–h), that were obtained from compounds 6a–h. The intermediate compounds 6a–h were obtained by the reaction of appropriate COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound2-aminopyridine/pyrimidine (4a,b) and bromoketones (5a–e) as shown in Scheme 1.
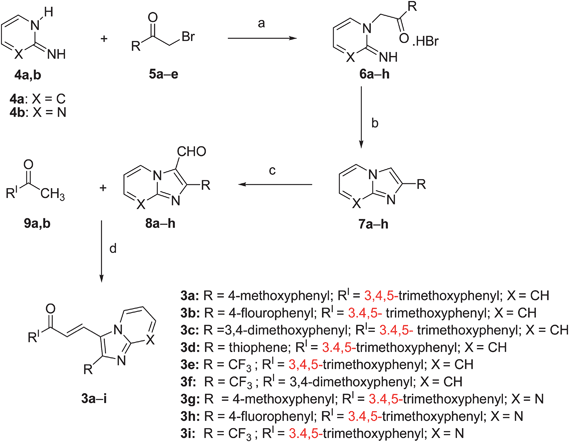 |
| | Scheme 1 Reagents and conditions: (a) COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundacetone, reflux, 6–8 h; (b) 2 N COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundHCl, reflux, 1 h, 85–92%; (c) POCl3, COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundDMF, reflux, 1 h, 75–80%; (d) 50% aq. KOH, 12 h, rt, 78–82%. | |
Biological evaluation
(A) Antiproliferative activity.
The representative compounds 3a and 3c–h were evaluated for in vitro antiproliferative activity in the standard 60 human cancer cell lines derived from nine cancer types (leukemia, non-small cell lung, colon, CNS, melanoma, ovarian, renal, prostate, and breast cancers). For each compound, dose–response curves for each cell line were measured at a minimum of five concentrations at 10-fold dilutions. A protocol of 48 h continuous drug exposure is used, and a sulforhodamine B (SRB) protein assay was used to estimate cell viability or growth. These compounds (3a and 3c–h) showed significant anticancer activity in a large number of cell lines with GI50 values ranging from 0.28 to 30.0 μM. Specifically, compound 3f exhibited promising antiproliferative activity with GI50 values less than 1 μM particularly against CCRF-CEM (0.44), K-562 (0.36), SR (0.44), RPMI-8226 (0.29) in leukemia cancer, NCI-H460 (0.68) in non small cell lung cancer, HCC-2298 (0.48), HCT-116 (0.84), KM12 (0.57) in colon cancer, IGROV1 (0.44) in ovarian cancer, DU-145 (0.65) in prostate cancer, MCF-7 (0.56) in breast cancer and LOXMVI (0.95), UACC-257 (0.62) in melanoma cancer cell lines as shown in Table 1. Compounds 3b (NSC: 750138) and 3i (NSC: 750339) did not exhibit significant activity in the one dose screening of the NCI.
Table 1 Antiproliferative activity of compounds 3a and 3c–h and in selected cancer cell linesa
| Cancer panel/cell line |
GI50/μM |
|
3a
|
3c
|
3d
|
3e
|
3f
|
3g
|
3h
|
|
Data obtained from NCI's in vitro anticancer activity cells screen.
|
|
Leukemia
|
| CCRF-CEM |
18.8 |
1.54 |
1.66 |
4.15 |
0.44 |
4.01 |
4.05 |
| HL-60(TB) |
34.0 |
2.53 |
2.01 |
4.12 |
1.52 |
2.52 |
3.33 |
| K-562 |
13.1 |
2.99 |
1.51 |
2.83 |
0.36 |
2.63 |
3.03 |
| MOLT-4 |
32.9 |
2.18 |
1.77 |
3.73 |
1.03 |
3.10 |
3.75 |
| SR |
3.71 |
1.90 |
1.23 |
2.91 |
0.44 |
1.53 |
2.28 |
| RPMI-8226 |
7.50 |
0.39 |
1.34 |
1.98 |
0.29 |
2.21 |
2.56 |
|
Non-small cell lung |
| A549/ATCC |
7.96 |
3.86 |
2.04 |
4.04 |
2.80 |
2.71 |
3.34 |
| EKVX |
7.42 |
1.71 |
3.12 |
2.37 |
3.74 |
1.61 |
2.71 |
| HOP-62 |
3.62 |
4.42 |
2.88 |
2.97 |
3.01 |
2.83 |
3.12 |
| HOP-92 |
2.36 |
— |
0.28 |
2.65 |
— |
1.43 |
1.77 |
| NCI-H226 |
24.1 |
6.10 |
2.04 |
3.55 |
4.55 |
2.28 |
3.08 |
| NCI-H23 |
3.41 |
3.78 |
2.20 |
1.51 |
2.37 |
1.35 |
1.58 |
| NCI-H322M |
4.63 |
2.08 |
3.02 |
1.66 |
3.15 |
1.76 |
2.07 |
| NCI-H460 |
4.09 |
2.99 |
1.42 |
2.01 |
0.68 |
1.43 |
1.59 |
| NCI-H522 |
3.12 |
2.74 |
1.10 |
1.77 |
5.73 |
1.66 |
2.87 |
|
Colon
|
| COLO 205 |
3.50 |
3.92 |
1.77 |
2.11 |
1.92 |
1.70 |
1.70 |
| HCC-2998 |
8.27 |
3.80 |
1.96 |
1.83 |
0.48 |
1.58 |
1.77 |
| HCT-116 |
2.26 |
2.62 |
1.63 |
1.26 |
0.84 |
1.17 |
1.53 |
| HCT-15 |
4.85 |
3.73 |
1.60 |
2.10 |
1.13 |
1.38 |
2.11 |
| HT29 |
4.80 |
3.20 |
2.09 |
1.93 |
1.48 |
2.19 |
2.52 |
| KM12 |
4.99 |
2.52 |
1.32 |
1.73 |
0.57 |
1.61 |
1.69 |
| SW-620 |
4.16 |
2.78 |
— |
1.68 |
— |
1.75 |
2.09 |
|
CNS
|
| SF-268 |
4.00 |
3.58 |
2.37 |
1.64 |
1.57 |
2.36 |
1.90 |
| SF-295 |
15.2 |
1.85 |
2.24 |
1.99 |
3.15 |
2.39 |
2.43 |
| SF-539 |
2.39 |
2.71 |
1.87 |
1.63 |
1.65 |
1.36 |
1.55 |
| SNB-19 |
25.2 |
4.91 |
2.10 |
1.93 |
1.59 |
3.01 |
3.12 |
| SNB-75 |
5.49 |
3.92 |
2.68 |
1.76 |
2.35 |
1.89 |
2.39 |
| U251 |
3.00 |
2.91 |
1.66 |
1.29 |
1.10 |
1.29 |
1.68 |
|
Ovarian
|
| IGROV1 |
8.86 |
3.03 |
1.89 |
2.23 |
0.44 |
2.20 |
1.93 |
| OVCAR-3 |
2.88 |
2.47 |
2.44 |
1.83 |
1.98 |
1.75 |
1.40 |
| OVCAR-4 |
5.72 |
2.43 |
3.13 |
4.10 |
2.49 |
2.91 |
2.93 |
| OVCAR-5 |
9.33 |
7.38 |
2.64 |
1.98 |
3.79 |
1.53 |
2.46 |
| OVCAR-8 |
3.92 |
2.83 |
1.68 |
3.39 |
1.31 |
2.72 |
2.69 |
| NCI/ADR-RES |
3.36 |
2.18 |
1.89 |
1.41 |
1.57 |
1.56 |
1.58 |
| SK-OV-3 |
14.8 |
4.92 |
3.01 |
4.04 |
2.49 |
2.50 |
2.64 |
|
Renal
|
| 786-0 |
3.90 |
3.74 |
2.31 |
1.72 |
2.87 |
1.84 |
2.76 |
| A498 |
10.4 |
2.93 |
0.98 |
2.40 |
2.45 |
2.13 |
2.20 |
| ACHN |
3.26 |
3.22 |
2.09 |
1.95 |
2.85 |
1.57 |
2.21 |
| CAKI-1 |
8.33 |
1.39 |
2.31 |
1.82 |
2.50 |
1.58 |
2.06 |
| RXF 393 |
2.94 |
— |
— |
1.87 |
— |
1.11 |
1.44 |
| SN12C |
11.4 |
4.10 |
2.44 |
2.09 |
1.91 |
2.56 |
2.26 |
| TK-10 |
4.16 |
3.28 |
3.78 |
2.09 |
3.78 |
2.88 |
3.25 |
| UO-31 |
1.31 |
0.55 |
1.07 |
1.49 |
1.44 |
0.51 |
0.73 |
|
Prostate
|
| PC-3 |
19.8 |
3.71 |
2.63 |
3.80 |
2.73 |
4.32 |
4.35 |
| DU-145 |
7.21 |
2.63 |
1.90 |
1.61 |
0.65 |
2.55 |
1.63 |
|
Breast
|
| MCF7 |
2.38 |
2.76 |
1.12 |
1.44 |
0.56 |
1.14 |
1.61 |
| MDA-MB-231/ATCC |
5.22 |
3.28 |
3.08 |
4.59 |
3.06 |
2.73 |
2.99 |
| HS 578T |
2.89 |
1.85 |
— |
3.27 |
— |
1.91 |
2.01 |
| BT-549 |
3.27 |
0.89 |
1.86 |
1.64 |
2.59 |
1.45 |
1.80 |
| T-47D |
2.59 |
3.01 |
1.72 |
3.52 |
1.43 |
2.53 |
2.09 |
| MDA-MB-468 |
11.9 |
2.28 |
1.72 |
2.12 |
1.90 |
1.81 |
2.75 |
|
Melanoma
|
| LOX IMVI |
6.05 |
2.26 |
1.87 |
1.66 |
0.95 |
1.46 |
1.57 |
| MALME-3M |
1.56 |
2.36 |
3.65 |
1.57 |
6.69 |
1.81 |
2.84 |
| M14 |
9.80 |
4.48 |
2.08 |
1.82 |
3.63 |
1.53 |
2.16 |
| MDAMB-435 |
5.76 |
1.78 |
2.49 |
2.26 |
2.04 |
2.28 |
2.36 |
| SK-MEL-2 |
4.95 |
2.37 |
2.27 |
2.43 |
1.72 |
1.87 |
2.38 |
| SK-MEL-28 |
7.10 |
3.30 |
— |
1.93 |
— |
1.65 |
3.68 |
| SK-MEL-5 |
2.72 |
1.50 |
1.33 |
1.62 |
1.63 |
1.52 |
2.22 |
| UACC-62 |
5.58 |
4.86 |
1.61 |
2.05 |
2.22 |
1.88 |
1.74 |
| UACC-257 |
5.85 |
3.24 |
1.88 |
1.66 |
0.62 |
1.82 |
3.73 |
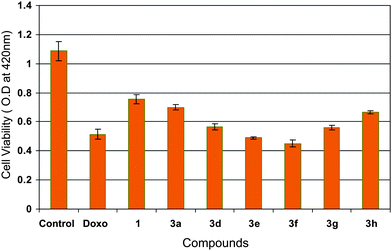
MTT assay was also carried out to identify the cytotoxic effect of some of these chalcone derivatives (3a and 3d–h) on MCF-7 cells at 4 μM concentration. Moreover, the anticancer activity of compound 1 (3,4,5-trimethoxy chalcone) and positive control COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compounddoxorubicin was examined to substantiate the anticancer activity of these chalcone derivatives (3a and 3d–h) and it is interesting to observe that these have shown a higher cytotoxicity than 1. Amongst all the derivatives the compound 3f was found to be the most significant one as shown in Fig. 2.
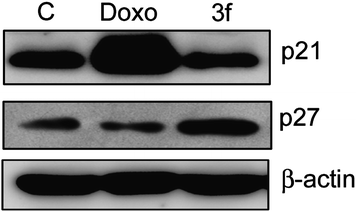
(D) Effect on protein expression of CDK inhibitors p21and p27.
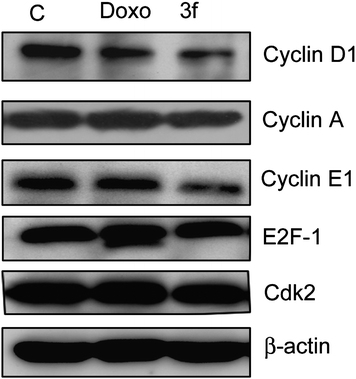
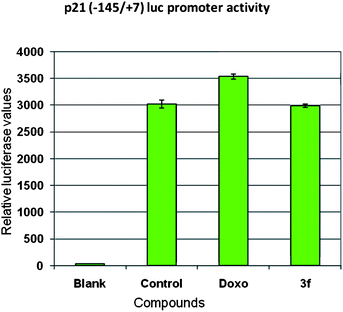
Previous studies have shown that CDK inhibitors regulate the progression of cells in G0/G1 phase and induction of CDKIs causes a blockade of G1/S transition thereby resulting in cell cycle arrest.22 The cip/kip family of CDK inhibitors, p21 and p27 impede cell cycle progression by inhibiting cyclin E-CDK2.23 Hence, we investigated the effect of compound 3f on the expression of p21 waf1/cip and p27 kip1 proteins in MCF-7 cells. Cells were treated over a period of 24 h and Western blot analysis was conducted with anti-p21 and p27 antibodies as shown in Fig. 5. Protein levels of p27 were upregulated, whereas p21 protein levels were not increased in relation to the control. In order to confirm this aspect promoter studies on p21 gene (P21 promoter −145/+8 with luciferase gene cloned downstream as reporter gene) were carried out. Ectopic expression of p21 promoter in MCF-7 cells by transfection method followed by compound treatment for 24 h resulted in no change in the levels of promoter activity in compound 3f whereas positive control COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compounddoxorubicin there was some upregulation of promoter activity (Fig. 6). This data clearly showed that compound 3f follows p27 (CDK inhibitor) dependent apoptotic pathway.
Conclusion
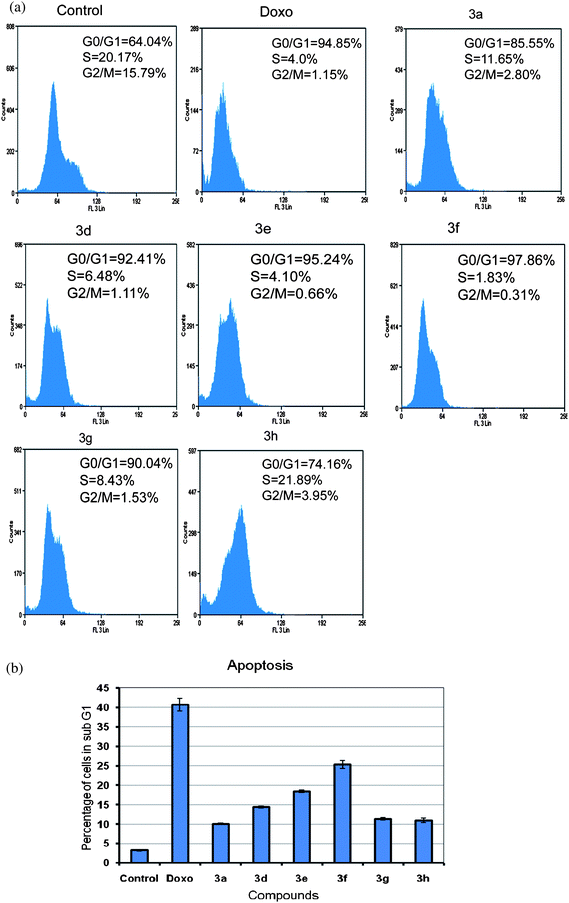
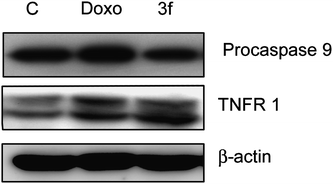
In conclusion, we have synthesized a new class of chalcones, wherein the B-ring has been replaced by an imidazo[2,1-b]pyridine/pyrimidine moiety. Compound 3f showed significant antiproliferative activity in the cell line panel assay of National Cancer Institute (NCI). From the MTT proliferation assay, it was observed that compound 3f is the most effective antiproliferative agent than the other compounds of the series in MCF-7 cells at 4 μM concentration. The FACS analysis also showed more population in sub-G1 phase indicating that these compounds have apoptotic inducing ability. From the results of the detailed biological assays, the down regulation of G1 phase cell cycle regulatory proteins such as cyclin D1, E1, and CDK2 was observed and G1/S check point associated tumor suppressor protein p27 was upregulated, thus suggesting that these proteins are responsible for cell cycle blockade at G1 phase in compound 3f. Moreover, procaspase-9 level was decreased and TNFR1 protein level was increased, thus indicating that compound 3f followed extrinsic apoptotic pathway. In this study an insight in the cell cycle regulatory role as well as apoptotic inducing ability of the compound 3f was elucidated. These studies suggest that compound 3f has the potential to be taken up for further in vivo studies, particularly against breast cancer.
Acknowledgements
We thank the National Cancer Institute, Bethesda, for in vitro anticancer assay in human cell lines. J.S.N.R., D.D.G., E.V.B and M.V.P.S are thankful to CSIR, New Delhi, for the award of research fellowships.
Notes and references
- Z. Nowakowska, Eur. J. Med. Chem., 2007, 42, 125–137 CrossRef CAS.
- T. Akihisa, H. Tokuda, D. Hasegawa, M. Ukiya, Y. Kimura and F. Enjo, J. Nat. Prod., 2006, 69, 38–42 CrossRef CAS.
- A. Modzelewska, C. Pettit, G. Achanta, N. E. Davidson, P. Huang and S. R. Khan, Bioorg. Med. Chem., 2006, 14, 3491–3495 CrossRef CAS.
- Y. L. Hsu, P. L. Kuo, W. S. Tzeng and C. C. Lin, Food Chem. Toxicol., 2006, 44, 704–713 CrossRef CAS.
- J. N. Dominguez, C. Leon, J. Rodrigues, N. Gamboa de Dominguez, J. Gut and P. J. Rosenthal, J. Med. Chem., 2005, 48, 3654–3658 CrossRef CAS.
- S. F. Nielsen, S. B. Christensen, G. Cruciani, A. Kharazmi and T. Liljefors, J. Med. Chem., 1998, 41, 4819–4832 CrossRef CAS.
- H. M. Yang, H. R. Shin, S. H. Cho, S. C. Bang, G. Y. Song and J. H. Ju, Bioorg. Med. Chem., 2007, 15, 104–111 CrossRef CAS.
- S. Cheenpracha, C. Karalai, C. Ponglimanont, S. Subhadhirasakul and S. Tewtrakul, Bioorg. Med. Chem., 2006, 14, 1710–1714 CrossRef CAS.
- L. Svetaz, A. Tapia, S. N. Lopez, R. L. F. Furlan, E. Petenatti and R. Pioli, J. Agric. Food Chem., 2004, 52, 3297–3000 CrossRef CAS.
- O. Nerya, R. Musa, S. Khatib, S. Tamir and J. Vaya, Phytochemistry, 2004, 65, 1389–1395 CrossRef CAS.
- R. de Vincenzo, G. Scambia and S. Mancuso, Anticancer Drug Des., 1995, 10, 481–490 CAS.
- S. Shibata, Stem Cells, 1994, 12, 44–52 Search PubMed.
- S. Yamamoto, E. Aizu, H. Jiang, T. Nakadate, I. Kiyoto, J. C. Wang and R. Kato, Carcinogenesis, 1991, 12, 317–323 CAS.
- C. Claude-Alain, L. Jean-Chritophe, T. Patrick, P. Christelle, H. Gerard, C. Albert-Jose and D. Jean-Luc, Anticancer Res., 2001, 21, 3949–3956.
- B. L. Wei, C. H. Teng, J. P. Wang, S. J. Won and C. N. Lin, Eur. J. Med. Chem., 2007, 42, 660–668 CrossRef CAS.
- T. Takahashi, N. Takasuka, M. Iigo, M. Baba, H. Nishino, H. Tsuda and T. Okuyama, Cancer Sci., 2004, 95, 448–453 CrossRef CAS.
- Q. C. Meng, L. Ni, K. J. Worsencroft, J. Ye, M. D. Weingarten, J. M. Simpson, J. W. Skudlarek, E. M. Marino, K. Suen, C. Kunsch, A. Souder, R. B. Howard, C. L. Sundell, M. A. Wasserman and J. A. Sikorski, J. Med. Chem., 2007, 50, 1304 CrossRef.
- D. Kumar, N. M. Kumar, K. Akamatsu, E. Kusaka, H. Harada and T. Ito, Bioorg. Med. Chem. Lett., 2010, 20, 3916–3919 CrossRef CAS.
- A. Andreani, S. Burnelli, M. Granaiola, A. Leoni, A. Locatelli, A. Morigi, M. Rambaldi, L. Varoli, N. Calonghi, C. Cappadone, G. Farruggia, M. Zini, C. Stefanelli, L. Masotti, S. R. Norman and H. S. Robert, J. Med. Chem., 2008, 51, 809–816 CrossRef CAS.
- A. Andreani, M. Granaiola, A. Leoni, A. Locatelli, R. Morigi, M. Rambaldi, V. Garaliene, W. Welsh, S. Arora, G. Farruggia and L. Masotti, J. Med. Chem., 2005, 48, 5604–5607 CrossRef CAS.
- C. Wang, Z. Li, M. Fu, T. Bouras and R. G. Pestell, Cancer Treatment and Research, 2004, 119, 217–237 Search PubMed.
- D. O. Morgan, Nature, 1995, 374, 131–314 CrossRef CAS.
- C. J. Sherr and J. M. Roberts, Genes Dev., 1999, 13, 1501–1512 CrossRef CAS.
- M. S. sheikh and Y. Huang, cell cycle, 2003, 6, 550–552 Search PubMed.
- M. O. Hengartner, Nature, 2000, 407, 770–776 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available: Spectral data of compounds 3a–i, 7a–h and 8a–h and experimental procedures for synthesis and biological evaluations. See DOI: 10.1039/c0md00116c |
|
| This journal is © The Royal Society of Chemistry 2010 |
Click here to see how this site uses Cookies. View our privacy policy here.