Salt-induced release of lipase from polyelectrolyte complex micelles
Saskia
Lindhoud
*a,
Renko
de Vries
a,
Ralf
Schweins
c,
Martien A.
Cohen Stuart
a and
Willem
Norde
ab
aLaboratory of Physical Chemistry and Colloid Science, Wageningen University, Dreijenplein 6, 6703 HB Wageningen, The Netherlands. E-mail: saskia.lindhoud@wur.nl
bDepartment of Biomedical Engineering, University Medical Center Groningen and University of Groningen, A. Deusinglaan 1, 9713 AV Groningen, The Netherlands
cLSS group, Institut Laue-Langevin, 6 rue Jules Horowitz BP 156, F-38042 Grenoble Cedex 9, France. E-mail: saskia.lindhoud@wur.nl
First published on 23rd October 2008
Abstract
With the aim to gain insight into the possible applicability of protein-filled polyelectrolyte complex micelles under physiological salt conditions, we studied the behavior of these micelles as a function of salt concentration. The micelles form by electrostatically driven co-assembly from strong cationic block copolymers poly(2-methyl vinyl pyridinium)41-block- poly(ethylene oxide)205, weak anionic homopolymers poly(acrylic acid)139, and negatively charged lipase molecules. The formation and disintegration of these micelles were studied with dynamic light scattering (DLS), by means of composition and salt titrations, respectively. The latter measurements revealed differences between disintegration of lipase-filled and normal polyelectrolyte complex micelles. These data, together with small angle neutron scattering (SANS) measurements provide indications that lipase is gradually released with increasing salt concentration. From the SANS data a linear relation between the intensity at q = 0 and the volume of the cores of the micelles at different salt concentrations was derived, indicating a loss of volume of the micelles due to the release of lipase molecules. It was estimated that beyond 0.12 M NaCl all lipase molecules are released.
1 Introduction
Functional ingredients, e.g., proteins may be encapsulated to enhance their stability or to prevent external attack. This is beneficial in various applications. In industry and food technology1,2 encapsulation might provide ways to protect enzymes against loss of activity or could even enhance the activity. Stabilization of enzymes might significantly increase the shelf-life of bioactive formulations. Moreover, it might enable recycling of the enzymes on industrial scale. In pharmaceutics one could think of controlled delivery and release of therapeutic proteins.3–6Several difficulties have to be overcome before proteins can usefully be incorporated in actual applications. The biological functioning of a protein is directly related to its molecular three-dimensional structure. This structure can very easily be destabilized, either physically, e.g., by changing the environmental conditions,7,8 or (bio)chemically, e.g., by proteolytic attack. Obtaining a self-assembled structure in which protein molecules can be safely stored is a complicated challenge. The main problem to be solved is how to design the architecture and structure of this protein containing particle. Difficulties arise from the requirement that the protein to be incorporated should not suffer from any irreversible damage either during the packaging procedure or due to interactions with the packaging material.
Because proteins have a structure that is very sensitive to changes in the environment, e.g., in temperature, polarity of the medium, pH, etc., use of organic reactions during the incorporation procedure may be hazardous. Alternatively, nearly all proteins have charges, therefore one could use oppositely charged polyelectrolytes as package material.9Protein–polyelectrolyte mixtures are known to form dense liquid-like phases, called complex coacervates.10 One advantage of using polyelectrolytes is that one can work in an aqueous environment and avoid the use of organic solvents that may damage the protein.
Depending on the charge ratio between the polyelectrolyte and the protein, either charged soluble complexes are formed or, at stoichiometric charge ratio, precipitation or complex coacervation occurs.11 Well-defined structures with specific distances between the protein molecules have been obtained by protein–polyelectrolyte complexation.12–15 However, not all polyelectrolyte–protein combinations will result in well-defined structures.
An extensively studied procedure for making encapsulating structures using polyelectrolytes is the layer-by-layer technique. Encapsulation of functional charged molecules can be achieved by exposing them in an alternating fashion to a solution containing polyanions and then to a solution containing polycations or vice versa. In this way layers of oppositely charged polymers are formed around the molecule. Since proteins are charged they can be encapsulated by this technique16 or they may be incorporated within these layers.17 Because of the electrostatic nature of the cohesive interactions, changes in ionic strength and/or pH can induce the disintegration of the structure. It is also possible to incorporate permeability in the layer-by-layer shell, thereby enabling protein molecules to leave the capsule.18
Depending on the type of application, the size of a structure is important. In food systems particles may have micrometre dimensions; for medical applications sizes from 10–100 nm are desired, since larger structures are known to be caught by the immune system (e.g., phagocytes) and accumulate in the liver and the spleen.19 For the nanopackaging and controlled delivery of hydrophobic drugs, promising results have been obtained20–22 by making use of micelles. Kataoka and co-workers, synthesized amphiphilic block copolymers having a folate group attached to the water soluble block. These folate-conjugated micelles were used to encapsulate hydrophobic anti-cancer drugs, by conjugating them to the hydrophobic part of the block copolymer linkers that are sensitive to acidic pH. The micelles thus obtained, showed lower in vivo toxicity and higher anti-tumor activity than conventional anti-cancer formulations.23–25
The use of micelles seems to be very promising for pharmaceutical applications, because micelles are nanostructures with a well-defined architecture. The core of micelles consisting of amphiphilic block copolymers is relatively hydrophobic which could have a damaging effect on the native structure of the protein. A gentle way to incorporate proteins in micellar structures is the use of polyelectrolyte complex micelles. These micelles consist of a diblock copolymer (having a charged and a neutral hydrophilic block) and an oppositely charged homopolymer or a diblock copolymer.26–28 At a stoichiometric charge ratio, micellar structures with a polyelectrolyte complex core are formed. These structures are stabilized by a corona made of the hydrophilic block of the block copolymers. Since proteins are charged at pHs other than their isoelectric point it is possible to incorporate them in the complex core by mixing them with an oppositely charged diblock copolymer.29,30
In our approach a protein solution is added to a solution containing like-charged homopolymers. This solution is subsequently mixed with a solution containing oppositely charged diblock copolymer.31 In this way micelles are obtained of which the stability against disintegration by salt and the number of protein molecules in the core can be controlled by changing the homopolymer-to-protein ratio (see Fig. 1 for a schematic illustration). It was found that stable micelles are formed when the homopolymer is in excess. This can be explained by the much higher charge density on the polyelectrolytes compared to that on the protein.
 | ||
| Fig. 1 A schematic illustration of the formation of a polyelectrolyte complex micelle with proteins. The aggregation number of the micelles is estimated from small angle neutron scattering data. | ||
In line with this, one could imagine that by reducing the strength of the electrostatic attraction, proteins are the first molecules to leave the micelle. It has been shown that it is possible to weaken electrostatic interactions between the proteins and oppositely charged polyelectrolytes by salt,32 pH,33,34 external electric field,35 and composition of the system. Hence, this would provide various ways to release protein molecules from their capsule.
To test whether proteins are indeed released from these polyelectrolyte complex micelles, we incorporated the (at pH 7) negatively charged lipase in micelles made of negatively charged homopolymer poly(acrylic acid)139 (PAA139) and the positively charged diblock copolymer poly(2-methylvinyl pyridinium)41-block-poly(ethylene oxide)205 (P2MVP41-PEO205). Lipase is a protein with a molar mass of 27![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 g mol−1. At pH 7 it has a net charge of −8e.36 Clearly, the number of charges on the protein is fairly low, especially compared to the number of charges on the homopolymer (−139e) and on the charged block of the diblock copolymer (+41e). In this study we try to make use of this difference in charge density for the controlled release of the proteins. The addition of salt causes screening of the charges on the (bio)polymers. Since lipase is a very big molecule compared to the polyelectrolytes and has only 8 charges and, hence, a low charge density, one would expect these molecules to be released from the micelles first because the electrostatic attraction between lipase and P2MVP41–PEO205 is much weaker than that between P2MVP41–PEO205 and PAA139.
000 g mol−1. At pH 7 it has a net charge of −8e.36 Clearly, the number of charges on the protein is fairly low, especially compared to the number of charges on the homopolymer (−139e) and on the charged block of the diblock copolymer (+41e). In this study we try to make use of this difference in charge density for the controlled release of the proteins. The addition of salt causes screening of the charges on the (bio)polymers. Since lipase is a very big molecule compared to the polyelectrolytes and has only 8 charges and, hence, a low charge density, one would expect these molecules to be released from the micelles first because the electrostatic attraction between lipase and P2MVP41–PEO205 is much weaker than that between P2MVP41–PEO205 and PAA139.
The topic of disintegration of polyelectrolyte complexes by addition of salt has already been addressed by other groups. In most cases the polyelectrolyte complex systems were not electroneutral, e.g., polyelectrolyte multilayers37,38 or non-stoichiometric complexes in solutions.39 The latter investigation by Zinchenko et al. was performed on systems containing highly aggregated complexes of nearly stoichiometric charge ratio and free excess polyelectrolyte. They describe in detail the disintegration process of the complexes and report a shift in the equilibrium between large aggregates and smaller soluble complexes towards the latter with increasing salt concentration.
Harada and Kataoka, have shown that it is possible to “unpack” proteins from polyelectrolyte complex micelles by the addition of salt. They made complexes with lysozyme and oppositely charged diblock copolymers and studied the enzymatic activity during and after encapsulation.32 Information about the disintegration of the micelles as a function of salt is very important for possible future applications. In our study lipase was chosen, because it is a commonly used industrial enzyme. This enzyme is e.g., used in detergent formulations. We are interested in the enzymatic activity of lipase in the micelles. To study the lipase activity, before, during and after nanopackaging, the salt concentration at which all lipase molecules are released has to be known. Therefore we investigate the behavior of the micelles as function of salt.
The techniques used to study the disintegration of the micelles as a function of salt were dynamic light scattering (DLS) and small angle neutron scattering (SANS). Two types of DLS-titrations were performed: composition and salt titrations. During a composition titration like charged molecules are titrated to oppositely charged molecules, and information is obtained on the hydrodynamic radius and the intensity as function of the composition of the micelles. From these titrations one can determine the preferred micellar composition (F−micelle). Solutions with this composition are used for the salt titration and the neutron-scattering experiments.
2 Experimental
Materials
The homopolymer used was poly(acrylic acid)139 (Polymer Source Inc., Canada), Mw = 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 g mol−1, PDI = 1.15, when fully charged this anion contains 139 charges. The lipase used in this study was Lipolase™, derived from the fungus Humicola Lanuginosa, and was a gift from Novozymes (Bagsvaerd, Denmark). The positively charged diblock copolymer poly(2-methylvinyl pyridinium iodide)41-block-poly(ethylene oxide)205, obtained by quarternization of poly(2-vinyl pyridinium)41-block-poly(ethylene oxide)205,) PDI = 1.05, purchased from Polymer Source Inc., Canada, and using the following protocol. In a typical reaction, 1 g of diblock copolymer containing P2VP was dissolved in 35 mL DMF. Iodomethane (3 mL) was added, and the reaction was stirred under nitrogen flux for 48 h at 60 °C. Ether (110 mL) was added to precipitate the polymer (added until no more precipitation occured). The precipitate was filtered and washed with ether (5 × 10 mL) to yield a light yellow powder. (After each washing step the polymeric product was less yellow, less sticky, and more powdery.) Subsequently, the polymer was placed in an oven at 50 °C to dry overnight. The mass after quarternization was 19
000 g mol−1, PDI = 1.15, when fully charged this anion contains 139 charges. The lipase used in this study was Lipolase™, derived from the fungus Humicola Lanuginosa, and was a gift from Novozymes (Bagsvaerd, Denmark). The positively charged diblock copolymer poly(2-methylvinyl pyridinium iodide)41-block-poly(ethylene oxide)205, obtained by quarternization of poly(2-vinyl pyridinium)41-block-poly(ethylene oxide)205,) PDI = 1.05, purchased from Polymer Source Inc., Canada, and using the following protocol. In a typical reaction, 1 g of diblock copolymer containing P2VP was dissolved in 35 mL DMF. Iodomethane (3 mL) was added, and the reaction was stirred under nitrogen flux for 48 h at 60 °C. Ether (110 mL) was added to precipitate the polymer (added until no more precipitation occured). The precipitate was filtered and washed with ether (5 × 10 mL) to yield a light yellow powder. (After each washing step the polymeric product was less yellow, less sticky, and more powdery.) Subsequently, the polymer was placed in an oven at 50 °C to dry overnight. The mass after quarternization was 19![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 g mol−1.
100 g mol−1.
Dynamic light scattering titrations
For the DLS-titrations we used an ALV5000 multiple tau digital correlator and an argon ion laser with a wavelength of 514.5 nm. All measurements were performed at a scattering angle of 90°. The temperature was kept constant at 25 °C by means of a Haake C35 thermostat, providing an accuracy of ±0.1 °C. The titrations were performed using a Schott-Geräte computer-controlled titration set-up to control the addition of titrant, the cell stirring, and delay times. The effective average hydrodynamic radii of the complexes were determined by analyzing the autocorrelation function using the methods of cumulants and using the Stokes–Einstein equation for spherical particles. From previous work we found that spherical particles are obtained when micelles contain more homopolymer molecules than protein molecules, although a certain polydispersity is expected.31In the composition titrations a mixture of negatively charged PAA and lipase were titrated to the positively charged P2MVP41-PEO205. The pH was kept constant by using a 3.5 mM sodium phosphate buffer of pH 7. The scattered intensity and hydrodynamic radius are typically presented as a function of the composition F−:
 | (1) |
For the salt titrations 9 mL solution (in 3.5 mM phosphate buffer, pH = 7) of micelles containing lipase and without lipase were prepared. The starting concentration of both solutions was 1 g L−1. Micelles with lipase were prepared in the following way: first lipase and PAA139 were mixed and then P2MVP41-PEO205 was added. This solution was kept in a refrigerator overnight. To this solution a 5 M NaCl solution was titrated in small steps of 20 µL. After every addition a delay time of 150 s (60 stirring, 90 s of waiting) was used.
Small angle neutron scattering
Small angle neutron scattering experiments were performed at the Institut Max von Laue-Paul Langevin (ILL), Grenoble, France, on the D22 beam line. Measurements were performed at two detector distances, 4 and 17.6 m, resulting in a q-range of 0.0023–0.137 Å−1. The wavelength was 8 Å. The spectra were treated according to ILL standard procedures, and put on an absolute scale using the known absolute scattering intensity of H2O.Samples were prepared from a stock solution of micelles at their preferred micellar composition F−micelle, in a 3.5 mM phosphate buffer in D2O, pH = 7 (pD = 6.56). After 6 h of equilibration of the micellar stock solution, the NaCl concentrations were set using a concentrated solution of NaCl in D2O, to give final NaCl concentrations in the samples of 0, 0.1, 0.3, 0.5 and 0.7 M, respectively, and a micelle concentration of 8 g l−1 (polymers and proteins combined).
For particles of volume Vpart and number density npart, the absolute scattering intensity, I(q), (cm−1) can be written as:
| I(q) = npartΔϱ2Vpart2P(q)S(q) | (2) |
From previous work31 we know that at low protein content the micelles are spherical, and rather monodisperse, hence we expect scattering curves to satisfy the P(q) for a sphere with radius R:
 | (3) |
At higher values of q, the internal structure of the polyelectrolyte core contributes to the form factor, and a simple sphere model no longer fits. Therefore, equation 3 is only used to fit the data up to q ≤ 0.024 Å−1 (which is sufficient to obtain a reliable estimate of the micellar core radius Rc). At higher salt concentrations, the scattering intensity drops rapidly. As a consequence, the statistics of the low-q data for the higher salt concentrations are not sufficient to allow for a form factor fit, even though we can still use the data for a rough estimate of the scattering intensity for q → 0.
The PEO corona of the polyelectrolyte core micelles is rather dilute (see Results and discussion) and may be expected to hardly contribute to the scattered intensity. Therefore, the scattering intensity is to a good approximation due to the polyelectrolyte complex core of the micelles, and the radius Rcore from the form factor fit is the radius of the micellar core. If we know both the scattering at zero angle and the core radius Rcore, we can estimate the degree of solvent-swelling of the micellar cores. The zero angle scattering I(0) is:
| I(0) = npart(Δϱ)2Vpart2 | (4) |
For a given global volume fraction Φ of swollen core material in the system, the number density of the micelles is:
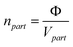 | (5) |
However, from the system composition we can only calculate the global volume fraction Φ0 of unswollen core material. The two are related by the local volume fraction φ of dry core material inside a swollen micelle,
| Φ0 = φΦ | (6) |
Swelling and deswelling also changes the contrast of the micellar cores, according to:
| Δϱ = φΔϱ0 | (7) |
| I(0) = Φ0φ(Δϱ0)2Vpart | (8) |
 , this equation can be used to obtain a rough estimate for φ, the total volume fraction of macromolecules in the micellar cores.
, this equation can be used to obtain a rough estimate for φ, the total volume fraction of macromolecules in the micellar cores.
3 Results and discussion
Dynamic light scattering composition titration
Previously it has been shown that DLS-titration is a useful tool to obtain information about polyelectrolyte complex micelle formation.40,41,31 During a composition titration like-charged molecules are titrated to molecules of opposite charge. This allows to simultaneously study the hydrodynamic radius and light-scattering intensity as function of the composition F− (equation 1). By changing the composition upon addition of titrant, the intensity increases until a maximum is found. The initial intensity increase is ascribed to the formation of soluble complexes, of which it is not possible to accurately determine the hydrodynamic radius. After reaching a certain composition micelles are formed. The maximum in the intensity corresponds to the preferred micellar composition F−micelle. Typically this maximum is at the electroneutral composition F− = 0.5 (equation 1).40 Maxima deviating from F− = 0.5 have been found in systems where polymers with a pH dependent charge or proteins are used.31The results of the DLS composition titration are shown in Fig. 2. In these measurements the starting solution contained P2MVP41-PEO205 to which a 9:1 mixture (molar ratio) of PAA139 and lipase was titrated. Fig. 1 shows a schematic representation of the formation of these micelles. The intensity (Fig. 2a) and hydrodynamic radius (Fig. 2b) of the micelles as function of the composition are plotted. According to equation 1 electroneutral particles are expected at F−micelle = 0.5. The hydrodynamic radius of the micelles consisting of PAA139, lipase and P2MVP41-PEO205 at F− = F−micelle is about 25 nm. From F−micelle the mixing ratio of the different components can be derived. Solutions with this optimal composition (for micelles with no extra salt added) are used in the salt titrations and the neutron scattering experiments. The intensity as function of the complex formation is asymmetric along F−. Similar asymmetry has been found in other systems,42,31 but there are also examples of symmetric intensity distributions around F− = 0.5.40,41
 | ||
| Fig. 2 DLS composition titration graphs. (a) Light-scattering intensity and (b) hydrodynamic radius as a function of the composition of solutions prepared with no salt (●) and 50 mM NaCl (○). Typically a 10 g L−1 solution of of PAA139 and lipase with a molar ratio of 9:1 titrated to a 1 g L−1 solution of P2MVP41-PEO205, pH = 7. | ||
The polyelectrolyte complex formation was studied at two different ionic strengths, 3.5 mM phosphate buffer with no extra salt added and 3.5 mM phosphate buffer with 50 mM NaCl, respectively. The light scattering intensity of the system without extra addition of salt is higher than the intensity of the system containing 50 mM NaCl, for all values of F−. The hydrodynamic radius is also larger. Anticipating the discussion of the DLSsalt titrations (Fig. 3) and the SANS data (Fig. 4), it is mentioned that, at 50 mM NaCl less protein molecules are incorporated, resulting in a lower scattered intensity and smaller hydrodynamic radius.
 | ||
| Fig. 3 DLS salt titration graphs. (a) Light-scattering intensity and (b) hydrodynamic radii of light-scattering titrations. Micelles with lipase (○) and micelles without lipase (×). The starting solution contained 9 mL of 1 g L−1 micelles, the titrant was 5 M NaCl, pH = 7, titration steps were 20 µL. | ||
 | ||
| Fig. 4 Small angle neutron scattering as a function of salt concentration: ● 0 M NaCl, ○ 0.1 M NaCl, ■ 0.3 M NaCl, ◇ 0.5 M NaCl and ▲ 0.7 M NaCl. The micellar concentration was 8 g L−1. The dashed lines of 0, 0.1 and 0.3 M NaCl are form factor fits for monodisperse spheres. The dashed lines of 0.5 and 0.7 indicate the estimated intensity at q = 0. | ||
There may be several reasons for the asymmetry of the intensity as function of the composition. Apparently, there is a difference in the formation of the micelles (F− < 0.5) and disintegration of the micelles (F− > 0.5). In the case of the complex formation between a homopolymer and diblock copolymer similar asymmetry was found when at least one of the components was a strong polyelectrolyte (having a charge which is pH independent). Symmetric curves have been found when both polyelectrolytes had a pH dependent charge. The titrations performed at 50 mM NaCl indeed suggest that the micelles more readily disintegrate (F− > 0.5), indicating that the differences in disintegration between the two systems are electrostatic in nature. At higher salt concentration electrostatic forces are weaker, allowing for faster rearrangements.
Another explanation for the asymmetry of Fig. 2a could be that the system itself is not symmetrical, because it contains three components. However, in our previous work we have seen that the intensity as function of the composition for a system containing only (positively charged) lysozyme and (negatively charged) PAA42PAAm417 was asymmetric and became more symmetric when the protein molecules were partly replaced by the positively charged homopolymer PDMAEMA150, yielding a symmetric curve for three component systems where homopolymer was in excess of the protein.31
The hydrodynamic radii as a function of the composition (Fig. 2b) show a minimum at F−micelle. At this composition electroneutral particles are obtained. The somewhat larger radii together with the lower light scattering intensity at compositions other than F−micelle indicate, larger less dense structures. Under these conditions the micelles (or soluble complexes) are expanded due to repulsion between the charged groups within these structures.
Dynamic light scattering titrations with salt
To study the disintegration of the micelles by weakening of electrostatic interactions a DLS-titration with salt was performed. During this measurement a concentrated NaCl (5 M) solution was titrated to a solution with composition F−micelle. In Fig. 3a and b the results of the salt titration are shown. In this figure, data for both the micelles with and without lipase are plotted. The composition of the normal micelles is the F−micelle (F− = 0.5) resulting from a titration with only PAA139 and P2MVP41-PEO205. Hence, these micelles are different structures to the lipase-filled micelles. At the same concentration of micelles and with no NaCl added, the light scattering intensity of the micelles with lipase is much higher than the light scattering intensity of the normal micelles.The scattering intensity is found to decrease with increasing salt concentration. This could be for several reasons. First, it may be that the addition of salt causes a shift in the equilibrium between small soluble complexes (consisting of a few polyelectrolytes) and micelles. Such a shift in equilibrium towards smaller soluble complexes was found to occur during the disintegration of non-stoichiometric polyelectrolyte complexes.39 Second, the number of polymer or protein molecules in the micelle i.e., aggregation number of the micelles, may be affected by salt. This will result in a population of micelles with a salt dependent aggregation number and free polyelectrolytes or proteins. This is most likely to be the case for micelles with proteins. The less densely charged protein molecules might be expelled from the micelles resulting in a different aggregation number. It is also possible that all these processes take place simultaneously.
Comparing the micelles with and without proteins reveals a few differences. The decrease in light scattering intensity with increasing NaCl concentration is more pronounced for micelles with lipase than for the micelles without lipase. The effect of salt on the hydrodynamic radius (Fig. 3b) is different for the micelles with and without lipase as well. For the micelles without protein the radius slightly increases between 0.2 and 0.5 M NaCl. This is consistent with swelling of the micelles because of screening of the charges. The hydrodynamic radius of the micelles with lipase decreases slightly upon the addition of salt. Above 0.5 M NaCl all the micelles, both with and without lipase, are disintegrated.
Since both systems show a decrease in light scattering intensity and a hydrodynamic radius that is approximately constant up to a salt concentration of 0.2 M NaCl, the mass of the micelles changes upon the addition of salt. The decrease in intensity of the micelles without lipase may result from an aggregation number which is dependent on the salt concentration. The decrease in intensity of the micelles with lipase is likely to be caused by either proteins or polyelectrolytes being expelled from the micelles. Previous work showed that complexes of lysozyme and PAA42-PAAm417 disintegrated at a salt concentration of 0.12 M NaCl.31 Harada and Kataoka used a salt concentration of 0.15 M NaCl to unpack lysozyme from polyelectrolyte complex micelles.32 So it could be that 0.12–0.15 M NaCl is a typical salt strength at which protein–polyelectrolyte complexes disintegrate.
One might expect that, when all proteins are released the micelles with and without lipase are similar, implying that both the intensity and hydrodynamic radius are the same at NaCl concentrations beyond 0.2 M. According to Fig. 3 this is clearly not the case and the dissimilarity may be explained as follows. The micelles without lipase are electroneutral structures. Lipase-release from the lipase-filled micelles will induce the formation of slightly charged micelles. However, since the charge density of a lipase molecule is much lower than the charge density of the poly(acrylic acid) the resulting micellar charge is negligibly small. The deviating characteristics of these micelles is most likely due to the fact that lipase has an influence on the packing of the micelles and, hence, on their internal structure. Lipase is rather big compared to the polyelectrolytes. By addition of salt, the electrostatic interaction between the lipase and positively charged diblock copolymer can easily be weakened, inducing the release of the lipase. The departing lipase molecules may leave behind some holes in the micellar core. It may be that rearrangement of the macromolecules in the micellar core is slow. Of course the addition of salt will enhance the rearrangement kinetics, but in Fig. 2 it is seen that their is only a small difference in terms of disintegration of the micelles between no salt and 50 mM NaCl. The normal micelles and micelles which have released all their lipase are two different structures, at least within the time frame of a salt titration measurement.
Small angle neutron scattering
The information, notably RH, derived from dynamic light scattering reflects the whole micelle, i.e., the core and corona. Because of our interest in what is happening during the disintegration of the micelles, it would be desired to get more information on the fate of the core of the micelle. To study the structure of the core of the micelles we used SANS. This technique provides information about the internal structure of the micelles, since it probes at a smaller scale than light scattering. Differences in contrast between the particles and the solvent are measured. At low scattering vectors, q, information about the size of the particles is obtained. At high q information about the internal structure of the micelles is derived. For micelles made of charged diblock copolymers and oppositely charged surfactant micelles a peak was found at high q, giving an insight into the distances between the surfactant micelles within the complexes.43 Similar peaks were also found in systems consisting of a protein and oppositely charged polyelectrolyte.12–14 In the following section, first the intensity at q = 0 will be discussed. This is followed by the fitting of the curves and the scattering at high q. In the next section a detailed analysis of the structure of the micelles and the release of lipase is presented.Fig. 4 shows that the intensity of the neutron scattering at q = 0 strongly decreases as function of the salt concentration. By either fitting (0, 0.1 and 0.3 M NaCl) or taking the intensity of the lowest accurate q-value (0.5 and 0.7 M NaCl) the intensity at q = 0 can be estimated. This intensity can directly be compared to the light scattering titration as function of salt. In Fig. 5 it is shown that the changes in light and neutron scattering intensity as a function of salt concentration are proportional; at the lower salt concentrations a decrease of the intensity as a function of salt concentration is found. From 0.5 to 0.9 M NaCl with both scattering techniques the intensity levels off to reach a constant value.
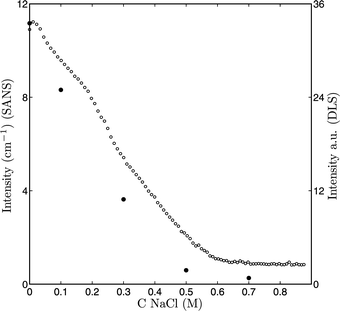 | ||
| Fig. 5 ● I(0) (SANS) and ○ I (light scattering) as function of the salt concentration. | ||
Knowledge of I(0) is also useful for another reason. In equation 8 it is shown that the intensity scales with the volume. Knowing the radius, which can be determined by fitting of the scattering curves, and calculation of the contrast (Δϱ0) and density (see Table 2), one can determine the mass and aggregation number of the particles. Since the only difference between the curves in Fig. 4 is the salt concentration, this figure directly suggests that the volume of the particles varies with the salt concentration. The signal-to-noise ratio for the scattering curves of the 0, 0.1 and 0.3 M NaCl samples allow for a detailed analysis, but the signal-to-noise ratio of the 0.5 and 0.7 M NaCl samples is not good enough for this purpose. The data quality could have been improved by increasing the concentration of the micelles or use a longer measuring time. However, from the DLS-titration with salt it became clear that at 0.5 and 0.7 M NaCl one is most likely dealing with soluble complexes and individual micellar building blocks rather than with micelles. A very detailed analysis of the curves obtained at these conditions is therefore less relevant for our understanding of the enzyme-filled micelles.
The scattering curves of 0, 0.1 and 0.3 M NaCl allow the characterization of the structure of the micelles. These scattering curves were fitted to the form factor for homogeneous spheres (equation 3). The dashed lines in Fig. 4 are the fits to the data (the dashed lines of the 0.5 and 0.7 M NaCl samples are simply the intensity at the lowest q-value measured). Scattering curves at 0, 0.1 and 0.3 M NaCl could only be fitted to q ≤ 0.024 Å−1. At higher q the internal structure of the micelles starts contributing to the scattering. Therefore this form factor fit could only be used to obtain an approximate core radius. Generalized indirect Fourier transformation (GIFT) is a model-free method to analyze small angle scattering data, which could be used over the total q-range. This method gave the same Rcore,44,45 indicating validity of our approach. Table 1 summarizes an overview of the radii determined by light- and neutron scattering. In the next section we will come back to these results.
As mentioned before, information about the internal structure of the micelles is extracted from the remaining part of the scattering curve at q-values > 0.024 Å−1. The five scattering curves cross each other around q = 0.05 Å−1 and the order of the scattering curves inverts at this point. Iso-scattering was also found during heat-induced gelation of globular proteins and was interpreted in terms of microphase separation between native proteins and aggregates.46 Such a point may be found when there are two distinct populations of different species present. Another reason for the existence of an iso-scattering point is a change in solvent quality for one or more components. For instance, it is known that the solubility of PEO decreases as function of salt concentration.47 Similar iso-scattering points are therefore also found in PEO-PPO-PEO micellar systems at different salt concentrations.48 Both explanations could be valid for our system. On one hand, two populations of species could be present in the system e.g. micelles versus single components. On the other hand, the lower solubility of PEO with increasing salt concentration could as well give rise to the iso-scattering point.
At higher q-values we expected to find a correlation peak due to scattering by the proteins.12–14,43 Correlation peaks are found when very distinct distances are present within a structure, e.g., protein molecules. In our data such peaks are not found, indicating that there is no well-defined characteristic distance between the protein molecules. Anticipating the discussion in the next section, the absence of such a characteristic distance and, hence, ordered packing of the protein molecules in the micellar core, could be due to a too small amount of protein in the micelles.
Lipase release from the micelles
In the Experimental section it has been described how to derive characteristics of the micelles from the neutron-scattering data. From the fitting of the SANS curves the radii of the cores (see Table 1) and I(0) have been determined. Using equation 8 and the values from Table 2, the total volume fraction of macromolecules in the core (φ) can be established. If we assume that all the protein molecules are encapsulated in the micelles when no salt is added, the scattering length density of the core material is Δϱ0 = 1.73 × 1010 cm−2 and the global volume fraction Φ0 of unswollen core material is 2.8 × 10−3. This results in φ ≈ 0.17 implying that 83% of the core of the micelles consist of water. This gives a molar mass of the core of approximately 1.2 × 106 g mol−1. Since the ratio between the various components is known, we can estimate the number of different molecules in the micelles: approximately 140 diblock copolymers, 40 homopolymers and 5 proteins. The area per block copolymer at the core–corona interface ( ) is about 17 nm2. The volume of five lipase molecules is about 2% of the core volume, and about 11% of its dry mass.
) is about 17 nm2. The volume of five lipase molecules is about 2% of the core volume, and about 11% of its dry mass.
It has been stated in the Experimental section that the PEO corona is rather dilute and is expected to hardly contribute to the neutron-scattering. Indeed, there is a clear difference between the radii of the micelles measured with light-and neutron-scattering, see Table 1. The hydrodynamic radius (RH) is much larger than the fitted SANS radius (Rcore). To estimate the corona contribution (r) to the neutron scattering we use the following expression:
 | (9) |
In this expression the scattering length densities of all the components and the solvent, as well as the densities of the components are needed in this equation (see Table 2 for these values). Furthermore, the volumes of (what is assumed to be) the core and the corona are required. The volume of the corona (Vcorona) was calculated in the following way: the core volume (Vcore =  ) was subtracted from the hydrodynamic volume (VH =
) was subtracted from the hydrodynamic volume (VH =  ). In equation 9φcorona is the volume fraction of PEO in the corona and Δϱ0,corona is the scattering length density of the corona. The corona contribution r to the scattering was less than 0.1% indicating that essentially the core radius is measured.
). In equation 9φcorona is the volume fraction of PEO in the corona and Δϱ0,corona is the scattering length density of the corona. The corona contribution r to the scattering was less than 0.1% indicating that essentially the core radius is measured.
As discussed above, the micelles with and without protein behave differently during the light scatteringsalt titrations (Fig. 3). For both systems the intensity decreases upon increasing salt, but for the lipase-filled micelles the influence of salt is more pronounced. Because the two systems are both electroneutral, the different sensitivities for salt mean that the systems are not the same. For the lipase-filled-micelles the charge of the proteins is needed to obtain electroneutrality, whereas the normal micelles at stoichiometric charge ratio consist of polyelectrolytes only. Also the hydrodynamic radii measured during light scattering titrations of micelles with and without lipase are slightly different. This could mean that lipase molecules are released, yielding smaller micelles because of the volume loss due to the released proteins. Please note, that the differences in hydrodynamic radii are masked by the large corona.
If upon the addition of salt only lipase molecules are released from the micelles, it would mean that the number of polyelectrolytes in the micelles remains constant. Changes in the volume of the micelles, the volume fractions of the polymers and the amount of water are expected to occur. Fig. 7 is an artistic impression of this process. The neutron-scattering curves of five systems at different salt concentrations show a reduced scattering intensity and radius of the micelles at higher salt concentration. In (neutron) scattering I(0) ∼ V2 (see also equation 2), because n Δϱ2 is approximately constant at 0. 0.1 and 0.3 M NaCl. This means that plotting I(0) as a function of V2core, where V2core is derived from the form factor fit (Vcore =  , see Table 1), should have a linear relationship if the core volume reduction is a function of salt concentration. In Fig. 6 it is seen that a linear relation between I(0) and V2core exists. Thus, the decrease in volume is likely to be caused by the release of the lipase.
, see Table 1), should have a linear relationship if the core volume reduction is a function of salt concentration. In Fig. 6 it is seen that a linear relation between I(0) and V2core exists. Thus, the decrease in volume is likely to be caused by the release of the lipase.
 | ||
| Fig. 6 ● I(0) (SANS) as function of V2core. Where Vcore was calculated from the fitted radii of the 0, 0.1 and 0.3 M NaCl scattering curves. | ||
 | ||
| Fig. 7 An artistic impression of the release of lipase from polyelectrolyte complex micelles as function of salt. | ||
A change in volume may also be caused by other components leaving the micelles. One could imagine that diblock copolymers or homopolymers or small soluble complexes consisting of both these polymers, are leaving the complex. However, considering the differences in charge density of the different components (−8 per lipase molecule versus −139 per PAA139 molecule) this scenario is rather unlikely. The charge density of the polymers is much higher than that of the proteins. Whenever a polymer would leave, the complex is no longer electroneutral and rearrangement of micelles has to occur, resulting in total re-organization of the system. This could lead to the formation of other complexes and influence the size and/or number of micelles in the system. Hence, in other scenarios a linear relation between I(0) and V2 is not necessarily the case. Moreover, to be electroneutral the charges of the lipase are not really needed, the poly(acrylic acid) is a weak polyelectrolyte and is therefore able to take up protons or release them. Micelles made of only polyelectrolytes seem to swell slightly upon the addition of salt and there is no indication of release of components from these micelles or a reduction in the number of micelles (see Fig. 3).
In conclusion, expulsion of polyelectrolytes from the complexes seems highly unlikely. Assuming that only lipase molecules are released, it should in principle be possible to estimate the number of incorporated proteins as function of the salt concentration. This can be calculated by defining ΔV = V0 − Vsalt and ΔV/Vlipase = number of free lipase molecules. (A lipase molecule is ellipsoidally shaped with dimensions 5.3 × 5.0 × 4.1 nm3.) If we assume that in a 3.5 mM phosphate buffer all the proteins are encapsulated, the number of protein molecules per micelle core can be derived from the neutron-scattering, using equation 8. Since the mass of the micelles can be calculated and the ratio between the components is known, the aggregation number of the micelles can be determined and, hence, the number of protein molecules per core. Furthermore, it is assumed that the number of polyelectrolytes in the micelles and therefore also the number of micelles remain unaltered. In this way it is possible to model the lipase release as a function of salt. Table 3 shows the results for different salt concentrations.
| Csalt/M | Number of lipases |
|---|---|
| 0 | 4.6 |
| 0.05 | 2.6 |
| 0.1 | 0.4 |
| 0.3 | 0 |
Interpolating the data in Table 3 reveals that at 0.12 M NaCl all the lipase molecules are released from the micelles. From previous DLS-titrations with salt we know that all complexes made of only diblock copolymer (PAA42PAAm417) and protein (lysozyme) are disintegrated at 0.12 M NaCl.31 Apparently, for these two systems the electrostatic interactions between a protein and oppositely charged polyelectrolyte vanish at this salt concentration. This salt concentration is lower than the physiological salt strength (150 mM), and therefore these systems are not suitable for drug-delivery applications.
4 Concluding remarks
This study shows a difference in disintegration between lipase-filled and normal polyelectrolyte complex micelles upon the addition of salt, studied with dynamic light scattering titrations. Combination of DLS measurements and small angle neutron scattering allowed for a structural analysis of the lipase-filled complexes. It was determined that without the addition of salt, five lipase molecules are incorporated in the micelles. Furthermore, the micellar core consists of 83% of water. This implies that the enzymes are probably quite accessible for substrate molecules.Increasing the salt concentration of a solution containing lipase-filled polyelectrolyte complex micelles, induces the release of lipase. This release can be explained in terms of charge density difference between the proteins and the polyelectrolytes. Around a salt concentration of 0.12 M all the lipase molecules are released. This is a rather low salt concentration and indicates that these micellar systems are not very useful for pharmaceutical applications, because they have released most of their proteins at physiological salt strength (150 mM). To make these systems applicable under these conditions, other interaction forces between the protein molecules and the polyelectrolytes have to be incorporated, such as, for instance, increasing the hydrophobic interactions.54 This may be achieved by designing polyelectrolytes that have repeating charged and hydrophobic groups, provided that the native three-dimensional structure of the protein is maintained. Another procedure could be to covalently attach the proteins to one of the polyelectrolytes and make this bond sensitive to an external trigger which induces the release of the protein molecules. One could for instance think of a similar kind of pH trigger that was used by Kataoka and co-workers, to release anti-cancer drugs from folate-conjugated micelles.23–25
In the future we intent to investigate the activity of lipase in the micelles. The knowledge that at 0.12 M NaCl all the enzymes have been released will enable us to study the activity of lipase before, during and after incorporation.
References
- A. Madene, M. Jacquot, J. Scher and S. Desobry, International Journal of Food Science and Technology, 2006, 41(1), 1–21 Search PubMed.
- C. Schmitt, C. Sanchez, S. Desobry-Banon and J. Hardy, Critical Reviews In Food Science And Nutrition, 1998, 38(8), 689–753 CrossRef CAS.
- K. Kataoka, A. Harada and Y. Nagasaki, Advanced Drug Delivery Reviews, 2001, 47(1), 113–131 Search PubMed.
- W. Wei, International Journal of Pharmaceutics, 1999, 185(2), 129–188 Search PubMed.
- A. Taluja, Y. S. Youn and Y. H. Bae, Journal of Materials Chemistry, 2007, 17, 4002–4014 Search PubMed.
- R. Solaro, Journal of Polymer Science Part a-Polymer Chemistry, 2008, 46, 1–11 Search PubMed.
- J. L. Silva and G. Weber, Annual Review of Physical Chemistry, 1993, 44, 89–113 Search PubMed.
- C. Scharnagl, M. Reif and J. Friedrich, Biochimica Et Biophysica Acta-Proteins and Proteomics, 2005, 1749(2), 187–213 Search PubMed.
- J. M. Park, B. B. Muhoberac, P. L. Dubin and J. L. Xia, Macromolecules, 1992, 25(1), 290–295 CrossRef CAS.
- H. Bungenberg de Jong, Complex colloid systems, Vol. 2 of Colloid Science, 1949 Search PubMed.
- C.G. de Kruif, F. Weinbreck and R. de Vries, Current Opinion In Colloid & Interface Science, 2004, 9(5), 340–349 CrossRef CAS.
- J. Gummel, F. Boue, B. Deme and F. Cousin, Journal of Physical Chemistry B, 2006, 110(49), 24837–24846 Search PubMed.
- F. Cousin, J. Gummel, D. Ung and F. Boue, Langmuir, 2005, 21(21), 9675–9688 CrossRef CAS.
- J. Gummel, F. Cousin and F. Boue, Journal of the American Chemical Society, 2007, 129, 5806–5807 CrossRef CAS.
- J. Gummel, F. Boue, D. Clemens and F. Cousin, Soft Matter, 2008, 4 Search PubMed 16-53-1664.
- H. Ai, S. A. Jones and Y. M. Lvov, Cell Biochemistry and Biophysics, 2003, 39(1), 23–43 Search PubMed.
- S. Ichikawa, S. Iwamoto and J. Watanabe, Bioscience Biotechnology and Biochemistry, 2005, 69(9), 1637–1642 Search PubMed.
- A. A. Antipov and G. B. Sukhorukov, Advances in Colloid and Interface Science, 2004, 111(1–2), 49–61 Search PubMed.
- M. Goldberg, R. Langer and X. Q. Jia, Journal of Biomaterials Science-Polymer Edition, 2007, 18(3), 241–268 Search PubMed.
- C. Allen, D. Maysinger and A. Eisenberg, Colloids and Surfaces B-Biointerfaces, 1999, 16(1–4), 3–27 CrossRef CAS.
- G. Gaucher, M. H. Dufresne, V. P. Sant, N. Kang, D. Maysinger and J. C. Leroux, Journal of Controlled Release, 2005, 109(1–3), 169–188 Search PubMed.
- N. Nishiyama and K. Kataoka, Pharmacology and Therapeutics, 2006, 112(3), 630–648 Search PubMed.
- Y. Bae, N. Nishiyama and K. Kataoka, Bioconjugate Chemistry, 2007, 18(4), 1131–1139 CrossRef CAS.
- Y. Bae, N. Nishiyama, S. Fukushima, H. Koyama, M. Yasuhiro and K. Kataoka, Bioconjugate Chemistry, 2005, 16(1), 122–130 CrossRef CAS.
- Y. Bae, W. D. Jang, N. Nishiyama, S. Fukushima and K. Kataoka, Molecular Biosystems, 2005, 1(3), 242–250 Search PubMed.
- M. A. Cohen Stuart, N. A. M. Besseling and R. G. Fokkink, Langmuir, 1998, 14(24), 6846–6849 CrossRef.
- A. Harada and K. Kataoka, Macromolecules, 1995, 28(15), 5294–5299 CrossRef CAS.
- A.V. Kabanov, T. K. Bronich, V. A. Kabanov, K. Yu and A. Eisenberg, Macromolecules, 1996, 29(21), 6797–6802 CrossRef CAS.
- A. Harada and K. Kataoka, Macromolecules, 1998, 31(2), 288–294 CrossRef CAS.
- A. Harada and K. Kataoka, Langmuir, 1999, 15(12), 4208–4212 CrossRef CAS.
- S. Lindhoud, R. de Vries, W. Norde and M. A. Cohen Stuart, Biomacromolecules, 2007, 8(7), 2219–2227 CrossRef CAS.
- A. Harada and K. Kataoka, Abstracts Of Papers Of The American Chemical Society, 2000, 219, U458–U458.
- M. Oishi, S. Sasaki, Y. Nagasaki and K. Kataoka, Biomacromolecules, 2003, 4(5), 1426–1432 CrossRef CAS.
- K. Kono, K. Kawakami, K. Morimoto and T. Takagishi, Journal of Applied Polymer Science, 1999, 72(13), 1763–1773 Search PubMed.
- A. Harada and K. Kataoka, Journal Of The American Chemical Society, 2003, 125(50), 15306–15307 CrossRef CAS.
- S. Duinhoven, R. Poort, G. Vandervoet, W. G. M. Agterof, W. Norde and J. Lyklema, Journal Of Colloid And Interface Science, 1995, 170(2), 351–357 Search PubMed.
- V. Izumrudov, E. Kharlampieva and S. A. Sukhishvili, Macromolecules, 2004, 37(22), 8400–8406 CrossRef CAS.
- H.W. Jomaa and J. B. Schlenoff, Macromolecules, 2005, 38(20), 8473–8480 CrossRef.
- A. Zintchenko, G. Rother and H. Dautzenberg, Langmuir, 2003, 19(6), 2507–2513 CrossRef CAS.
- S. van der Burgh, A. de Keizer and M. A. Cohen Stuart, Langmuir, 2004, 20(4), 1073–1084 CrossRef CAS.
- B. Hofs, I. K. Voets, A. d. Keizer and M. A. Cohen Stuart, Physical Chemistry Chemical Physics, 2006, 8, 4242–4251 RSC.
- B. Hofs, A. de Keizer and M. A. Cohen Stuart, Journal of Physical Chemistry B, 2007, 111(20), 5621–5627 Search PubMed.
- J. F. Berret, P. Herve, O. Aguerre-Chariol and J. Oberdisse, Journal Of Physical Chemistry B, 2003, 107(32), 8111–8118 Search PubMed.
- O. Glatter, Journal Of Applied Crystallography, 1977, 10(OCT1), 415–421 Search PubMed.
- O. Glatter, Journal Of Applied Crystallography, 1979, 12(APR), 166–175 Search PubMed.
- T. Nicolai, M. Pouzot, D. Durand, M. Weijers and R. W. Visschers, Europhysics Letters, 2006, 73(2), 299–305 Search PubMed.
- B.R. Postmus, F. A. M. Leermakers and M. A. Cohen Stuart, Langmuir, 2008, 24, 1930–1942 CrossRef CAS.
- V. K. Aswal, P. S. Goyal, J. Kohlbrecher and P. Bahadur, Chemical Physics Letters, 2001, 349(5–6), 458–462 CrossRef CAS.
- K. Hiraoka, H. Shin and T. Yokoyama, Polymer Bulletin, 1982, 8(7–8), 303–309 CAS.
- H. Fischer, I. Polikarpov and A. F. Craievich, Protein Science, 2004, 13(10), 2825–2828 CrossRef CAS.
- J.F. Berret, K. Yokota, M. Morvan and R. Schweins, Journal of Physical Chemistry B, 2006, 110(39), 19140–19146 Search PubMed.
- I.K. Voets, S. van der Burgh, B. Farago, R. Fokkink, D. Kovacevic, T. Hellweg, A. de Keizer and M. A. Cohen Stuart, Macromolecules, 2007, 40, 8476–8482 CrossRef CAS.
- D. Pignol, L. Ayvazian, B. Kerfelec, P. Timmins, I. Crenon, J. Hermoso, J. C. Fontecilla-Camps and C. Chapus, Journal Of Biological Chemistry, 2000, 275(6), 4220–4224 Search PubMed.
- X. F. Yuan, A. Harada, Y. Yamasaki and K. Kataoka, Langmuir, 2005, 21(7), 2668–2674 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2009 |
