Development of a DNA-dosimeter system for monitoring the effects of solar-ultraviolet radiation
André Passaglia Schucha, Rodrigo da Silva Galhardoa, Keronninn Moreno de Lima-Bessaa, Nelson Jorge Schuchb and Carlos Frederico Martins Menck*a
aDepartment of Microbiology, Institute of Biomedical Sciences, University of São Paulo, Av. Prof. Lineu Prestes, 1374, São Paulo, SP 05508-900, Brazil. E-mail: cfmmenck@usp.br; Fax: 55.11.3091-7354; Tel: 55.11.3091-7499
bSouthern Regional Space Research Center, CRSPE/INPE–MCT, Santa Maria, RS, Brazil
First published on 20th November 2008
Abstract
Solar radiation sustains and affects all life forms on Earth. In recent years, the increase in environmental levels of solar-UV radiation due to depletion of the stratospheric ozone layer, as a result of anthropogenic emission of destructive chemicals, has highlighted serious issues of social concern. This becomes still more dramatic in tropical and subtropical regions, where the intensity of solar radiation is higher. To better understand the impact of the harmful effects of solar-UV radiation on the DNA molecule, we developed a reliable biological monitoring system based on the exposure of plasmid DNA to artificial UV lamps and sunlight. The determination and quantification of different types of UV photoproducts were performed through the use of specific DNA repair enzymes and antibodies. As expected, a significant number of CPDs and 6-4PPs was observed when the DNA-dosimeter system was exposed to increasing doses of UVB radiation. Moreover, CPDs could also be clearly detected in plasmid DNA when this system was exposed to either UVA or directly to sunlight. Interestingly, although less abundant, 6-4PPs and oxidative DNA damage were also generated after exposure to both UVA and sunlight. These results confirm the genotoxic potential of sunlight, reveal that UVA may also produce CPDs and 6-4PPs directly in naked DNA and demonstrate the applicability of a DNA-dosimeter system for monitoring the biological effects of solar-UV radiation.
Introduction
On a global scale, besides CO2 accumulation other environmental variables such as temperature and UV radiation are increasing by various degrees at different latitudes.1 The increase in environmental levels of solar-UV radiation observed over the last decades has been addressed to depletion of the stratospheric ozone layer as a result of anthropogenic emission of destructive chemicals into the atmosphere.2The proportion of total UV radiation currently reaching the Earth's surface is only 6.1% of the solar electromagnetic spectrum. Most of this corresponds to UVA radiation (320 to 400 nm), which comprises 5.6% of the solar spectrum, and reaches the ground without significant absorption by the atmosphere. UVB radiation (280 to 320 nm) constitutes about 0.5% of the solar spectrum, even though ozone layer depletion can cause changes in its spectral distribution. Ozone and oxygen completely absorb UVC radiation (<280 nm), thus these wavelengths do not reach the Earth's surface.3,4 Although UV radiation represents only a small part of the solar spectrum, it has enormous importance on the structure of the atmosphere, as well as a critical impact on the biosphere.5 Its main biological relevance is due to its capacity to induce DNA damage in different aquatic and terrestrial life forms.6
The chemical nature and efficiency in the formation of DNA lesions greatly depend on the wavelength of the incident photons. A striking biological observation is that DNA strongly absorbs UV light with an absorption spectrum paralleling that of ozone, thus providing a perfect umbrella to shield DNA from the damages of solar-UV radiation.3,7
Photocarcinogenesis is most commonly thought to be the result of a chain of events that involve induction of DNA damage and subsequent mutation formation following exposure to UV light.8 Bipyrimidine photoproducts, including Cis-syn cyclobutane pyrimidine dimers (CPDs) and pyrimidine-(6-4)-pyrimidone photoproducts (6-4PPs), are the main lesions induced by UVB radiation, and support for the involvement of these lesions in photo-carcinogenesis being provided by the overwhelming proportion of mutations detected at bipyrimidine sites in human skin-tumors.9,10 CPDs are formed between bonds 5 and 6 of two adjacent pyrimidine bases, whereas 6-4PPs are characterized by a stable bond between positions 6 and 4 of two neighboring pyrimidines. The energy of these short-wavelength UVB photons is high enough to generate the formation of CPDs or 6-4PPs through direct absorption by DNA.4,11–13
The absorption of DNA is rather weak in the less energetic, long-wavelength UV range, and it is evident that UVA radiation is far less efficient in producing direct photo-lesions.4 Oxidized bases have been shown to be induced in a naked DNA solution and in cells by UVA radiation.14 On the other hand, recent research employing cultured cells challenges the hypothesis that UVA-induced DNA damage is mostly mediated by oxidative stress, showing that CPDs are more efficiently generated by UVA and are more biologically relevant than oxidatively induced DNA lesions.15,16 These results confirm previous observations by Tyrrell et al.,17 who found that UVA produces CPDs in naked and phage DNA.
Although the induction of 6-4PPs by UVA has been reported as undetectable,13,15,16,18 wavelengths close to 320 nm are capable of inducing its photo-isomerization into Dewar isomers.10,16,19 It has been demonstrated that Dewar photoproducts (DewarPP, a valence isomer of 6-4PP) are removed from the genome of mammalian cells with kinetics and efficiency similar to those of CPD, a similarity of response thereby suggesting that this type of DNA photoproduct may also contribute significantly to the mutagenesis that occurs after exposure to sunlight.19
In an attempt to evaluate the biological effects of solar-UV radiation, efforts are being made worldwide to develop dosimetric systems through the use of biological material.6,20,21 Since DNA is the main UV target in living organisms, it is quite natural to employ DNA as a molecular dosimeter for the detection of damage. In fact, a UVB DNA dosimeter was developed based on minidots of dried bacteriophage λ DNA placed on a UV-transparent polymer film. The photoinduced DNA damages block synthesis during polymerase chain reactions (PCR) reducing the amount of amplified product of UV-exposed DNA compared to the control DNA.22 The possibility to evaluate the integration of DNA-absorbed solar energy was considered through the irradiation of populations of radiation sensitive spores (B. subtilis UVSSP), vegetative bacteria (E. coli K12-AB2480) and bacteriophage (E. coli phage T4vx) with monochromatic far- and near-UV and sunlight.23 Four DNA-based biological dosimeters (B. subtilis spores,6,24 DLR-biofilm developed specifically from B. subtilis spore,25 bacteriophage T726 and polycristalline uracil thin layer27) were also evaluated in laboratory and field measurements for their biological effectiveness as UV detector.28 It has been demonstrated that these types of dosimeters can be employed in different field applications, including the measurement of extraterrestrial solar radiation.29
In this work, we describe the development of a versatile and reliable system with the aim of evaluating the effects of direct solar-UV radiation on DNA. The DNA-dosimeter system is based on the exposure of a plasmid DNA solution to each UV region of the terrestrial solar spectrum (UVB and UVA) and also directly to sunlight. In order to discriminate the different types of DNA photolesions after exposure to UV lamps and sunlight, the E. coli formamidopyrimidine-DNA glycosylase (Fpg - recognizes mainly oxidative damages in purines), the T4 bacteriophage endonuclease V (T4-endo V - recognizes manly CPDs), and the yeast DNA repair enzyme Ultraviolet Damage Endonuclease (UVDE - recognizes mainly large distortions in DNA double helix like CPDs, 6-4PPs, as well as DewarPPs), were employed. Apurinic/apyrimidinic (AP) sites and 6-4PPs were also identified by alkali treatment of the irradiated DNA. Moreover, specific antibodies were used in immunoblot assays to confirm the induction of CPDs and 6-4PPs after UV exposure. We were able to demonstrate the biological relevance of this DNA-dosimeter through the quantification of oxidative DNA damage, CPDs, 6-4PPs and other distorting lesions, possibly DewarPPs, in plasmid DNA samples exposed to solar radiation. The ratio of the different types of lesions was also described for each specific radiation condition, the data indicating that, as expected, sunlight induces lesions that resemble UVB, in addition to oxidative DNA damages.
Materials and methods
Plasmid
The E. coli strain DH10b was made electro-competent30 and transformed with pCMUT vector (1762 bp) (C - cloramphenicol resistance, and MUT - supF mutation target gene). The plasmid was derived from pAC189 plasmid31 as described in Fig. 1. Purification of DNA samples was prepared by using Qiagen maxi kits (Valencia, USA), and stored in TE buffer (10 mM Tris-HCl [pH 8.0], 1 mM EDTA).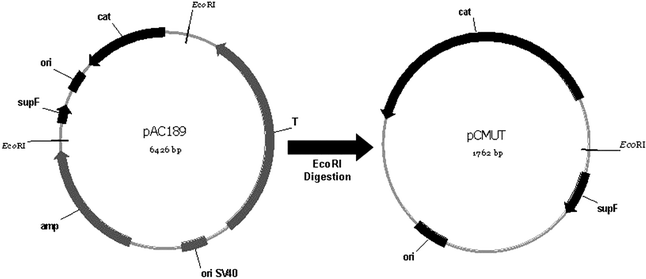 | ||
| Fig. 1 Scheme for the genetic map of the plasmid pCMUT. This plasmid was obtained through EcoRI digestion of pAC189. The main sites are indicated and the plasmids are not in scale. | ||
UV sources and irradiation of the DNA-dosimeter system
UVB radiation was carried out with a Vilber Loumart T15M 15 W lamp and, in this case, the DNA-dosimeter system was covered with a polycarbonate sheet to filter UVC wavelengths. For UVA radiation, samples in the DNA-dosimeter system were irradiated by an Osram Ultramed FDA KY10s 1000 W lamp filtered with a Schott BG39 glass filter 3 mm thickness (Schott Glass, Germany). The dose rates of UVB and UVA lamps were measured with a UV radiometer VLX 3 W (Vilber Lourmat, Torcy, France), whereby values obtained were 4.8 J m−2 s−1, and 87 J m−2 s−1, respectively. The irradiance spectra for the two lamps with and without filter, according to Gróf et al.,32 are presented in Fig. 2. The amount of UVC contamination for UVB light, as well as, UVC and UVB for UVA was below detection limits. | ||
| Fig. 2 Irradiance spectra of UVB and UVA lamps. A. Irradiance spectra of a Vilber Loumart T15M 15 W UVB lamp filtered with a polycarbonate sheet. B. Irradiance spectra of an Osram Ultramed FDA KY10s 1000 W lamp filtered with a Schott BG39 glass filter 3 mm thickness (Schott Glass, Germany). These spectra were obtained by using an Ocean Optics 1560 Spectroradiometer HR4000. The environmental temperature and relative air humidity were measured by a Visomes LV12168/06 Thermohygrometer, varying between 21 °C–22 °C and 60%–70%, respectively. | ||
Sunlight exposures were performed on a platform placed at the Southern Space Observatory (SSO), São Martinho da Serra, located in the central region of the southernmost Brazilian State, Rio Grande do Sul (Southern region of Brazil, 29.44 °S, 53.8 °W) in parallel to radiometric photometry observations in December 2006 (27th and 29th) and January 2007 (4th). On each day, exposures of the DNA-dosimeter system were performed for 4 continuous hours from 10:00 am to 2:00 pm together with unexposed controls (the samples were covered with aluminum foils). The UVB and UVA doses were obtained from integrations of irradiance values measured by continuous UVB and UVA meters (UVB and UVA Radiometers, EKO Instruments Trading Co., Ltd., Japan) installed at SSO.
DNA photoproducts quantification
To determine the average number of DNA photoproducts generated by UV lamps and sunlight, the relative amounts of supercoiled (FI) and circle (FII) plasmid DNA forms were measured after separation by 0.8% agarose gel electrophoresis through densitometry analyses (Image Quant 300, GE Healthcare, USA). Samples with 200 ng of DNA were pre-incubated with 0.8 U of E. coli Formamidopyrimidine-DNA glycosylase (Fpg protein, from New England Biolabs, Ipswich, USA), 70 ng of T4 bacteriophage endonuclease V (T4-endo V, produced in this laboratory) and 250 ng of yeast DNA repair enzyme Ultraviolet Damage Endonuclease (UVDE, from Trevigen, Gaithersburg, USA), to discriminate the different types of DNA lesions. These samples were incubated at 37 °C (for Fpg and T4-endo V) and 30 °C (for UVDE) for 30 min. T4-endo V and Fpg buffers and incubation conditions were previously described.4 UVDE application was performed according to manufacturer's instructions. The enzymes were tested up to saturation, and were used in amounts where no specific cleavage is observed.The number of enzyme sensitive sites per Kbp of plasmid DNA was calculated, assuming a Poisson distribution adapted to this technique, by the following equation:
| X = −ln (1.4 × FI/1.4 × FI + FII)/1.8 |
Immunoblot assays
Plasmid DNA samples obtained after each exposure were boiled for 10 min and then spotted onto a nitrocellulose membrane (BIO-RAD, Hercules, CA, USA) by using a slot-blot apparatus. The membrane was subsequently incubated with 5 X SSC (750 mM NaCl, 75 mM sodium citrate) for 15 min at 37 °C, dried at room temperature and then baked for 2 hours at 80 °C. Blocking was performed in 5% milk for 18 h, whereupon the membrane was incubated with the respective primary antibodies (MBL International Corporation, Nakaku Nagoya, Japan) for immuno-detection of CPDs and 6-4PPs. Anti-mouse IgG HRP conjugate (R & D Systems, Minneapolis, MN, USA) was used as a secondary antibody. Blots were developed by using a chemiluminescence detection system (Image Quant 300, GE Healthcare).Identification of AP sites and 6-4 photoproducts through alkali treatment
The direct quantification of AP sites and 6-4PPs was also performed by an adaptation of the technique described by Higurashi et al.34 Plasmid DNA samples exposed to UVA light were dissolved in 50 mM of sodium phosphate (pH 12.0), and the mixtures were incubated at 60 °C for 30 min (mild treatment) to detect AP sites and 4 h for 6-4PPs. After neutralization with HCl 0.1 M, the relative amounts of supercoiled (FI) and circle (FII) plasmid DNA forms (200 ng) were measured after separation by 0.8% agarose gel electrophoresis through densitometry analyses (Image Quant 300, GE Healthcare), and the number of lesions per Kbp calculated as described above.Results
Development of the DNA-dosimeter system
To develop a reliable system for the exposure of DNA samples to sunlight, we searched for material that would present the most adequate features: (i) high transmittance to UVB and UVA wavelengths; (ii) resistance to environmental adversities, thereby avoiding tanning; (iii) the possibility of framing the shape of the template according to the aim of the experiment; and (iv) low costs. We chose the Elastomer Sylgard 184™ Kit (Dow Corning Corporation, USA) for constructing the DNA-dosimeter system, since it was the one which best fits these requirements. Both the DNA-dosimeter system developed to perform exposures to sunlight, as well as its transmittance spectrum, are shown in Fig. 3. As indicated in Fig. 3A, DNA samples were applied in triplicate in the DNA-dosimeter system for performing the desired exposures. For unexposed samples, the DNA-dosimeter system was covered with an aluminum foil and submitted to the same conditions as those exposed to sunlight. As shown in Fig. 3B, this elastomer provides high transmittance for wavelengths above 240 nm.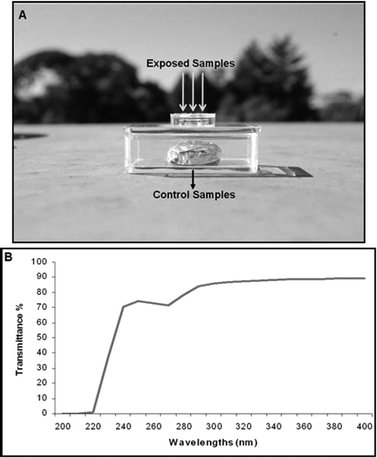 | ||
| Fig. 3 The DNA-dosimeter system. A. The DNA-dosimeter system was developed by employing the Kit Elastomer Sylgard 184™. This system was used for exposures of DNA samples to sunlight, as indicated by arrows. Control samples were covered with an aluminum foil. B. Transmittance spectrum of the Kit Elastomer Sylgard 184™. | ||
Quantification of DNA lesions induced by UVB and UVA radiation
Initially, the generation of plasmid DNA damage was defined after exposure to artificial UV sources. Plasmid DNA samples were irradiated by UVB or UVA lamps, and then treated with Fpg, T4-endo V and UVDE DNA repair enzymes to quantify specific DNA lesions induced by each UV band. Samples were analyzed by electrophoresis migration in agarose gels to discriminate supercoiled DNA (form I) from open-circle relaxed DNA presenting breaks or nicks caused by enzymatic cleavage (form II). Fig. 4 illustrates one of these experiments. A summary of the results is presented in Fig. 5. | ||
| Fig. 4 Representative example of DNA photolesions determination after the DNA exposure in the DNA-dosimeter system to UVB (A) and UVA radiation (B). Plasmid DNA samples were exposed to increasing doses of UVB and UVA radiation and then treated with Fpg, T4-endo V, and UVDE enzymes; no indicates similar treatment with no enzyme. FI indicates the supercoiled DNA form and FII the relaxed DNA form. | ||
 | ||
| Fig. 5 Induction of DNA photoproducts after exposures to UVB (A) and UVA (B) lamps. The results represent the average and standard deviation of three independent experiments for each condition of exposure. SSB - single strand breaks; Fpg-SS - Fpg sensitive sites; T4-endo V-SS - T4-endo V sensitive sites; UVDE-SS - UVDE sensitive sites. | ||
There was a dose-dependent increase of circle-relaxed DNA (form II), when UVB or UVA-irradiated DNA samples were incubated with the respective DNA repair enzymes, as clearly shown in Fig. 4. In addition, as shown in Fig. 5, the increase in the frequency of DNA lesions was linear according to the UV dose employed. Few or no direct breaks (single-strand breaks - SSBs) were detected for any of the types of radiation and doses performed. As expected, the induction of Fpg-sensitive sites (Fpg-SS - oxidative DNA damage) were significantly higher (compared to the other photoproducts) after UVA, in relation to UVB radiation. T4-endo V-sensitive sites (T4-endo V-SS) were highly induced by UVB, but the higher number of UVDE-sensitive sites (UVDE-SS) was most probably due to the formation of 6-4PPs besides CPDs. In addition, although at a lower frequency when compared to UVB, T4-endo V-SSs were also detected after UVA radiation. Surprisingly, the induction of UVDE-SS was more frequent than T4-endo V-SS, thereby indicating the presence of other distorting lesions in addition to CPDs in these DNA samples, possibly 6-4PPs and/or DewarPPs.
Quantification of DNA lesions induced by sunlight
Environmental exposures using the DNA-dosimeter system were carried out on a platform situated at the Southern Space Observatory (SSO). Exposures were performed during two cloudless (December 27th and 29th, 2006), and one cloudy (January 4th, 2007) days, for four continuous hours, from 10:00 am to 2:00 pm. In parallel, solar UVB and UVA radiations were measured by solar-UV radiometers. UV radiation doses and the average temperature during exposure periods are presented in Table 1. The induction of DNA photoproducts direct by sunlight was measured as described above, and the data are shown in Fig. 6.| Sunlight exposures SSO Brazil | SSO 10:00 am–14:00 pm UVA KJ m−2 315–400 nm | SSO 10:00 am–14:00 pm UVB KJ m−2 280–315 nm | Average Temperature °C |
|---|---|---|---|
| 12/27/2006 | 191.1 | 5.2 | 22.6 |
| 12/29/2006 | 176.7 | 4.8 | 25.9 |
| 01/04/2007 | 81.4 | 2.3 | 24.2 |
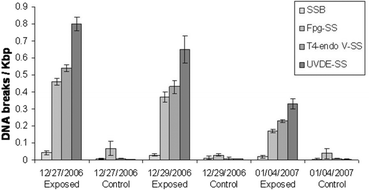 | ||
| Fig. 6 Induction of DNA lesions after exposures to sunlight at SSO using the DNA-dosimeter system. The results represent the average and standard deviation of experiments performed in triplicate for each day of exposure. Control samples were also exposed, but the DNA-dosimeter system was covered with aluminum foil. SSB- single strand breaks; Fpg-SS - Fpg sensitive sites; T4-endo V-SS - T4-endo V sensitive sites; UVDE-SS - UVDE sensitive sites. | ||
Although the frequency of DNA photoproducts varied according to radiation dose received daily (Table 1), the relative proportion of the types of DNA lesions was basically similar for the three days. The frequency of observed direct breaks (SSBs) was very low, although significantly higher for DNA exposed 12/27/2006.
A direct correlation between UVB and UVA doses measured during sunlight exposures (Table 1) with irradiation performed in the laboratory is difficult, as the dosimeters employed were different, and solar-UV represents in fact a combination of light irradiation, with a different incident spectrum when compared to those emitted by artificial lamps. However, it is interesting to note that for the same intensity, the frequency of T4-endo V-SS produced after sunlight exposure was lower than that observed for independent UVB and UVA radiation.
Different from what was observed after irradiation with artificial lamps, after exposure to sunlight, Fpg-SS levels were higher and very close to those of T4-endo V-SS. In fact, the ratio of T4-endo V-SS/Fpg-SS was close to 1.2 after sunlight, when compared to 2.2 after UVA and 9.2 after UVB radiation (Table 2).
| UVB Doses/KJ m−2 | T4-endo V-SS/Fpg-SS | T4-endo V-SS/UVDE-SS–T4-endo V-SS |
|---|---|---|
| 2 | 12.1 | 3.2 |
| 4 | 8.7 | 2.3 |
| 6 | 6.8 | 2.8 |
| UVA Doses/KJ m−2 | ||
| 100 | 2.5 | 4.0 |
| 200 | 2.1 | 3.6 |
| 300 | 2.0 | 3.0 |
| Sunlight exposures | ||
| 2006/12/27 | 1.2 | 3.1 |
| 2006/12/29 | 1.3 | 2.9 |
| 2007/01/04 | 1.3 | 3.1 |
Interestingly, a further increase was observed for UVDE-SS, probably due to the induction of CPDs, 6-4PPs and DewarPPs in plasmid DNA samples exposed to sunlight. Considering that T4-endonuclease V recognizes mainly CPDs and that the extra DNA lesions recognized as UVDE-SS are composed of 6-4PPs and DewarPPs, we also calculated the ratio of CPDs/(6-4PPs + DewarPPs) (T4-endo V-SS/UVDE-SS - T4-endo V-SS), in order to compare the different types of irradiation. The data are presented in Table 2. While CPDs were the most frequent DNA photoproducts observed following UVB, UVA and sunlight, the ratio of CPDs/(6-4PPs + DewarPPs) was slightly higher for UVA (3.0 to 4.0), when compared to UVB (2.3 to 3.2) or sunlight exposures (2.9 to 3.1).
Detection of DNA photoproducts through immunoblot assays
Immunoblot assays using specific antibodies were carried out with the purpose of confirming and comparing the efficiency of induction of CPDs and 6-4PPs, after exposure of DNA samples to UVB, UVA, and sunlight radiation. The induction of these photoproducts by artificial light is shown in Fig. 7.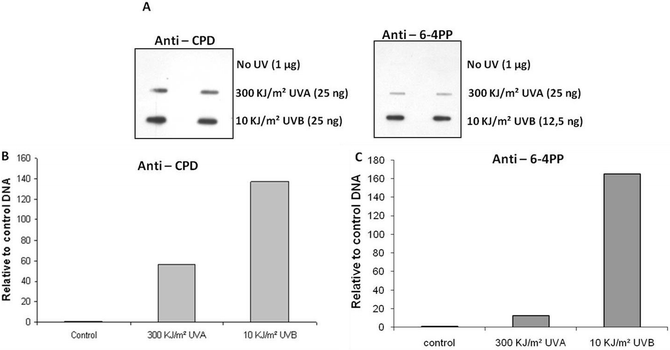 | ||
| Fig. 7 Immunoblot assay with plasmid DNA samples exposed to UVB and UVA radiation. A. Detection through the use of antibodies anti-CPD and anti-6-4PP. B. Quantification of CPD induction relative to DNA control samples (fold increase in relation to non-exposed control samples - no UV). C. Quantification of 6-4PP induction relative to DNA control samples (fold increase in relation to non-exposed control samples - no UV). | ||
As expected, the induction of CPDs after UVB and UVA exposures was very efficient. Confirming the data with UVDE-SS, 6-4PPs were also detected by the specific antibody when DNA samples were exposed to UVB or UVA radiation. However, contrary to what was observed for UVDE-SS and T4-endo V-SS, 6-4PPs were much less efficiently generated by UVA than by UVB.
A similar experiment was performed with the DNA samples exposed to sunlight. Results are shown in Fig. 8. As indicated by experiments with enzymes, the induction of both types of lesions (CPDs and 6-4PPs) could be observed in DNA samples exposed to sunlight.
 | ||
| Fig. 8 Immunoblot assay with plasmid DNA samples exposed to UVA (25 ng), UVB (25 ng), and sunlight at SSO (200 ng). A. CPDs and 6-4PPs detection by using anti-CPD and anti-6-4PP antibodies. B. Quantification of CPDs detection by using an anti-CPD antibody after exposure to sunlight (fold increase in relation to non-exposed control samples for each day). C. Quantification of 6-4PPs detection by using an anti-6-4PP antibody after exposure to sunlight (fold increase in relation to non-exposed control samples for each day). | ||
Detection of UVA-induced AP sites and 6-4PPs by alkali treatment
The data above indicated that direct UVA radiation of plasmid DNA led to the formation of 6-4PP lesions. This was further tested using a simple hot alkali treatment, as this method was shown to identify these lesions, as well as AP sites, after mild treatment.34 As shown in Fig. 9, AP sites were almost undetectable using this methodology, but a clear dose-dependent increase of 6-4PPs was observed after UVA radiation and hot alkali treatment. | ||
| Fig. 9 Detection of AP sites and 6-4PPs by alkali treatment of UVA-exposed DNA samples. A. Representative example of the determination of AP sites and 6-4PPs in DNA by hot alkali treatment after exposure to UVA irradiation. The conditions to discriminate between these two lesions are described in Material and Methods, adapted from previous work.34B. Quantification of AP sites and 6-4PPs induced after exposures to UVA light at the indicated doses and hot alkali treatment. | ||
Discussion
There is overwhelming evidence indicating that sunlight is a human carcinogen, and in terms of absolute numbers, is one of the most significant to which populations are exposed. Thinning of the stratospheric ozone-shield and the resulting increase in UVB radiation reaching the Earth are expected to directly affect human health, as the incidence of all kinds of skin cancer increases with higher exposure to UVB radiation.12,13,16In order to better understand the respective contributions of each UV portion of the solar spectrum, Kuluncsics et al.4 established profiles of DNA damage induced in plasmid DNA by UVC, UVB, and UVA radiation, in comparison with the profile obtained with simulated sunlight. From that study, it was clear that UVC was more efficient in inducing CPD than UVB, whereas UVA has a very low, if any, capacity to cause this kind of DNA lesion. In comparison, simulated sunlight was 10-fold more effective in generating CPDs than UVA radiation, this reflecting the contribution of UVB wavelengths.4
Through the utilization of DNA repair enzymes and DNA sequencing methods, Yoon et al.18 observed the induction of CPDs when DNA or cells were irradiated with simulated sunlight. It was also found that 6-4PPs were formed at almost undetectable levels under conditions of irradiation for up to 5 h with a solar-UV simulator. The conclusion was that CPDs are induced at least 20 to 40 times more frequently than other DNA photoproducts after exposure to simulated sunlight.18
Using HPLC associated with tandem-mass spectrometry, Courdavault et al.10 studied the induction and repair of CPDs, 6-4PPs, and DewarPPs in primary cultures of human keratinocytes. They concluded that UVB radiation is capable of inducing CPDs and 6-4PPs, although no Dewar valence isomer was detected. On the other hand, UVA was found to efficiently convert 6-4PPs into their respective DewarPPs in a dose-dependent way. In addition, the induction of TT-CPDs was almost the only bipyrimidine photoproduct produced upon UVA radiation.10 Using the same methodology, Mouret et al.16 determined both type and yield in the formation of DNA damage throughout an entire patch of human skin when exposed to UVB or UVA radiation. The results also demonstrated that CPDs were produced in a higher yield than oxidatively generated DNA lesions, when the skin was exposed to UVA radiation. This recent, novel information challenges the hypothesis that UVA-induced DNA damages are mostly mediated by oxidative stress. Furthermore, no other photoproducts, such as 6-4PPs and Dewar valence isomers, could be detected in human skin exposed to UVA light.16
Through the application of an immuno-dot-blot quantification assay calibrated with DNA repair enzymes, Perdiz et al.19 examined the absolute amount of each type of UV photoproduct induced by UVC, UVB, UVA and simulated sunlight in plasmid DNA and genomic DNA from CHO cells. CPD induction was confirmed after all kinds of treatment, and 6-4PPs were readily produced by UVC, UVB and simulated sunlight but not by UVA. DewarPPs were also not detected after UVC and UVA exposure. On the other hand, these were relatively more frequent in cellular DNA subject to simulated sunlight exposure than UVB radiation. From these data, the authors concluded that the yield of photoproducts induced by simulated sunlight was 6 CPDs for each 6-4PP produced.19
The relative induction of CPDs and 6-4PPs (and recently DewarPPs) by solar-UV radiation and their contribution in biological process have been a matter for debate for more than 20 years. As is shown in the articles cited above, there is an enormous discrepancy among studies on the efficiency of induction of these photoproducts, when these are undertaken in an attempt to simulate environmental UV doses. In this work, we examined the induction of DNA photoproducts when subject to sunlight during three days under varying weather conditions, and then compared the results to those obtained after UVA and UVB radiation.
For this type of work, we developed a new exposure method for DNA samples with a DNA-dosimeter system (Intellectual Propriety requested), whereby all wavelengths of solar-UV radiation reach the sample with almost the original intensity. Through the use of specific DNA repair enzymes and antibodies, it was possible to access the types of DNA lesions by using relatively small amounts of DNA samples.
After exposure of the DNA-dosimeter system to UVB and UVA, as well as directly to solar-UV radiation, we detected the induction of T4-endo V-SS in all DNA-irradiated samples, thus suggesting the formation of CPDs (Fig. 5 and 6). In agreement with several previous reports,4,10,13,15,16,19,35 we not only present herein evidence that CPDs are generated by UVA radiation, but also show that this is the most induced type of DNA photoproduct after UVA treatment (Fig. 5B).
Consistent with other previously published results,4,14,36,37 exposure to UVA radiation generated a significant amount of oxidized bases, as indicated by Fpg-SS (Fig. 5B). These Fpg-SS in plasmid DNA following UVA radiation might have been induced by an indirect mechanism through hydroxyl radicals in a Fenton reaction, due to the presence of iron bounded to DNA molecules.4 The UVA-DNA absorption spectra, as well as, UVA-DNA damage profile in plasmid DNA purified by CsCl gradient38 was very similar than those observed with DNA samples purified with Quiagen maxi Kit (see Supplementary Data, Figure 1S), making highly unlikely the possibility that some type of photosensitizer contaminating DNA preparations could cause these photoproducts.
In order to evaluate the use of the DNA-dosimeter system to quantify DNA damage induced by direct sunlight, environmental exposures were carried out at SSO, in the South of Brazil. The results presented in Fig. 6 show that oxidative damage (Fpg-SS) was clearly induced, although CPDs (T4-endo V-SS) arose as the most abundant kind of DNA lesion after exposure to sunlight at this latitude. Curiously, the relative amount of Fpg-SS was higher after exposure to sunlight than after UVA radiation (Table 2).
Interestingly, more UVDE-SS than T4-endo V-SS were detected in all the conditions of irradiation tested. These data suggest that other distorting lesions (such as 6-4PPs) are also produced by UVB, UVA and sunlight. In order to confirm the induction of CPDs and 6-4PPs by UVA radiation and sunlight, we evaluated the occurrence of these DNA lesions by means of immunoblot assays, using antibodies specific for each kind of photoproduct. Our results clearly confirm the induction of 6-4PPs after exposure of plasmid DNA samples in the DNA-dosimeter system to UVB, UVA and sunlight (Fig. 7 and 8). Nevertheless, in the case of 6-4PPs in UVA irradiated DNA, the incidence was much lower than that detected for UVB radiation, when compared with data obtained through the use of DNA repair enzymes.
It is important to note that the two antibodies which recognize the lesions are different and the experiments are independent, thus the direct ratio of the types of DNA lesion produced (CPDs and 6-4PPs) cannot be determined by this methodology alone. In addition, the low specificity of UVDE when compared to a highly specific antibody against 6-4PP may explain the difference in the relative amount of this lesion. However, if this is the case, the results with UVDE may, in fact, be explained by the presence of a third and frequent lesion induced mainly by exposure to UVA and sunlight. Considering that the conversion of 6-4PPs to DewarPPs by UVA was previously observed,19 it is reasonable to propose that this other type of lesion is DewarPPs, also known to be a substract for UVDE.39
We also used a third methodology, hot alkali treatment, to confirm the induction of 6-4PPs by UVA light. This method was adapted from the work of Higurashi et al.34, who strongly suggest that this treatment results in degradation of 6-4PPs, or AP sites for mild treatment, as monitored by HPLC, NMR spectroscopy, and mass spectroscopy. Exposure of plasmid DNA to UVA light, followed by hot alkali treatment, revealed a dose-dependent increase of DNA cleavage (Fig. 9). This cleavage is most likely due to the degradation of 6-4PPs, although it should be pointed that previous work have also revealed that DewarPPs are also cleaved by hot alkali (piperidine) treatment,40,41 but this still needs confirmation with the alkali conditions employed in this work.
Anyway, the amount of these lesions (Fig. 9B) corresponds to approximately half of the lesions detected by UVDE enzyme, thus also pointing to the possible formation of other lesions that could be recognized by this enzyme. UVDE also cleaves DNA containing AP sites,39 thus we tested this possibility in DNA exposed to UVA light. Mild alkali treatment34 failed to detect any significant amount of AP sites in this DNA (Fig. 9). We further tested this DNA with the E. coli endonuclease IV (product of the nfo gene), but the results indicated that a very low, if any, AP site is induced by UVA light in our experimental conditions (Supplementary Data Figure 2S). The results with hot alkali treatment and Nfo incubation confirm the enzymatic and immunological data, giving support to the idea that UVA radiation is able to induce 6-4PPs and isomerizes it in DewarPPs concomitantly, increasing the amount of UVDE-SS in relation to T4-endo V-SS.
Discrepancies observed here and those presented in the literature may simply reflect the different methodologies applied. The techniques employed in this work are very sensitive, although they are not as specific as HPLC/MS.10,16 Alternatively, the differences may also be due to the manner in which samples are exposed to artificial UV lamps, in this DNA dosimeter system. In fact, when DNA samples were directly exposed to UVA light inside 30 mm polycarbonate plates, only trace amounts of 6-4PPs were detected by anti-6-4PP antibodies (data not shown). Also, induction of Fpg-SS was even higher than T4-endo V-SS after UVA treatments, this indicating an alteration in the profile of DNA damage incurred by these wavelengths (data not shown). Therefore, the use of high transmittance material such as that employed in the present work should be indicated in exposures to artificial UV lamps, since there are no guidelines giving an exact definition of the transmission curve of physical radiometers for UV measurements. Consequently, every manufacturer uses filters with different spectral curves for each UV band. Thus, measurements of a specific broad-band UV dose are not similar according to the models of both the radiometer and the lighting-source used, as well as to the material of apparatus employed to expose biological samples.
Conclusions and perspectives
One of the most important criteria for the validity of a biological dosimeter is the relevance of the photobiological/photochemical process to the biological effects. In many of the biological consequences, the key process is DNA damage due to UV radiation. Thus DNA-based biological dosimeters have genuine biological relevance.28 The results obtained in this work show that the DNA-dosimeter system is efficient for indicating the amount and nature of specific DNA damage induced by sunlight. Moreover, a comparison between the efficiency of induction of both types of photoproducts after exposure to UVB, UVA, and solar-UV radiation, reveals a considerable amount of CPDs in plasmid DNA samples exposed to UVA doses. The induction of 6-4PPs after exposure to UVA light was clear, although lower than after UVB radiation. The mechanisms involved in the generation of such lesions by UVA light (CPDs and 6-4PPs) and in oxidative DNA damage, are unknown, as DNA was irradiated in the absence of any photosensitizer and no UVB trace was detected with the filters used (Fig. 2). This question remains open and is worthy of further investigation. Nonetheless, considering environmental sunlight exposure, the DNA lesion profile obtained significantly indicated that the contribution of UVB is very high, in addition to the generation of a high frequency of oxidative DNA lesions.Currently, we are initiating the measurement of solar UVB and UVA radiation on a parallel with an evaluation of its interaction with the DNA molecule, by exposing DNA samples to sunlight and using the DNA-dosimeter system in the city of São Paulo, Brazil. Thus, the plasmid vector employed can be transformed into bacteria, thereby providing the means of evaluating the genetic effects of sunlight. In fact, shortly the biological effects of sunlight will be evaluated through an analysis of the DNA inactivation rate, mutagenesis frequency and the mutation spectra of the supF gene of pCMUT plasmid, after replication in bacterial strains and by using simple techniques.42
Therefore, we are developing a suitable system capable of fulfilling the basic concept of DNA-based biological dosimetry28 ((i) use of DNA-containing systems assuming them as models of DNA damage in the chromosomes; (ii) quantification of the photobiological/photochemical effect induced by UV radiation; (iii) extrapolation of the measured biological effect from a simple system of a biological dosimeter to a more complex biological effect), providing a wide comprehension of the biological effectiveness of solar-UV radiation upon the DNA molecule. Comparing the results obtained in several places world-wide at different latitudes and periods of the year may reveal important specific light influence in the living organisms.
Acknowledgements
We wish to thank Dr Marcelo Barcellos da Rosa and Pabulo Henrique Rampelotto for assistance during the environmental exposures at SSO. This work was supported by FAPESP (São Paulo, Brazil) and CNPq (Brasília, Brazil).References
- M. M. Caldwell, J. F. Bornman, C. L. Ballare, S. D. Flint and G. Kulandaivelu, Terrestrial ecosystems, increased solar ultraviolet radiation, and interactions with other climate change factors, Photochem. Photobiol. Sci., 2007, 6, 252–266 RSC.
- M. Norval, A. P. Cullen, F. R. de Gruijl, J. Longstreth, Y. Takizawa, R. M. Lucas, F. P. Noonan and J. C. van der Leun, The effects on human health from stratospheric ozone depletion and its interactions with climate change, Photochem. Photobiol. Sci., 2007, 6, 232–251 RSC.
- A. Takahashi and T. Ohnishi, The significance of the study about the biological effects of solar ultraviolet radiation using the Exposed Facility on the International Space Station, Biol. Sci. Space, 2004, 18, 255–260 Search PubMed.
- Z. Kuluncsics, D. Perdiz, E. Brulay, B. Muel and E. Sage, Wavelength dependence of ultraviolet-induced DNA damage distribution: involvement of direct or indirect mechanisms and possible artefacts, J. Photochem. Photobiol., B, 1999, 49, 71–80 CrossRef CAS.
- J. E. Frederick and D. Lubin, Solar ultraviolet irradiance at Palmer Station, Antarctica. Ultraviolet Radiation in Antarctica: Measurements and Biological Effects, 1994 Search PubMed.
- N. Munakata, S. Cornain, M. Kanoko, K. Mulyadi, S. Lestari, W. Wirohadidjojo, D. Bolsee, S. Kazadzis, V. Meyer-Rochow, N. Schuch, C. Casiccia, M. Kaneko, C. M. Liu, K. Jimbow, T. Saida, C. Nishigori, K. Ogata, K. Inafuku, K. Hieda and M. Ichihashi, Biological monitoring of solar UV radiation at 17 sites in Asia, Europe and South America from 1999 to 2004, Photochem. Photobiol., 2006, 82, 689–694 CrossRef CAS.
- J. Rozema, P. Boelen and P. Blokker, Depletion of stratospheric ozone over the Antartic and Arctic: Responses of plants of polar terrestrial ecosystems to enhanced UV-B, an overview, Environ. Poll., 2005, 137, 428–442 CrossRef CAS.
- T. M. Rünger, How different wavelengths of the ultraviolet spectrum contribute to skin carcinogenesis: The role of cellular damage responses, Journal of Investigative Dermatology, 2007, 127, 2103–2105 Search PubMed.
- L. Daya-Grosjean and A. Sarasin, The role of UV induced lesions in skin carcinogenesis: an overview of oncogene and tumor suppressor gene modifications in xeroderma pigmentosum skin tumors, Mutat. Res., 2005, 571, 43–56 CrossRef CAS.
- S. Courdavault, C. Baudouin, M. Charveron, B. Canguilhem, A. Favier, J. Cadet and T. Douki, Repair of the three main types of bipyrimidine DNA photoproducts in human keratinocytes exposed to UVB and UVA radiations, DNA Repair (Amst), 2005, 4, 836–844 CrossRef CAS.
- S. E. Freeman, H. Hacham, R. W. Gange, D. J. Maytum, J. C. Sutherland and B. M. Sutherland, Wavelength dependence of pyrimidine dimer formation in DNA of human skin irradiated in situ with ultraviolet light, Proc. Natl. Acad. Sci. USA, 1989, 86, 5605–5609 CrossRef CAS.
- S. I. Tsilimigaki, N. Messini-Nikolaki, M. Kanariou and S. M. Piperakis, A study on the effects of seasonal solar radiation on exposed populations, Mutagenesis, 2003, 18, 139–143 CrossRef CAS.
- G. P. Pfeifer, Y. H. You and A. Besaratinia, Mutations induced by ultraviolet light, Mutat. Res., 2005, 571, 19–31 CrossRef CAS.
- J. M. Song, J. R. Milligan and B. M. Sutherland, Bistranded oxidized purine damage clusters: induced in DNA by long-wavelength ultraviolet (290–400 nm) radiation, Biochemistry, 2002, 41, 8683–8688 CrossRef CAS.
- T. Douki, A. Reynaud-Angelin, J. Cadet and E. Sage, Bipyrimidine photoproducts rather than oxidative lesions are the main type of DNA damage involved in the genotoxic effect of solar UVA radiation, Biochemistry, 2003, 42, 9221–9226 CrossRef CAS.
- S. Mouret, C. Baudouin, M. Charveron, A. Favier, J. Cadet and T. Douki, Cyclobutane pyrimidine dimers are predominant DNA lesions in whole human skin exposed to UVA radiation, Proc. Natl. Acad. Sci. USA, 2006, 103, 13765–13770 CrossRef CAS.
- R. M. Tyrrell, Induction of pyrimidine dimers in bacterial DNA by 365 nm radiation, Photochem. Photobiol., 1973, 17, 69–73 CrossRef CAS.
- J. H. Yoon, C. S. Lee, T. R. O'Connor, A. Yasui and G. P. Pfeifer, The DNA damage spectrum produced by simulated sunlight, J. Mol. Biol., 2000, 299, 681–693 CrossRef CAS.
- D. Perdiz, P. Grof, M. Mezzina, O. Nikaido, E. Moustacchi and E. Sage, Distribution and repair of bipyrimidine photoproducts in solar UV-irradiated mammalian cells. Possible role of Dewar photoproducts in solar mutagenesis, J. Biol. Chem., 2000, 275, 26732–26742 CAS.
- P. Rettberg and C. S. Cockell, Biological UV dosimetry using the DLR-biofilm, Photochem. Photobiol. Sci., 2004, 3, 781–787 RSC.
- A. George, H. Peat and A. Buma, Evaluation of DNA dosimetry to asses ozone-mediated variability of biologically harmful radiation in Antartica., Photochem. Photobiol., 2002, 76, 274–280 CrossRef CAS.
- H. Yoshida and J. D. Regan, Solar UVB dosimetry by amplification of short and long segments in phage lambda DNA, Photochem. Photobiol., 1997, 66, 672–675 CrossRef CAS.
- R. M. Tyrrell, Solar dosimetry with repair deficient bacterial spores: action spectra, photoproduct measurements and a comparison with other biological systems, Photochem. Photobiol., 1978, 27, 571–579 CrossRef CAS.
- N. Munakata and C. S. Rupert, Genetically controlled removal of “spore photoproduct” from deoxyribonucleic acid of ultraviolet-irradiated Bacillus subtilis spores, J. Bacteriol., 1972, 111, 192–198 CAS.
- M. Puskeppeleit, L. E. Quintern, S. El Naggar, J. U. Schott, U. Eschweiler, G. Horneck and H. Bucker, Long-Term Dosimetry of Solar UV Radiation in Antarctica with Spores of Bacillus subtilis, Appl. Environ. Microbiol., 1992, 58, 2355–2359 CAS.
- G. Ronto, S. Gaspar and A. Berces, Phage T7 in biological UV dose measurement, J. Photochem. Photobiol., B, 1992, 12, 285–294 CrossRef CAS.
- P. Grof, S. Gaspar and G. Ronto, Use of uracil thin layer for measuring biologically effective UV dose, Photochem. Photobiol., 1996, 64, 800–806 CrossRef CAS.
- A. Berces, A. Fekete, S. Gaspar, P. Grof, P. Rettberg, G. Horneck and G. Ronto, Biological UV dosimeters in the assessment of the biological hazard from environmental radiation, J. Photochem. Photobiol., B, 1999, 53, 36–43 CrossRef CAS.
- G. Horneck, P. Rettberg, E. Rabbow, W. Strauch, G. Seckmeyer, R. Facius, G. Reitz, K. Strauch and J. U. Schott, Biological dosimetry of solar radiation for different simulated ozone column thicknesses, J. Photochem. Photobiol., B, 1996, 32, 189–196 CrossRef CAS.
- K. A. Datsenko and B. L. Wanner, One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products, Proc. Natl. Acad. Sci. USA, 2000, 97, 6640–6645 CrossRef CAS.
- L. F. Agnez-Lima, P. Di Mascio, R. L. Napolitano, R. P. Fuchs and C. F. Menck, Mutation spectrum induced by singlet oxygen in Escherichia coli deficient in exonuclease III, Photochem. Photobiol., 1999, 70, 505–511 CrossRef CAS.
- P. Grof, G. Ronto and E. Sage, A computational study of physical and biological characterization of common UV sources and filters, and their relevance for substituting sunlight, J. Photochem. Photobiol., B, 2002, 68, 53–59 CrossRef CAS.
- P. Di Mascio, H. Wefers, H. P. Do-Thi, M. V. Lafleur and H. Sies, Singlet molecular oxygen causes loss of biological activity in plasmid and bacteriophage DNA and induces single-strand breaks, Biochim. Biophys. Acta, 1989, 1007, 151–157 CAS.
- M. Higurashi, T. Ohtsuki, A. Inase, R. Kusumoto, C. Masutani, F. Hanaoka and S. Iwai, Identification and characterization of an intermediate in the alkali degradation of (6-4) photoproduct-containing DNA, J. Biol. Chem., 2003, 278, 51968–51973 CrossRef CAS.
- R. M. Tyrrell, R. D. Ley and R. B. Webb, Induction of single-strand breaks (alkali-labile bonds) in bacterial and phage DNA by near UV (365 nm) radiation, Photochem. Photobiol., 1974, 20, 395–398 CrossRef CAS.
- J. Cadet, E. Sage and T. Douki, Ultraviolet radiation-mediated damage to cellular DNA, Mutat. Res., 2005, 571, 3–17 CrossRef CAS.
- C. Kielbassa, L. Roza and B. Epe, Wavelength dependence of oxidative DNA damage induced by UV and visible light, Carcinogenesis, 1997, 18, 811–816 CrossRef CAS.
- J. Sambrook, E. F. Fritsch and T. Maniats, Cold Spring Harbor Laboratory Press, New York, 2nd edn., 1989, vol. 1, ch. 1, pp. 1.42-41.46.
- S. Kanno, S. Iwai, M. Takao and A. Yasui, Repair of apurinic/apyrimidinic sites by UV damage endonuclease; a repair protein for UV and oxidative damage, Nucleic Acids Res., 1999, 27, 3096–3103 CrossRef CAS.
- L. S. Kan, L. Voituriez and J. Cadet, The Dewar valence isomer of the (6-4) photoadduct of thymidylyl-(3′-5′)-thymidine monophosphate: formation, alkaline lability and conformational properties, J. Photochem. Photobiol., B, 1992, 12, 339–357 CrossRef CAS.
- C. A. Smith and J. S. Taylor, Preparation and characterization of a set of deoxyoligonucleotide 49-mers containing site-specific cis-syn, trans-syn-I, (6-4), and Dewar photoproducts of thymidylyl(3′–>5′)-thymidine, J. Biol. Chem., 1993, 268, 11143–11151 CAS.
- R. R. Ariza, T. Roldan-Arjona, C. Hera and C. Pueyo, A method for selection of forward mutations in supF gene carried by shuttle-vector plasmids, Carcinogenesis, 1993, 14, 303–305 CrossRef CAS.
| This journal is © The Royal Society of Chemistry and Owner Societies 2009 |
