Feasibility of a capillary LC/ESI-Q-TOF MS method for the detection of milk allergens in an incurred model food matrix
Linda
Monaci
*ab,
Jørgen V.
Nørgaard
a and
Arjon J.
van Hengel
a
aEuropean Commission, Directorate General Joint Research Centre, Institute for Reference Materials and Measurements, Retieseweg 111, B-2440, Geel, Belgium. E-mail: linda.monaci@ispa.cnr.it; Fax: +39 080 5929374; Tel: +39 080 5929343
bCNR, Institute of Sciences of Food Production (ISPA), National Research Council,
Via Amendola 122/O, 70126, Bari, Italy
First published on 6th May 2010
Abstract
The capability of a capillary LC-Q-TOF mass spectrometer system as a qualitative tool for the identification and confirmation of milk allergens in thermally processed food was investigated. Milk powder incurred cookies were produced in-house and chosen as the model food matrix. To unequivocally assess the presence of milk allergens, samples testing positive to ELISA were analysed by a capillary LC-Q-TOF MS/MS method in order to identify specific peptides that can be used as markers for milk allergens. Results show that α-S1 casein was the protein identified with the highest score (in 100 μg g−1 milk powder incurred cookies) and its identity was confirmed by detection of the peptides m/z 692.86 and 634.34 and their specific MS/MS ions providing a fingerprint for α-S1 casein. Besides that, other milk proteins were highlighted by performing database searching. The proteomic MS-based method employing a capillary LC-Q-TOF system proved to be a valuable tool to carry out qualitative and confirmative analysis to trace contamination of milk allergens in processed food matrices.
Introduction
Milk proteins are commonly utilised as food ingredients in the manufacture of several food products. As the majority of milk proteins are considered food allergens,1–3 the occurrence of non declared milk proteins in foods may pose a risk to the health of sensitive individuals.4In order to protect allergic consumers, legislation on food labelling has been put in place in Europe and the recent Directive 2007/68/EC requires mandatory labelling of 14 allergenic food ingredients (milk included) on the relative food product.5 The hazard of hidden allergens occurring in the food chain has generated a high demand for sensitive methods tracing food allergens in different commodities to be implemented by food industries. In this regard, a number of immunoassay methods are to date available for the detection of milk allergens in different commodities for screening purposes,6–8 but owing to the possibility of false positives and cross-reactivity phenomena9 confirmative methods are urgently demanded. Notably, drawbacks of immunoassay methods might be encountered when thermally processed milk allergen-containing foods are investigated, probably due to the lack of antibody recognition of the allergen.9–13 Confirmatory methods that are independent from the type of processing the food has undergone are then required to corroborate ELISA results.
Mass spectrometry is a powerful technique currently used for protein identification and characterization.14 MS-based proteomic methods developed for food allergen detection in foods have been recently reviewed and only a few methods are presented so far in the literature.15 Hybrid systems linking a quadrupole to an orthogonal acceleration time-of-flight (TOF) mass spectrometer (Q-TOF MS) show an increased mass accuracy and resolution compared to MALDI-TOF instruments and can be exploited to search for allergen markers.14 In general, to improve confidence in protein identification, MS/MS data from individual peptides are needed. This information along with sophisticated software for database searching, lead to an accurate and unambiguous identification of the original protein. As far as mass spectrometric methods for detection of milk allergens are concerning, Weber et al.16 developed a method using a nanoLC-MS/MS system for the detection of milk allergen traces in spiked cookies. Although the method was sensitive, the authors limited their study to the analysis of cookies to which powdered milk was added after baking and they did not investigate the effect of food processing on the detection of milk allergens by MS. This aspect is instead addressed in the present work. Besides, allergen extraction represents a critical issue, whose yield depends on both the characteristics of the allergenic protein and the processing to which the allergen containing food has undergone. Due to the heating applied, strong interactions with matrix components and/or a decrease of solubility of proteins in aqueous buffers are likely to occur.13 Besides that, it has been recently shown that food processing can also have an influence on the detection of allergen sequence tags by MS.17 For all these reasons the original protein can be very difficult to be extracted, digested and eventually detected in thermally treated food matrices. In this article the method developed by Weber et al.16 was duly modified, optimized and finally applied to detect stable milk peptides in fortified home-made baked cookies. Allergen-free and allergen-containing cookies were produced in-house and fortified with milk powder either before (incurred) or after (spiked) baking at 10, 100 and 1000 μg g−1. A first evaluation was performed by means of milk allergen ELISA kits in order to investigate the effect of baking on milk allergen detection.18 The same batches of cookies were extracted with different extraction solutions in order to find the most suitable one, and successively submitted to enzymatic reaction prior to injection into the capillary LC-Q-TOF apparatus. A fast method using capillary LC-Q-TOF detection was therefore optimized and applied to identify reliable markers in milk-incurred home-made cookies. The method proved to be suitable as a confirmatory method for milk allergen detection.
Experimental
Chemicals
All chemicals used for sample preparation were provided from VWR International (West Chester, PA) and were of analytical grade. Water from a Milli-Q water system was used. RapiGest was purchased from Waters (Manchester, UK) and sequencing grade trypsin was from Merck (Darmstadt, Germany). Formic acid and HPLC grade acetonitrile were purchased from Sigma-Aldrich (St. Louis, MO). Protein purification was performed by using a 2-D clean up kit and protein quantification by using 2-D quantification kit from GE Healthcare (Uppsala, Sweden).Skimmed milk powder was purchased from Sigma-Aldrich (St. Louis, MO, USA). Veratox ELISA kits for total milk allergens were purchased from Neogen (Lansing, MI, USA).
Preparation of model food matrix
Blank and incurred cookies were prepared in-house. The following recipe was employed to be sure to avoid any trace of milk in the blank. Medium eggs: 3; wheat flour (ANCO Flour Patisserie) 190 g; oil 20 ml; sugar 90 g; distilled water (blank samples) or milk powder solution (incurred samples): 50 ml. Total weight: 500 g. Incurred cookies were prepared as following: 50 ml of milk powder solution (original milk powder solutions: 10 g of powder/l of water for 1000 μg g−1 spike and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 or 1
10 or 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 dilution for 100 and 10 μg g−1 spikes) was added to the original dough with the aim to obtain the desired concentration in the final dough before baking.
100 dilution for 100 and 10 μg g−1 spikes) was added to the original dough with the aim to obtain the desired concentration in the final dough before baking.
20 g of dough was spread into each cookie tin and baked at a fixed temperature of 180 °C for 9 min, as that was a good compromise between baking time and the browning of cookies. The final cookies were ground in liquid nitrogen to obtain a fine powder.18
Sample extraction and pre-treatment
The following buffers were prepared and tested to find the optimum for milk protein extraction: A) guanidine-HCl 1 M, dithiothreitol 20 mM, tri-sodium citrate 10 mM final pH = 6.75; B) Trizma-base 20 mM, Tween 0.1% final pH = 10.20 and C) phosphate buffer final pH = 7.4 provided with the ELISA kit. Milk powder incurred cookies at 1000 μg g−1 level were separately extracted with the three different buffers under the same conditions and submitted to ELISA and MS screening for milk protein detection. Briefly, 1 gram of milled cookies was extracted using 25 ml of buffer and shaken for 30 min. After centrifugation at 8000 g for 10 min an aliquot of 100 μl of the supernatant was withdrawn from the mixture and cleaned-up with the 2-D clean-up kit. The obtained pellet was then re-suspended in 50 μl of 0.2% RapiGest™ solution (0.2% in 50 mM ammonium bicarbonate pH 8.6) and put in a vortex for 2 min. Reduction was performed by adding 5 μl of 50 mM DTT solution and incubating the mixture in the water bath at 60 °C for 30 min. For alkylation, 10 μl of 100 mM iodoacetamide solution was added and the mixture placed in the dark for 30 min. Successively, 40 μl of trypsin was added (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50 protease: protein ratio) and the tube gently flicked to mix before incubation at 37 °C for 2 h. Ultimately, 10 μl of 500 mM HCl solution was added to the digested sample to stop the enzymatic reaction, then incubated for 15 min at room temperature and centrifuged at 16100 g for 10 min. The supernatant was carefully transferred to another microcentrifuge tube and stored at −20 °C before LC-MS analysis.
50 protease: protein ratio) and the tube gently flicked to mix before incubation at 37 °C for 2 h. Ultimately, 10 μl of 500 mM HCl solution was added to the digested sample to stop the enzymatic reaction, then incubated for 15 min at room temperature and centrifuged at 16100 g for 10 min. The supernatant was carefully transferred to another microcentrifuge tube and stored at −20 °C before LC-MS analysis.
Capillary high-performance liquid chromatography, microESI/Q-TOF tandem mass spectrometry and protein identification
Capillary LC-Q-TOF analyses were performed on a capillary flow-liquid chromatography system (CapLC, Waters-Micromass, Manchester, UK). A pre-column Acurate micro-flow splitting system (LC Packings, Sunnyvale, CA, USA) was used. The binary CapLC pump was operated at a flow rate of 10 μl min−1 and a flow rate of 1 μl min−1 was obtained after a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 split. An auxiliary pump for sample injection was operated at a flow rate of 10 μl min−1.
10 split. An auxiliary pump for sample injection was operated at a flow rate of 10 μl min−1.
Peptide mixtures from the trypsin digest were injected by the autosampler and loaded at a flow rate of 10 μl min−1 (using the auxiliary pump) on a LC Packings C18 PepMap trap column (Dionex; 300 μm id, 5 mm length, 5 μm particles size) for desalting and pre-concentration for 5 min. A post-injection switch valve was activated and a binary gradient was generated using the binary pump. Peptides were eluted from the trap column with increasing organic phase (acetonitrile), refocused and separated on a Prosphere C18 capillary column (Alltech; 150 μm id, 150 mm length, 3 μm particles size) (flow rate of 1.0 μl min−1). The eluents used were: A) 1% HCOOH in milli-Q H2O and B) 1% HCOOH in CH3CN.
The linear gradient used to achieve the separation was as follows: 0–5 min: 5% B; 5–70 min: 5–100% B; 70–120 min: 100% B; 120–140 min: 100–5% B. The column was re-equilibrated for 15 min with 5% B prior to the next injection.
Microelectrospray experiments were conducted on a quadrupole-time-of-flight mass spectrometer (Q-TOF Ultima Global, Waters, Manchester, U.K.) equipped with a nanoelectrospray Z spray source. The operating conditions of the Q-TOF mass spectrometer were as follows: capillary voltage: 3.2 kV, sample cone voltage: 100 V, source temperature: 80 °C. The instrument was operated in the positive ion mode. Time-of-flight (TOF) was performed in a continuous extraction mode. The TOF analyser was calibrated in MS/MS mode on a daily basis using Glu1-Fibrinopeptide B (Sigma-Aldrich, St Louis, MO, USA) in the range m/z 70–1500. Full scans (MS mode) were performed over the m/z range 200–1500 with a scan time of 0.9 s and inter scan time of 0.1 s.
For peptide fragmentation the instrument was operated in “MS survey scan” mode (MS/MS) with the quadrupole mass filter low mass (LM) and high mass (HM) resolution settings at 10 and 10. The cone voltage was set at 35 V. MS to MS/MS switch criteria were as follows: threshold 10 counts/s, charge states: +1, +2, +3, +4. MS/MS to MS switch occurs when TIC is falling below 5 counts/s or after a defined time of 8 s. For the analysis of incurred and blank cookies, ions originated from specific peptides were selected by the first quadrupole and the “MS/MS scan” mode was used in order to confirm the identity of the peptides. Identification of the protein was achieved by using the survey/MS/MS data.
The software package used was MassLynx-ProteinLynx Global Server v2.1 (Waters) which includes a search engine. Data directed analysis acquired in survey mode were processed by MassLynx (version 4.0, Waters) to create peak lists also containing MS/MS data to be searched against Swiss-Prot/TremBL and a reduced database of known milk allergens. Auto de novo sequencing (Auto mode) with combined database searching was also performed using the Protein Lynx Global Server with a peptide tolerance of 100 ppm and fragment mass tolerance of 0.05 Da. The enzyme entry was set for trypsin and the maximum number of missed cleavages with trypsin was set to 3. A certain number of peptide modifications were considered (e.g. alkylation C, oxidation M) for searching. Search results were validated when at least three consecutive measured fragment ions of a peptide matched theoretical b- or y-fragment ions of a known protein sequence tag. The probability for each identified protein was calculated and gave a score reported as natural logs.
Results and discussion
Protein extraction
Baking is known to affect the extractability of allergenic proteins as well as of total proteins.11–13 Different buffer composition and extraction temperatures were tested in order to find the one achieving the highest protein extraction yield. Cookies were extracted with three different buffers and shaken for 30 min at 40 °C and 60 °C, as reported in “Materials and methods”. and the final extracts submitted to MS detection after ELISA analysis. Results show that cookies extracted with buffer B at the temperature of 60 °C displayed a more intense signal and a higher number of milk peptides analysed by MS compared to the other solutions tested, therefore these conditions were used for subsequent analysis. In order to corroborate ELISA results confirming the presence of milk allergens and in order to avoid false positives, samples testing positive to ELISA were successively submitted to LC-MS analysis. The feasibility of the ESI-Q-TOF system for the detection of milk markers was then investigated.First, a standard solution of skimmed milk powder at the concentration of 10 μg g−1 was digested with trypsin and analysed, and a pool of candidate peptide markers was identified and further searched in the processed food model matrix by using a modification of the method developed by Weber et al.16
Detection of biomarker by LC-Q-TOF analysis and peptides identification
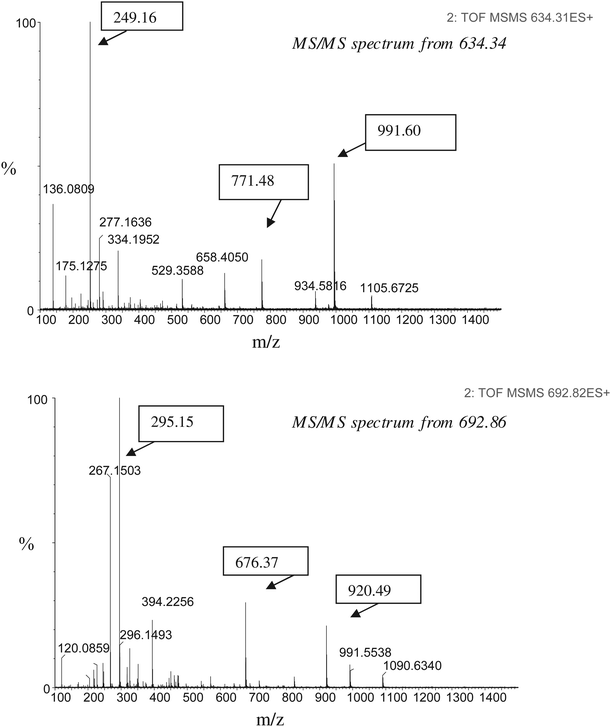
The extracted and precipitated protein fraction was submitted to tryptic digestion according to the protocol already described in the “Material and methods” and the resulting peptide mixture was enriched using a precolumn, separated by a micro LC column and detected by mass spectrometry. Fig. 1 reports an overview of chromatograms of a cookie incurred with milk powder at 100 μg g−1. The upper part shows the reconstructed ion chromatogram (RIC) filtering the total ion current (TIC) on two peptide masses m/z 692.86 (+2) (FFVAPFPEVFGK) and 634.34 (+2) (YLGYLEQLLR) eluted later in the run. These peptides were the same found in the milk powder digest previously analysed and can be considered as stable markers from caseins. By contrast, the bottom of the figure shows a typical chromatogram acquired in full scan mode. When the two peptides were fragmented they provided MS/MS spectra that mirrored those obtained (with similar relative abundances) from the analysis of the same peptides found in milk powder digests. The MS/MS most abundant fragments were m/z 295.14, 676.36, 920.48 from peptide m/z 692.86, and m/z 249.16, 771.47, 991.59 from peptide m/z 634.34 respectively, as depicted in Fig. 2. To allow the unambiguous identification of the offending allergen in the food, the MS/MS fragments of the selected peptides can be used as confirmative markers. This is particularly useful in the case of high protein samples where it is difficult to discriminate the signal from the matrix background. Guidelines on protein identification have been drawn up with the aim to drastically reduce the number of false positives. Together with increasing the stringency of the database search and the criteria for its acceptance it is likewise advisable to use more peptides for the identification of a protein and when possible include MS/MS data coming from peptide fragmentation. Fig. 3 shows a RIC filtering the specific masses (m/z 295.14 and 249.16 that provided the highest relative abundance in the spectrum) from the MS/MS total ion chromatogram of the selected peptides. Recording the specified MS/MS transitions reduced the complexity of the chromatograms, thus enabling the final confirmation of the allergen in the extract. | ||
| Fig. 1 Comparison of chromatograms relative to milk powder incurred cookie 100 μg g−1. On the top, a reconstructed ion chromatogram filtering the TIC on the peptide masses 692.86 and 634.34. At the bottom, a typical chromatogram acquired in full scan mode. | ||
 | ||
| Fig. 3 RIC filtering the specific masses. | ||
After databank searching and sequence analysis each identified peptide was assigned a “peptide score” by a probability based score algorithm, which is an indication for the reliability of the peptide identification (scale 1–100). Our results show that four peptides (m/z = 634.34, 692.86, 844.37, 865.06) were identified as potential markers of α-S1 casein in a 100 μg g−1 incurred cookies providing a good protein coverage. Some of those peptides were already pointed out by Weber et al. in a previous work.16 The probability bar of correct peptide assignment was 100% for peptide m/z 634.34, 48% for peptide 692.86, 28% for peptide m/z 844.37 and 22% for peptide m/z 865.06. As the first two peptides showed the highest score of peptide assignment and provided the best fragmentation pattern with the most intense ion fragments m/z 249.16 and m/z 295.14 in the MS/MS spectrum, they were selected as candidate markers. The software also provided insights about the degree of modification and substitution. While peptides m/z 634.34 and 692.86 did not show any modification, peptide m/z 844.37 was glycated and peptide m/z 865.06 showed both pyrrolidone carboxylic acid modification and a substitution. These two modified peptides were not found in cookies fortified after heating, which proves that the peptides that can be detected in baked cookies differ from those that can be detected in milk proteins that have not undergone the baking process. It has to be pointed out that in blank cookie extracts the two peptides marker and their MS/MS fragments were always absent. Finally by performing a database search a list of other milk protein were identified. Table 1 reports an overview of all milk proteins hits, the peptides matching and the % of protein coverage relative to incurred cookies at the levels of 100 and 1000 μg g−1.
Summarising, two peptides m/z 634.34 and 692.86 have been confirmed as markers in non processed as well as thermally processed milk spiked cookies since they correlate well with the presence of milk allergens. This is in agreement with that found by Weber et al.16 in cookies spiked with milk powder after baking. Moreover MS/MS fragments together with their parents peptides can be used as a fingerprint of the protein increasing the confidence of the analytical result. MS/MS markers can be particularly useful in the case of complex matrices under investigation where interfering compounds may contribute to a masking effect of the allergen then lowering the sensitivity of the technique. The lowest concentration of milk allergens detectable with this method corresponded to incurred cookies at 100 μg g−1, which was roughly 1 order of magnitude higher then what was detected in cookies fortified after baking. Moreover thanks to the powerful bioinformatics tools, a number of milk proteins can be identified with a high score although more of them appeared modified, probably due to the heating applied. It is worthy to remark that using the bioinformatics approach the analysis of cookies that were incurred with a higher amount of milk powder lead to a better milk protein coverage and a higher number of proteins identified. Although bioinformatic represents a valuable tool for skimming out the results, it is recommended that reliable peptides markers are used together with database searching tools to confirm the presence of the offending allergen in the samples analysed. These results confirm that this approach using a preconcentration operated by the precolumn combined with a microLC-ESI-Q TOF system is a feasible technique for the identification/confirmation of milk allergens in baked cookies.
Conclusions
A method based on capillary LC/Q-TOF was optimized and applied for the identification of potential markers in milk powder incurred home-made cookies. Different extraction solutions were tested in order to find the most suitable for MS analysis and finally a Trizma-base solution with Tween 0.1% was selected as the best for our purposes. Samples tested positive to ELISA were submitted to microLC-ESI-Q-TOF analysis and a few peptides were found to be clearly detectable both before and after baking. Two of them coming from α-S1 casein were selected as the best in terms of peptide score assignment and intensity of their fragments. The selected peptides along with their MS/MS fragments proved to be reliable markers to trace down the presence of milk allergens in baked cookies and can therefore be proposed as markers for prescreening and/or confirmatory purposes. Moreover by performing database searching, a number of milk proteins were identified with a high score.Acknowledgements
The authors acknowledge Dr Franz Ulberth for his valuable help in reviewing the manuscript and Marcel Broheé for his help in the production of cookie food material.References
- J. M. Wal, Allergy, 2001, 56, 35–38 CrossRef.
- L. Monaci, V. Tregoat, A. van Hengel and E. Anklam, Eur. Food Res. Technol., 2006, 223, 149–179 CrossRef CAS.
- M. Natale, C. Bisson, G. Monti, G. Peltran, L. P. Garoffo, S. Valentini, C. Fabris, E. Bertino, A. Coscia and A. Conti, Mol. Nutr. Food Res., 2004, 48, 363–369 CrossRef CAS.
- J. M. Wal, Ann. Allergy Asthma Immunol., 2004, 93, S2–S11 Search PubMed.
- Commission of the European Communities, Official J. Europ. Union, 2007 Search PubMed , L310/11-12.
- J. van Hengel, Anal. Bioanal. Chem., 2007, 389, 111–118 CrossRef CAS.
- P. Schubert-Ullrich, J. Rudolf, P. Ansari, B. Galler, M. Fuhrer, A. Molinelli and S. Baumgartner, Anal. Bioanal. Chem., 2009, 395, 69–81 CrossRef CAS.
- L. Monaci and A. Visconti, Tr. Food Sci. Technol., 2010 DOI:10.1016/j.tifs.2010.02.003.
- R. Rozenfeld, G. H. Docena, M. C. Aňón and C. A. Fossati, Clin. Exp. Immunol., 2002, 130, 49–58 CrossRef.
- C. Diaz-Amigo, Food Anal. Methods, 2009 DOI:10.1007/s12161-009-9095-y.
- S. L. Taylor, J. A. Nordlee, L. M. Niemann and D. M. Lambrecht, Anal. Bioanal. Chem., 2009, 395, 83–92 CrossRef CAS.
- R. Poms and E. Anklam, J. AOAC, 2004, 87, 1466–1473 CAS.
- S. K. Sathe and G. M. Sharma, Mol. Nutr. Food Res., 2009, 53, 970–978 CrossRef CAS.
- B. Domon and R. Aebersold, Science, 2006, 312, 212–217 CrossRef CAS.
- L. Monaci and A. Visconti, Trends Anal. Chem., 2009, 28, 581–591 CrossRef CAS.
- D. Weber, P. Raymond, S. Ben-Rejeb and B. Lau, J. Agric. Food Chem., 2006, 54, 1604–1610 CrossRef CAS.
- H. Chassaigne, J. V. Nørgaard and A. J. van Hengel, J. Agric. Food Chem., 2007, 55, 4461–4473 CrossRef CAS.
- L. Monaci, M. Brohèe, V. Tregoat, A. van Hengel, Food Chem., submitted Search PubMed.
| This journal is © The Royal Society of Chemistry 2010 |