Very long-chain fatty tails for enhanced transfection†
Received 9th September 2008, Accepted 15th October 2008
First published on 26th November 2008
Abstract
The long chain saturated fatty acids, arachidic (C20) and lignoceric (C24), are found as components of phospholipids within mammalian cellular membranes. Although these lipids have rarely been used as components of transfection reagents, we recently demonstrated that elongation of the fatty tail beyond C18 provide a means of increasing the transfection efficiency of cationic lipids. To investigate this effect further, a new library of single-chained cationic lipids consisting of mono-, di- or tri-arginine residues, a range of amino acid spacers and these long-chain saturated fatty tails were synthesised using an Fmoc solid-phase strategy, which allowed the preparation of 18 compounds, some with remarkable transfection abilities.
Introduction
To enable gene therapy to become a practical therapeutic option, chemistry tools need to be developed to “carry” and “release” genetic material into the host cell and, if required, mediate delivery into the nucleus, while being benign to cellular functionality. Vectors based upon viral systems (adenoviruses, retroviruses, etc.)1 have been found to be highly efficient for delivering DNA into cells but have a number of major issues (e.g. toxicity, immunogencity, limited capacity, difficulties associated with production, purification and storage) that have raised concerns about their medical application.2,3There have been many efforts to provide efficient chemical substitutes to viral carriers, that have resulted in the development of a broad range of delivery systems.4 Cationic polymers and cationic lipids, in particular have been targeted, and although less efficient than viruses, have inherent advantages over viral vectors such as a larger packing capacity of nucleic acids and generally low immunogenicity, while being readily synthesisable.5
Since 1987, when Felgner and co-workers6 pioneered gene delivery with cationic lipids, a large number of cationic lipids have been developed, including DOTMA61, DOSPA72, and DOGS83 (see Fig. 1), which have been commercialized under a variety of names. In general, the structure of any cationic lipid consist of three parts: (i) a positively-charged polar head-group, (ii) a “spacer group”, and (iii) a non-polar component. Usually, polar domains are nitrogen-based motifs while the hydrophobic part typically comprises two fatty tails or one cholesteryl moiety. The amphiphilic nature of the cationic lipids imparts on the ability to interact electrostatically with DNA to form a complex known as a lipoplex and allows the generation of “membrane-like” structures in water. The best carriers are able to produce lipoplexes that can be engulfed by cells via non-specific endocytosis and then release the endosomally-trapped plasmid DNA into the cytoplasm.9 The addition of neutral lipids (so-called “helper” lipids) such as DOPE10,11 or cholesterol,12,13 improve the transfection abilities of the complexes by promoting the escape of DNA from the early endosome.14
 |
| | Fig. 1 Structure of cationic lipids. | |
The design of cationic lipids makes them excellent candidates for chemistry studies, allowing an investigation of structure–activity relationships within families of reagents.15 In recent years libraries of potential transfection reagents have been reported,16–21 with single-chained lipids containing one17, two18 or three21 cationic head groups (compounds 4, 5 and 6, Fig. 1) showing high activity. It was observed21 that elongation of the fatty tail beyond C18 could be an attractive means of increasing the transfection efficiency of cationic lipids. To investigate the effect mediated by these under-utilised lipid moieties on the transfection abilities of arginine-lipid conjugates,17 a new library of single-chained cationic lipids consisting of very long-chain saturated fatty tails (>C18) were synthesized. The library was prepared using a solid-phase approach and combined a range of amino acid spacers, arginine residues, and arachidic and lignoceric acid, giving 18 mono-, di- or tri-arginine lipid conjugates.
Results and discussion
1. Synthesis of cationic lipids
Single-chained cationic lipids were assembled on aminomethyl polystyrene resin (loading 1.01 mmol g−1, 1% DVB) functionalized with a Rink amide linker using an Fmoc-based solid-phase strategy.22,23 Fmoc-Arg(Pbf)-OH was coupled onto resin 7 using HOBt–DIC followed by Fmoc-deprotection to give 8 (see Scheme 1). Repetition of the synthetic steps gave di- or tri-arginine scaffolds 9 and 10, respectively. Fmoc-protected amino acid (glycine, β-alanine, γ-aminobutyric acid and 6-aminohexanoic acid) were coupled onto each of the three arginine scaffold resins, and subsequently, either arachidic (C20) or lignoceric (C24) acid were attached onto the corresponding scaffolds and then cleaved from the resin under acidic conditions using TFA–TIS–H2O (95 : 2.5 : 2.5) mixture to give rise to the final TFA salts (see Table 1 and Scheme 1). |
| | Scheme 1 Reagents and conditions: (i) Fmoc-Arg(Pbf)-OH (3 eq.), DIC (3 eq.), HOBt (3 eq), DCM–DMF (2 : 1, 4.5 ml); (ii) 20% piperidine in DMF; (iii) Fmoc-protected spacer (3 eq.), DIC (3 eq.), HOBt (3 eq.), DCM–DMF (2 : 1, 4.5 ml); (iv) arachidic or lignoceric acid (2 eq.), DIC (2 eq.), HOBt (2 eq.), DMF–DCM–THF (2 : 1 : 1, 4.5 ml); (v) TFA–TIS–H2O (95 : 2.5 : 2.5). | |
Table 1 List of cationic lipids synthesised
| Compound | Fatty tail | Spacera | Arginine no. |
|---|
| Gly = glycine; βAla = β-alanine; Abu = 4-aminobutyric acid; Ahx = 6-aminohexanoic acid. |
|---|
| 11a | C20 | Gly | 1 |
| 11b | C20 | βAla | 1 |
| 11c | C20 | Abu | 1 |
| 11d | C20 | Ahx | 1 |
| 11e | C24 | Gly | 1 |
| 11f | C24 | βAla | 1 |
| 11g | C24 | Abu | 1 |
| 12a | C20 | Gly | 2 |
| 12b | C20 | βAla | 2 |
| 12c | C20 | Ahx | 2 |
| 12d | C24 | Gly | 2 |
| 12e | C24 | βAla | 2 |
| 12f | C24 | Abu | 2 |
| 13a | C20 | Gly | 3 |
| 13b | C20 | Abu | 3 |
| 13c | C20 | Ahx | 3 |
| 13d | C24 | βAla | 3 |
| 13e | C24 | Ahx | 3 |
2. Gel retardation assay
The binding affinities of the transfection compounds for DNA were studied by agarose gel retardation assays. As the main interactions between cationic lipids and DNA are ionic, these two components were mixed as a function of N/P charge ratios.24 Representative electrophoretic gel patterns of cationic lipid–DNA complexes are shown in Fig. 2. Analysis showed that the number of arginine residues and the length of hydrophobic tail had a major effect on the ability of the compounds to retard DNA mobility, while the type of spacer had no significant effect. Compounds containing one arginine head-group and the arachidyl tail (11a–d) displayed a poor ability to complex DNA, while lipids 12a–c and 13a–c (two or three arginine residues and arachidyl moiety) showed a high capacity to inhibit plasmid mobility regardless of the spacer. None of the compounds containing the lignoceryl tail (11f–h, 12d–f, 13d–e), which is the longest and most hydrophobic tail used, was able to fully prevent DNA movement regardless of the number of arginine residues and the type of spacer. However, as it will be shown below, gel retardation appears to offer little relevance to cellular DNA delivery.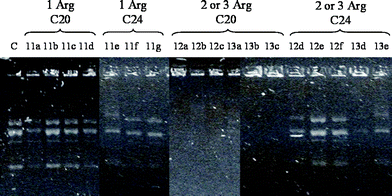 |
| | Fig. 2 Gel retardation assay. Compounds were complexed with pEGFP-C1 at a N/P charge ratio of 10, loaded in an agarose gel (1% agarose, 1 μg mL−1 ethidium bromide in water), and run at 100 V for 30 min. C = naked pEGFP-C1. | |
3. Screening of transfection abilities
The transfection abilities of the cationic lipids were studied using a variety of cell lines (human ovarian cancer (HeLa), human embryonic kidney (HEK293T) and mouse melanoma (B16F10), a difficult-to-transfect cell line).25 The lipoplexes were formulated at two N/P ratios (5 and 10), using DOPE as a helper lipid (molar ratio of 1 : 1 and 1 : 2 cationic-lipid–DOPE) with pEGFP-C1 as a GFP-reporter.26 All experiments were performed in serum-containing media, tested in triplicate and compared with two commercially available compounds: Effectene (Qiagen) and Lipofectamine 2000 (Invitrogen). Cellular fluorescence was determined using a fluorescence microplate reader (excitation 488 nm) after 48 h incubation and confirmed by microscopy and flow cytometry.As expected, analysis showed that gene transfer abilities were strongly dependent on the length of the hydrophobic tail and the number of arginine residues. Multi-arginine derivatives were significantly more active than those compounds comprising a single arginine residue (Fig. 3). Cells treated with most of the mono-arginine derivatives showed no transfection, with the exception of 11e (1 Arg, Gly, C24), which showed significant transfection efficacy in all three cell lines tested. Remarkable gene expression was detected in HeLa and HEK293T cells with the di- and tri-arginine derivatives at various N/P ratios (see Fig. 3A,B), highlighting the transfection efficacy demonstrated by compound 12e (2 Arg, βAla, C24) when formulated with DOPE at a 1 : 1 molar ratio and complexed with pEGFP-C1 at N/P ratio 5. Although most of the compounds showed low transfection results when tested with B16F10 cells, compound 12e was very efficient and significantly better than the positive controls Effectene and Lipofectamine 2000 (see Fig. 3C).
 |
| | Fig. 3 Analysis of cell fluorescence (arbitrary units) 48 h after transfection of pEGFP-C1 (0.2 μg per well) with all library members and positive controls: (A) HeLa, (B) HEK 293T, (C) B16F10 cells. | |
Transfection efficiency of compound 12e in B16F10 cells was analyzed via flow cytometry and compared with Effectene and Lipofectamine 2000. Compounds 4 (the best transfection reagent from previous work in the group),1714 (the C18 counterpart of 12e)17 and 6 (the C24 tripodal cationic lipid)21 were also assayed in order to compare the transfection efficiency of 12e with structurally related materials. Lipoplexes formulated with 12e : DOPE (1 : 1) at N/P 5 again demonstrated the greatest transfection levels, with 65% of the cells having fluorescence above background (Fig. 4).19 Interestingly, the C18 counterpart and derivative 14, showed very low transfection abilities (5%).
 |
| | Fig. 4 (A) Percentage of the B16F10 cell population expressing eGFP. Cell fluorescence was analyzed via flow cytometry. (B,C) Flow cytometry histograms of B16F10 cells: (B) control and (C) after transfection with compound 12e–DOPE (1 : 1) at N/P 5. The histogram shows cell number (y axis) relative to fluorescence (x axis). | |
4. Transfection cytotoxicity
The cytotoxicity of the cationic lipid formulations was examined by measuring changes in the cell metabolic activity (MTT assay)27 of the cells after 4 hours incubation with the compounds. As illustrated in Fig 5, compounds showed very low toxicity in all the cell lines used with the exception of compound 13a when formulated with N/P 10.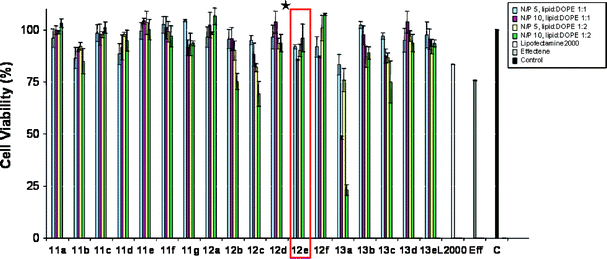 |
| | Fig. 5 MTT cell viability assays of the library members with B16F10 cells. *12e the most active compound. | |
Conclusions
A Fmoc solid phase strategy was applied to the preparation of an 18-membered library of arginine lipid conjugates consisting of very long-chain saturated fatty tails. The transfection ability of the library members was tested with a set of mammalian cell lines, with some of the derivatives showing highly motivating transfection properties. In particular, derivative 12e (consisting of 2 arginine headgroups, aminobutyryl spacer and lignoceryl tail) had remarkable gene delivery abilities, leading to 65% transfection with B16F10 cells (a difficult-to-transfect cell line). Once again, these results point to the importance of the lipid moiety for the transfection abilities of cationic lipids and underline the great potential of using long-chain saturated fatty acids in order to improve the transfection properties of existing amphiphilic molecules.Experimental section
1.1. General information
All commercially available chemicals were reagent grade and were used without further purification. Unless otherwise indicated, all the reactions were performed at room temperature. NMR spectra were recorded using a Bruker AC250 spectrometer operating at 250 MHz for 1H at 298 K. Chemical shifts are reported in ppm and were referenced to residual protio solvents resonances. All coupling constants (J values) were measured in Hz. IR spectra were recorded on a Bruker Tensor 27 FT-IR with a golden SPECAC gate accessory with neat compounds. Low resolution mass spectra (LRMS) were recorded using a VG Platform Quadrupole Electrospray Ionisation (ES+) mass spectrometer. High-resolution fast-atom bombardment (FAB+) mass spectrometry was recorded on KRATOS MS50TC.1.2. Synthesis of resin with the Rink linker (7)
The Fmoc-Rink-amide linker (3 eq., 0.6 mmol) was dissolved in DCM–DMF (2 : 1, 4.5 mL). DIC (3 eq., 0.6 mmol) and HOBt (3 eq., 0.6 mmol) were added and the mixture was left to stir for 5 min. This solution was added to aminomethyl polystyrene resin (1.01 mmol g−1, 200 mg) and shaken overnight. The resulting resin was washed with DCM, DMF, MeOH, DMF and DCM (3 × 5 mL each) and then treated with a solution of 20% piperidine in DMF (2 × 10 min) followed by washing with DCM, DMF, MeOH, DMF and DCM (3 × 5 mL each).1.3. Synthesis of arginine scaffold resin (7–9)
The mono-arginine scaffold resin 8 was synthesized by the coupling of the resin 7 with a solution of Fmoc-L-arginine(Pbf)-OH (3 eq., 0.6 mmol), DIC (3 eq., 0.6 mmol) and HOBt (3 eq., 0.6 mmol) in DMF–DCM (2 : 1, 4.5 mL). The mixture was shaken overnight. The resulting resin was washed with DCM, DMF, MeOH, DMF and DCM (3 × 5 mL each) and then treated with a solution of 20% piperidine in DMF (2 × 10 min). The resulting resin 8 was washed with DCM, DMF, MeOH, DMF and DCM (3 × 5 mL each). Repetition of this synthetic method once or twice afforded di- and tri-arginine scaffold resins 9 and 10.1.4. Synthesis of arginine scaffold resin with spacers
The arginine scaffold resins 8–10 were reacted with Fmoc-protected glycine, β-alanine, 4-aminobutyric acid or 6-aminohexanoic acid spacers (3 eq., 0.6 mmol) using DIC (3 eq., 0.6 mmol) and HOBt (3 eq., 0.6 mmol) in DMF–DCM (2 : 1, 4.5 mL). The suspensions were shaken for 2 hours and washed with DCM, DMF, MeOH, DMF and DCM (3 × 5 mL each). The resulting resins were treated with a solution of 20% piperidine in DMF (2 × 10 min) and washed with DCM, DMF, MeOH, DMF and DCM (3 × 5 mL each).1.5. Coupling of carboxylic acids
A stock solution (4.5 mL), consisting of the corresponding carboxylic acid (2 eq, 0.4 mmol), DIC (2 eq, 0.4 mmol) and HOBt (2 eq, 0.4 mmol) in DMF–DCM–THF, was added to the resins and the suspensions were shaken for 6 h. The resulting resins were washed with DCM, DMF, MeOH, DMF and DCM (3 × 5 mL each) and dried under vacuum.1.6. Cleavage of the product from the resin
The resins were pre-swollen for 15 min in DCM and filtered. A solution of TFA–TIS–H2O (95 : 2.5 : 2.5, 3 mL) was added to the resins and the suspensions were shaken for 2 h. The solvents were removed in vacuo. The resulting products were redissolved in DCM and precipitated with Et2O. The solutions were centrifuged and the solvent was removed using a pipette. The desired products were further dried under vacuum for 2 h.N-(N′-Eicosanoyl-2-aminoacetyl)-L-argininamide, TFA salt (11a). Yield: 82%. 1H NMR (250 MHz, DMSO) δ: 0.85 (t, J = 7, 3H); 1.08–1.85 (m, 38H); 2.11 (t, J = 7, 2H); 3.02–3.14 (m, 2H); 3.62–3.80 (m, 2H); 4.11–4.31 (m, 1H); 7.1 (br. s, 1H); 7.37 (br. s 1H); 7.92 (d, J = 8, 1H); 8.01–8.15 (m, 1H). IR ν: 3304, 3193 (N–H); 2916 (C–H); 1648 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O); δ: 1530 (N–H); 1471 (C–H) cm−1. MS (ES+): m/z (%): 525.5 (100) [M + H]+. HRMS calcd for C28H57N6O3 525.4486 ([M + H]+), mass found m/z: 525.4492.
O); δ: 1530 (N–H); 1471 (C–H) cm−1. MS (ES+): m/z (%): 525.5 (100) [M + H]+. HRMS calcd for C28H57N6O3 525.4486 ([M + H]+), mass found m/z: 525.4492. N-(N′-Eicosanoyl-3-aminopropanoyl)-L-argininamide, TFA salt (11b). Yield: 91%. 1H NMR (250 MHz, DMSO) δ = 0.85 (t, J 7, 3H); 1.09–1.7 (m, 38H); 2.11 (t, J 7, 2H); 3.01–3.16 (m, 4H); 3.28–3.4 (m, 2H); 4.12–4.22 (m, 1H); 7.03 (br. s, 1H; COHNH); 7.33 (br. s 1H); 7.85 (d, J 8, 1H). IR ν: 3298, 3197 (N–H); 2917 (C–H); 1641 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O); δ: 1540 (N–H), 1471 (C–H) cm−1. MS (ES+): m/z (%): 539.5 (100) [M + H]+. HRMS calcd for C29H59N6O3 539.4643 ([M + H]+), mass found m/z: 539.4647.
O); δ: 1540 (N–H), 1471 (C–H) cm−1. MS (ES+): m/z (%): 539.5 (100) [M + H]+. HRMS calcd for C29H59N6O3 539.4643 ([M + H]+), mass found m/z: 539.4647. N-(N′-Eicosanoyl-4-aminobutanoyl)-L-argininamide, TFA salt (11c). Yield: 63%. 1H NMR (250 MHz, DMSO) δ: 0.85 (t, J = 7, 3H); 1.12–1.72 (m, 40H); 2.03 (t, J = 7, 2H); 2.13 (t, J = 7, 2H); 2.92–3.15 (m, 4H); 4.11–4.25 (m, 1H); 7.03 (br. s, 1H); 7.37 (br. s 1H); 7.50–7.62 (m, 1H); 7.77 (t, J = 6, 1H); 7.91 (d, J = 8, 1H). IR ν: 3296, 3195 (N–H); 2918 (C–H); 1639 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O); δ: 1543 (N–H); 1463 (C–H) cm−1. MS (ES+): m/z (%): 553.6 (100) [M + H]+. HRMS calcd for C30H61N6O3 553.4799 ([M + H]+), mass found m/z: 553.4806.
O); δ: 1543 (N–H); 1463 (C–H) cm−1. MS (ES+): m/z (%): 553.6 (100) [M + H]+. HRMS calcd for C30H61N6O3 553.4799 ([M + H]+), mass found m/z: 553.4806. N-(N′-Eicosanoyl-6-aminohexanoyl)-L-argininamide, TFA salt (11d). Yield: 90%. 1H NMR (250 MHz, DMSO) δ: 0.85 (t, J = 7, 3H); 1.11–1.57 (m, 44H); 2.01 (t, J = 7, 2H); 2.10 (t, J = 7, 2H); 2.98 (dd, J = 7, 13 2H); 3.05–3.16 (m, 2H); 4.11–4.25 (m, 1H); 6.93 (br. s. 1H); 7.13 (br. s. 1H); 7.33 (br. s. 1H), 7.57 (t, J = 6, 1H); 7.74 (t, J = 6, 1H); 8.08 (d, J = 8, 1H). IR ν: 3288, 3191 (N–H); 2917 (C–H); 1619 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O); δ: 1542 (N–H); 1464 (C–H) cm−1. MS (ES+): m/z (%): 581.5 (100) [M + H]+. HRMS calcd for C32H64N6O3 581.5112 ([M + H]+), mass found m/z: 581.5112.
O); δ: 1542 (N–H); 1464 (C–H) cm−1. MS (ES+): m/z (%): 581.5 (100) [M + H]+. HRMS calcd for C32H64N6O3 581.5112 ([M + H]+), mass found m/z: 581.5112. N-(N′-Tetracosanoyl-2-aminoacetyl)-L-argininamide, TFA salt (11e). Yield: 92%. 1H NMR (250 MHz, DMSO) δ: 0.8–0.9 (m, 3H); 1.05–1.76 (m, 46H); 2.06–2.2 (m, 2H); 3.0–3.15 (m, 2H); 3.69–3.85 (m, 2H); 4.09–4.29 (m, 1H); 7.07 (br. s, 1H); 7.13 (br. s 1H); 7.55–7.65 (m, 1H); 7.84–8.05 (m, 1H). IR ν: 3295, 3193 (N–H); 2915 (C–H); 1649 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O); δ: 1535 (N–H); 1471 (C–H) cm−1. MS (ES+): m/z (%): 581.6 (100) [M + H]+. HRMS calcd for C32H65N6O3 581.5112 ([M + H]+), mass found m/z: 581.5111.
O); δ: 1535 (N–H); 1471 (C–H) cm−1. MS (ES+): m/z (%): 581.6 (100) [M + H]+. HRMS calcd for C32H65N6O3 581.5112 ([M + H]+), mass found m/z: 581.5111. N-(N′-Tetracosanoyl-3-aminopropanoyl)-L-argininamide, TFA salt (11f). Yield: 65%. 1H NMR (250 MHz, DMSO) δ: 0.82–0.92 (m, 3H); 1.27–1.73 (m, 46H); 1.98–2.1 (m, 2H); 2.28–2.37 (m, 2H); 3.03–3.17 (m, 2H); 3.20–3.42 (m, 2H); 4.12–4.3 (m, 1H); 7.09 (br. s, 1H); 7.33 (br. s 1H); 7.44 (d, J = 8, 1H); 7.78–8.23 (m, 1H). IR ν: 3295, 3188 (N–H); 1639 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O); 2919 (C–H); δ: 1542 (N–H); 1472 (C–H) cm−1. MS (ES+): m/z (%): 595.5 (75) [M + H]+. HRMS calcd for C33H67N6O3 595.5269 ([M + H]+), mass found m/z: 595.5265.
O); 2919 (C–H); δ: 1542 (N–H); 1472 (C–H) cm−1. MS (ES+): m/z (%): 595.5 (75) [M + H]+. HRMS calcd for C33H67N6O3 595.5269 ([M + H]+), mass found m/z: 595.5265. N-(N′-Tetracosanoyl-4-aminobutanoyl)-L-argininamide, TFA salt (11g). Yield: 81%. 1H NMR (250 MHz, DMSO) δ: 0.85 (t, J = 7, 3H); 1.11–1.51 (m, 48H); 2.04–2.15 (m, 4H); 3.02–3.14 (m, 4H); 4.15–4.25 (m, 1H); 7.09 (br. s, 1H); 7.30 (br. s 1H); 7.42–7.50 (m, 1H); 7.83 (d, J = 8, 1H); 8.02 (d, J = 7, 1H). IR: ν: 3293, 3190 (N–H); 2915 (C–H); 1643 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O); δ: 1542 (N–H); 1471 (C–H) cm−1. MS (ES+): m/z (%): 609.6 (100) [M + H]+. HRMS calcd for C34H69N6O3 609.5425 ([M + H]+), mass found m/z: 609.5412.
O); δ: 1542 (N–H); 1471 (C–H) cm−1. MS (ES+): m/z (%): 609.6 (100) [M + H]+. HRMS calcd for C34H69N6O3 609.5425 ([M + H]+), mass found m/z: 609.5412. N-[N′-(N″-Eicosanoyl-(2-aminocaetyl))-L-argininyl]-L-argininamide, TFA salt (12a). Yield: 62%. 1H NMR (250 MHz, DMSO) δ: 0.85 (t, J = 6, 3H); 1.11–1.77 (m, 42H); 2.13 (t, J = 7, 2H); 3.02–3.15 (m, 4H); 3.6–3.85 (m, 2H); 4.05–4.22 (m, 2H); 7.0–7.29 (m, 8H); 7.98–8.16 (m, 3H); 8.28 (t, J = 6, 1H); 8.49 (d, J = 6, 1H). IR ν: 3278, 3182 (N–H); 2917 (C–H); 1650 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O); δ: 1536 (N–H); 1466 (C–H) cm−1. MS (ES+): m/z (%): 341.4 (100) [M + 2H]2+, 681.7 (15) [M + H]+. HRMS calcd for C34H69N10O4 681.5497 ([M + H]+), mass found m/z: 681.5491.
O); δ: 1536 (N–H); 1466 (C–H) cm−1. MS (ES+): m/z (%): 341.4 (100) [M + 2H]2+, 681.7 (15) [M + H]+. HRMS calcd for C34H69N10O4 681.5497 ([M + H]+), mass found m/z: 681.5491. N-[N′-(N″-Eicosanoyl-(3-aminopropanoyl))-L-argininyl]-L-argininamide, TFA salt (12b). Yield: 92%. 1H NMR (250 MHz, DMSO) δ: 0.85 (t, J = 7, 3H); 1.13–1.75 (m, 42H); 2.02 (t, J = 7, 2H); 2.32 (t, J = 7, 2H); 3.0–3.13 (m, 4H); 3.18–3.29 (m, 2H); 4.07–4.22 (m, 2H); 7.02–7.45 (m, 8H); 7.75–7.94 (m, 3H); 8.0–8.08 (m, 1H); 8.33–8.42 (m, 1H). IR ν: 3280, 3187 (N–H); 2917 (C–H); 1630 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O); δ: 1538 (N–H); 1467 (C–H) cm−1. MS (ES+): m/z (%): 348.4 (100) [M + 2H]2+. HRMS calcd for C35H71N10O4 695.5654 ([M + H]+), mass found m/z: 695.5652.
O); δ: 1538 (N–H); 1467 (C–H) cm−1. MS (ES+): m/z (%): 348.4 (100) [M + 2H]2+. HRMS calcd for C35H71N10O4 695.5654 ([M + H]+), mass found m/z: 695.5652. N-[N′-(N″-Eicosanoyl-(6-aminohexanoyl))-L-argininyl]-L-argininamide, TFA salt (12c). Yield: 91%. 1H NMR (250 MHz, DMSO) δ: 1H NMR (250 MHz, DMSO) δ = 0.85 (t, J 7, 3H); 1.15–1.75 (m, 48H); 2.02 (t, J 7, 2H); 2.1–2.2 (m, 2H); 2.94–3.15 (m, 6H); 4.01–4.24 (m, 2H); 7.05–7.38 (m, 8H); 7.72–7.83 (m, 3H); 7.93–8.05 (m, 1H); 8.15–8.23 (m, 1H). IR ν: 3275, 3187 (N–H); 2917 (C–H); 1630 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O); δ: 1542 (N–H); 1466 (C–H) cm−1. MS (ES+): m/z (%): 369.4 (100) [M + 2H]2+. HRMS calcd for C38H77N10O4 737.6123 ([M + H]+), mass found m/z: 737.6107.
O); δ: 1542 (N–H); 1466 (C–H) cm−1. MS (ES+): m/z (%): 369.4 (100) [M + 2H]2+. HRMS calcd for C38H77N10O4 737.6123 ([M + H]+), mass found m/z: 737.6107. N-[N′-(N″-Tetracosanoyl-(2-aminoacetyl))-L-argininyl]-L-argininamide, TFA salt (12d). Yield: 74%. 1H NMR (250 MHz, DMSO) δ: 0.84 (t, J = 7, 3H); 1.12–1.72 (m, 50H); 2.10 (t, J = 7, 2H); 3.02–3.15 (m, 8H); 3.67–3.72 (m, 2H); 6.92–7.35 (m, 2H); 7.50–7.60 (m, 3H); 7.98 (d, J = 8, 1H); 8.11 (dd, J = 7, 15 3H). IR ν: 3282, 3183 (N–H); 2916 (C–H); 1650 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O); δ: 1539 (N–H); 1466 (C–H) cm−1. MS (ES+): m/z (%): 369.4 (100) [M + 2H]2+. HRMS calcd for C38H77N10O4 737.6123 ([M + H]+), mass found m/z: 737.6122.
O); δ: 1539 (N–H); 1466 (C–H) cm−1. MS (ES+): m/z (%): 369.4 (100) [M + 2H]2+. HRMS calcd for C38H77N10O4 737.6123 ([M + H]+), mass found m/z: 737.6122. N-[N′-(N″-Tetracosanoyl-(3-aminopropanoyl))-L-argininyl]-L-argininamide, TFA salt (12e). Yield: 80%. 1H NMR (250 MHz, DMSO) δ: 0.85 (t, J 6, 3H); 1.11–1.78 (m, 50H); 2.02 (t, J = 7, 2H); 2.30 (t, J = 7, 2H); 3.0–3.15 (m, 6H); 3.18–3.52 (m, 8H); 4.18–4.28 (m, 2H); 7.31 (br. s, 1H); 7.54 (br. s, 1H); 7.75–7.85 (m, 1H); 7.94–7.82 (m, 1H); 8.15–8.23 (m, 1H). IR ν: 3274, 3185 (N–H); 2916 (C–H); 1636 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O); δ: 1541 (N–H); 1468 (C–H) cm−1. MS (ES+): m/z (%): 376.4 (100) [M + 2H]2+. HRMS calcd for C39H79N10O4 751.6280 ([M + H]+), mass found m/z: 751.6280.
O); δ: 1541 (N–H); 1468 (C–H) cm−1. MS (ES+): m/z (%): 376.4 (100) [M + 2H]2+. HRMS calcd for C39H79N10O4 751.6280 ([M + H]+), mass found m/z: 751.6280. N-[N′-(N″-Tetracosanoyl-(4-aminobutanoyl))-L-argininyl]-L-argininamide, TFA salt (12f). Yield: 80%. 1H NMR (250 MHz, DMSO) δ: 0.84 (t, J = 7, 3H); 1.12–1.66 (m, 54H); 2.03 (t, J = 7, 2H); 2.13 (t, J = 7, 2H); 2.95–3.15 (m, 6H); 4.1–4.25 (m, 2H); 7.31 (br. s, 1H); 7.54 (br. s, 1H); 6.86–7.32 (m, 8H); 7.52–7.62 (m, 1H); 7.73–85 (m, 1H); 7.92–8.05 (m, 1H). IR ν: 3286, 3187 (N–H); 2917 (C–H); 1633 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O); δ: 1539 (N–H); 1467 (C–H) cm−1. MS (ES+): m/z (%): 383.5 (100) [M + 2H]2+. HRMS calcd for C40H81N10O4 765.6436 ([M + H]+), mass found m/z: 765.6406.
O); δ: 1539 (N–H); 1467 (C–H) cm−1. MS (ES+): m/z (%): 383.5 (100) [M + 2H]2+. HRMS calcd for C40H81N10O4 765.6436 ([M + H]+), mass found m/z: 765.6406. N-[N′-(N″-[N‴-Eicosanoyl-(2-aminoacetyl)]-L-argininyl)-L-argininyl]-L-argininamide, TFA salt (13a). Yield: 56%. 1H NMR (250 MHz, DMSO) δ: 0.8–0.9 (m 3H); 1.13–1.78 (m, 46H); 2.08–2.17 (m, 2H); 2.95–3.19 (m, 6H); 3.28–3.41 (m, 2H); 4.0–4.35 (m, 3H); 6.98–7.21 (m, 12H); 7.72–9.15 (m, 6H). IR ν: 3273, 3178 (N-H); 2920 (C–H); 1644 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O); δ: 1536 (N–H); 1454 (C–H) cm−1. MS (ES+): m/z (%): 280.1 (100) [M + 3H]3+, 419.8 (50) [M + 2H]2+. HRMS calcd for C40H81N14O5 837.6508 ([M + H]+), mass found m/z: 837.6547.
O); δ: 1536 (N–H); 1454 (C–H) cm−1. MS (ES+): m/z (%): 280.1 (100) [M + 3H]3+, 419.8 (50) [M + 2H]2+. HRMS calcd for C40H81N14O5 837.6508 ([M + H]+), mass found m/z: 837.6547. N-[N′-(N″-[N‴-Eicosanoyl-(4-aminobutanoyl)]-L-argininyl)-L-argininyl]-L-argininamide, TFA salt (13b). Yield: 67%. 1H NMR (250 MHz, DMSO) δ: 0.84 (t, J = 7, 3H); 1.12–1.9 (m, 48H); 2.03 (t, J = 7, 2H); 2.14 (t, J = 7, 2H); 2.92–3.14 (m, 8H); 4.04–4.29 (m, 3H); 6.88–7.37 (m, 12H); 7.77–7.9 (m, 1H); 8.02 (br. s, 2H); 8.25–8.32 (m, 2H); 8.59–8.69 (m, 1H). IR ν: 3294, 3188 (N–H); 2917 (C–H); 1642 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O); δ: 1538 (N–H); 1466 (C–H) cm−1. MS (ES+): m/z (%): 289.4 (100) [M + 3H]3+, 433.5 (30) [M + 2H]2+. HRMS calcd for C42H85N14O5 865.6822 ([M + H]+), mass found m/z: 865.6822.
O); δ: 1538 (N–H); 1466 (C–H) cm−1. MS (ES+): m/z (%): 289.4 (100) [M + 3H]3+, 433.5 (30) [M + 2H]2+. HRMS calcd for C42H85N14O5 865.6822 ([M + H]+), mass found m/z: 865.6822. N-[N′-(N″-[N‴-Eicosanoyl-(6-aminohexanoyl)]-L-argininyl)-L-argininyl]-L-argininamide, TFA salt (13c). Yield: 70%. 1H NMR (250 MHz, DMSO) δ: 0.84 (t, J = 7, 3H); 1.1–1.8 (m, 52H); 2.09 (t, J = 7, 2H); 2.3 (t, J = 7, 2H); 3.0–3.18 (m, 6H); 3.2–3.3 (m, 2H); 4.04–4.29 (m, 2H); 6.88–7.36 (m, 12H); 7.77–8.0 (m, 3H); 8.18–8.26 (m, 2H); 8.42–8.52 (m, 1H). IR ν: 3272, 3176 (N–H); 2919 (C–H); 1630 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O); δ: 1542 (N–H); 1454 (C–H) cm−1. MS (ES+): m/z (%): 298.71 (100) [M + 3H]3+, 447.5 (35) [M + 2H]2+. HRMS calcd for C44H89N14O5 893.7135 ([M + H]+), mass found m/z: 893.7135.
O); δ: 1542 (N–H); 1454 (C–H) cm−1. MS (ES+): m/z (%): 298.71 (100) [M + 3H]3+, 447.5 (35) [M + 2H]2+. HRMS calcd for C44H89N14O5 893.7135 ([M + H]+), mass found m/z: 893.7135. N-[N′-(N″-[N‴-Tetracosanoyl-(3-aminopropanoyl)]-L-argininyl)-L-argininyl]-L-argininamide, TFA salt (13d). Yield: 85%. 1H NMR (250 MHz, DMSO) δ: 0.78–0.96 (m, 3H); 0.9–1.81 (m, 54H); 2.1–2.4 (m, 4H); 2.92–3.2 (m, 6H); 3.92–3.2 (m, 6H); 3.65–3.84 (m, 2H); 4.02–4.3 (m, 3H); 6.82–7.24 (m, 4H); 7.73–8.6 (m, 6H). IR ν: 3303, 3192 (N–H); 2916 (C–H); 1637 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O); δ: 1541 (N–H); 1462 (C–H) cm−1. MS (ES+): m/z (%): 303.1 (100) [M + 3H]3+. HRMS calcd for C45H90N14O5 907.7291 ([M + H]+), mass found m/z: 907.7288.
O); δ: 1541 (N–H); 1462 (C–H) cm−1. MS (ES+): m/z (%): 303.1 (100) [M + 3H]3+. HRMS calcd for C45H90N14O5 907.7291 ([M + H]+), mass found m/z: 907.7288. N-[N′-(N″-[N‴-Tetracosanoyl-(6-aminohexanoyl)]-L-argininyl)-L-argininyl]-l-argininamide, TFA salt (13e). Yield: 71%. 1H NMR (250 MHz, DMSO) δ: 0.84 (t, J = 7, 3H); 1.2–1.78 (m, 54H); 2.01 (t, J = 7, 2H); 2.11 (t, J = 7, 2H); 2.92–3.18 (m, 8H); 4.07–4.27 (m, 2H); 6.88–7.50 (m, 12H); 7.68–7.97 (m, 3H); 8.05–8.15 (m, 2H); 8.28–8.32 (m, 1H). IR ν: 3273, 3180 (N–H); 2917 (C–H); 1632 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O); δ: 1539 (N–H); 1466 (C–H) cm−1. MS (ES+): m/z (%): 317.5 (100) [M + 3H]3+, 475.5 (30) [M + 2H]2+. HRMS calcd for C48H97N14O5 949.7761 ([M + H]+), mass found m/z: 949.7733.
O); δ: 1539 (N–H); 1466 (C–H) cm−1. MS (ES+): m/z (%): 317.5 (100) [M + 3H]3+, 475.5 (30) [M + 2H]2+. HRMS calcd for C48H97N14O5 949.7761 ([M + H]+), mass found m/z: 949.7733. 2. Lipoplex preparation
Cationic liposomes were prepared by mixing the compounds (1 mM in ethanol) with DOPE (1 mM in methanol). The solvents were then evaporated in the oven at 37 °C overnight and the resulting films were re-suspended in phosphate buffered saline (PBS). The mixtures were vortexed for 5 s and incubated for 30 min at room temperature before adding the plasmid. Lipoplexes were then prepared by mixing the corresponding quantities of each formulation with 0.2 μg of pEGFP-C1 (1 mg mL−1 in water) at two charge ratios (5 : 1 and 10 : 1 reagent–DNA). Finally, the formulations were mixed by vortexing for 5 s and incubated for 30 min at room temperature before use.3. Cell culture
Media, sera and antibiotics were purchased from Gibco or Sigma-Aldrich. Cell culture was performed in a 5% CO2 atmosphere at 37 °C in a SteriCult 200 (Hucoa-Erloss) incubator. Cells were cultured in media (DMEM for HEK293T and B16F10, and RPMI for HeLa) supplemented with 10% fetal bovine serum (FBS), glutamine (4 mM) and antibiotics (penicillin and streptomycin, 100 units per mL). The day before transfection, the cells were washed with phosphate buffered saline (PBS), detached with trypsin–EDTA, counted, and diluted with media to a final concentration of 1 × 104 cells per mL. 100 μL of this dilution was added per well on a 96-well plate and incubated overnight. The lipoplexes were added to the cells and incubated for 5 h (37 °C and 5% CO2). After that the culture medium was exchanged for fresh medium and incubated for 43 h (37 °C and 5% CO2).Effectene® and Lipofectamine™ 2000 were mixed with pEGFP-C1 following the procedure recommended by the suppliers.
4. Analysis of transfection efficiency
The medium was removed and a solution of trypan blue in PBS (0.04%) added to decrease the extracellular background. Cellular fluorescence was analysed using a BioTek microplate reader with excitation at 488 nm. Fluorescence values were expressed as mean fluorescence (arbitrary units).5. Cell viability assay
Lipids (N/P ratio of 5 : 0.91 mM, 0.45 mM, 0.3 mM for 11a–g, 12a–f and 13a–e respectively; N/P ratio of 10 : 1.82 mM, 0.91 mM, 0.6 mM for 11a–g, 12a–f and 13a–e respectively) were complexed with DOPE (same and double concentration as for lipid) and pEGFP-C1 (200 ng per well) as described previously and added to the cells. Cells were incubated with complexes at 37 °C and 5% CO2 for 12 h. After incubation, media were removed, the cells washed with 100 μL of PBS and then a MTT solution (5 mg mL−1 of MTT dissolved in phenol red free medium)27 was added to each well and incubated for 3 h. After incubation, the resulting formazan crystals were dissolved with a solution of 100 μL of 10% Triton X-100 (v/v) in anhydrous isopropanol. Absorbances were measured at a wavelength of 570 nm using a Bio-Rad Benchmark microplate reader and converted to percentage of cell viability (relative to control cells).Abbreviations
| DOTMA | N-[1-(2,3-dioleyloxy)propyl]-N,N,N-trimethylammonium chloride |
| DOSPA | 2.3-dioleyloxy-N(2(sperminecarboxamido)ethyl)-N,N-dimethyl-1-propanaminium trifluoroacetate |
| DOGS | dioctadecylamidoglycylspermine tetratrifluoroacetate |
| DOPE | dioleoylphosphatidylethanolamine |
| Pbf | 2,2,4,6,7-pentamethyldihydrobenzofuran-5-sulfonyl |
| HOBt | 1-hydroxybenzotriazole |
| DIC | N,N′-diisopropylcarbodiimide |
| TFA | trifluoroacetic acid |
| TIS | triisopropyl silane |
| TAMTAT | N-[tris(3-(amino)propyl)methyl]tetraeicosanamide trihydrochloride |
| GFP | green fluorescence protein |
| MTT | 3-(4,5-dimethylthiazol-2-yl)2,5-diphenyltetrazolium bromide |
Acknowledgements
We thank the MRC for funding. We thank Dr John White for biochemical assistance.Notes and references
- M. L. Edelstein, M. R. Abedi and J. Wixon, J. Gene Med., 2007, 9, 833–842 CrossRef.
- A. Rolland and P. L. Felgner, Adv. Drug Delivery Rev., 1998, 30, 1–3 CrossRef CAS.
- R. G. Amado and I. S. Y. Chen, Science, 1999, 285, 674–676 CrossRef CAS.
- M. D. Brown, A. G. Schätzlein and I. F. Uchegbu, Int. J. Pharm., 2001, 229, 1–21 CrossRef CAS.
- P. P. Karmali and A. Chaudhuri, Med. Res. Rev., 2007, 27, 696–722 CrossRef CAS.
- P. L. Felgner, T. R. Gadek, M. Holm, R. Roman, H. W. Chan, M. Wenz, J. P. Northrop, G. M. Ringold and M. Danielsen, Proc. Natl. Acad. Sci. USA, 1987, 84, 7413–7417.
- H. E. J. Hofland, L. Shephard and S. M. Sullivan, Proc. Natl. Acad. Sci. USA, 1996, 93, 7305–7309 CrossRef CAS.
- B. Demeneix, J. P. Loeffler and J. Perez-Mutul, Proc. Natl. Acad. Sci. USA, 1989, 86, 6982–6986 CAS.
- Y. Xu and F. C. Szoka, Jr., Biochemistry, 1996, 35, 5616–5623 CrossRef CAS.
- H. Farhood, N. Serbina and L. Huang, Biochim. Biophys. Acta, 1995, 1235, 289–295 CrossRef CAS.
- A. D. Miller, Angew. Chem., Int. Ed., 1998, 37, 1768–1785 CrossRef.
- S. C. Semple, A. Chonn and P. R. Cullis, Biochemistry, 1996, 35, 2521–2525 CrossRef CAS.
- Y. Liu, L. C. Mounkes, H. D. Liggitt, C. S. Brown, I. Solodin, T. D. Heath and R. J. Debs, Nat. Biotechnol., 1997, 15, 167–173 CrossRef CAS.
- I. Koltover, T. Salditt, J. O. Rädler and C. R. Safinya, Science, 1998, 281, 78–81 CrossRef CAS.
- B. Martin, M. Sainlos, A. Aissaoui, N. Oudrhiri, M. Hauchecorne, J.-P. Vigneron, J.-M. Lehn and P. Lehn, Curr. Pharm. Des., 2005, 11, 375–394 CrossRef CAS.
- S. E. How, B. Yingyongnarongkul, M. A. Fara, J. J. Diaz-Mochon, S. Mittoo and M. Bradley, Comb. Chem. High Throughput Screen., 2004, 7, 423–430 CAS.
- B. Yingyongnarongkul, M. Howarth, T. Elliott and M. Bradley, J. Comb. Chem., 2004, 6, 453–460.
- B. Yingyongnarongkul, M. Howarth, T. Elliott and M. Bradley, Chem.–Eur. J., 2004, 10, 463–473 CrossRef CAS.
- S.-E. How, A. Unciti-Broceta, R. M. Sánchez-Martín and M. Bradley, Org. Biomol. Chem., 2008, 6, 2266–2269 RSC.
- J. J. Diaz-Mochon, M. A. Fara, R. M. Sanchez-Martin and M. Bradley, Tetrahedron Lett., 2008, 49, 923–926 CrossRef CAS.
- A. Unciti-Broceta, E. Holder, L. J. Jones, B. Stevenson, A. R. Turner, D. J. Porteous, A. C. Boyd and M. Bradley, J. Med. Chem., 2008, 51, 4076–4084 CrossRef CAS.
- M. A. Fara, J. J. Diaz-Mochon and M. Bradley, Tetrahedron Lett, 2006, 47, 1011–1014 CrossRef CAS.
- A. Unciti-Broceta, F. Diezmann, C. Y. Ou-Yang, M. A. Fara and M. Bradley, Bioorg. Med. Chem, 2008 DOI:10.1016/j.bmc.2008.02.068.
- For a full description of the charge ratio see ref. 22.
- J. M. Weiss, R. Shivakumar, S. Feller, L.-H. Li, A. Hanson, W. E. Fogler, J. C. Fratantoni and L. N. Liu, Cancer Gene Ther., 2004, 11, 346–353 CrossRef CAS.
- R. Y. Tsien, Annu. Rev. Biochem., 1998, 67, 509–544 CrossRef CAS.
- T. Mosmann, J. Immun. Meth., 1983, 65, 55–63 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: 1H-NMR spectra of 11f–h, 12d–f, 13d–e. See DOI: 10.1039/b815733b |
|
| This journal is © The Royal Society of Chemistry 2009 |
Click here to see how this site uses Cookies. View our privacy policy here. 





![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O); δ: 1530 (N–H); 1471 (C–H) cm−1. MS (ES+): m/z (%): 525.5 (100) [M + H]+. HRMS calcd for C28H57N6O3 525.4486 ([M + H]+), mass found m/z: 525.4492.
O); δ: 1530 (N–H); 1471 (C–H) cm−1. MS (ES+): m/z (%): 525.5 (100) [M + H]+. HRMS calcd for C28H57N6O3 525.4486 ([M + H]+), mass found m/z: 525.4492.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O); δ: 1540 (N–H), 1471 (C–H) cm−1. MS (ES+): m/z (%): 539.5 (100) [M + H]+. HRMS calcd for C29H59N6O3 539.4643 ([M + H]+), mass found m/z: 539.4647.
O); δ: 1540 (N–H), 1471 (C–H) cm−1. MS (ES+): m/z (%): 539.5 (100) [M + H]+. HRMS calcd for C29H59N6O3 539.4643 ([M + H]+), mass found m/z: 539.4647.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O); δ: 1543 (N–H); 1463 (C–H) cm−1. MS (ES+): m/z (%): 553.6 (100) [M + H]+. HRMS calcd for C30H61N6O3 553.4799 ([M + H]+), mass found m/z: 553.4806.
O); δ: 1543 (N–H); 1463 (C–H) cm−1. MS (ES+): m/z (%): 553.6 (100) [M + H]+. HRMS calcd for C30H61N6O3 553.4799 ([M + H]+), mass found m/z: 553.4806.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O); δ: 1542 (N–H); 1464 (C–H) cm−1. MS (ES+): m/z (%): 581.5 (100) [M + H]+. HRMS calcd for C32H64N6O3 581.5112 ([M + H]+), mass found m/z: 581.5112.
O); δ: 1542 (N–H); 1464 (C–H) cm−1. MS (ES+): m/z (%): 581.5 (100) [M + H]+. HRMS calcd for C32H64N6O3 581.5112 ([M + H]+), mass found m/z: 581.5112.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O); δ: 1535 (N–H); 1471 (C–H) cm−1. MS (ES+): m/z (%): 581.6 (100) [M + H]+. HRMS calcd for C32H65N6O3 581.5112 ([M + H]+), mass found m/z: 581.5111.
O); δ: 1535 (N–H); 1471 (C–H) cm−1. MS (ES+): m/z (%): 581.6 (100) [M + H]+. HRMS calcd for C32H65N6O3 581.5112 ([M + H]+), mass found m/z: 581.5111.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O); 2919 (C–H); δ: 1542 (N–H); 1472 (C–H) cm−1. MS (ES+): m/z (%): 595.5 (75) [M + H]+. HRMS calcd for C33H67N6O3 595.5269 ([M + H]+), mass found m/z: 595.5265.
O); 2919 (C–H); δ: 1542 (N–H); 1472 (C–H) cm−1. MS (ES+): m/z (%): 595.5 (75) [M + H]+. HRMS calcd for C33H67N6O3 595.5269 ([M + H]+), mass found m/z: 595.5265.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O); δ: 1542 (N–H); 1471 (C–H) cm−1. MS (ES+): m/z (%): 609.6 (100) [M + H]+. HRMS calcd for C34H69N6O3 609.5425 ([M + H]+), mass found m/z: 609.5412.
O); δ: 1542 (N–H); 1471 (C–H) cm−1. MS (ES+): m/z (%): 609.6 (100) [M + H]+. HRMS calcd for C34H69N6O3 609.5425 ([M + H]+), mass found m/z: 609.5412.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O); δ: 1536 (N–H); 1466 (C–H) cm−1. MS (ES+): m/z (%): 341.4 (100) [M + 2H]2+, 681.7 (15) [M + H]+. HRMS calcd for C34H69N10O4 681.5497 ([M + H]+), mass found m/z: 681.5491.
O); δ: 1536 (N–H); 1466 (C–H) cm−1. MS (ES+): m/z (%): 341.4 (100) [M + 2H]2+, 681.7 (15) [M + H]+. HRMS calcd for C34H69N10O4 681.5497 ([M + H]+), mass found m/z: 681.5491.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O); δ: 1538 (N–H); 1467 (C–H) cm−1. MS (ES+): m/z (%): 348.4 (100) [M + 2H]2+. HRMS calcd for C35H71N10O4 695.5654 ([M + H]+), mass found m/z: 695.5652.
O); δ: 1538 (N–H); 1467 (C–H) cm−1. MS (ES+): m/z (%): 348.4 (100) [M + 2H]2+. HRMS calcd for C35H71N10O4 695.5654 ([M + H]+), mass found m/z: 695.5652.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O); δ: 1542 (N–H); 1466 (C–H) cm−1. MS (ES+): m/z (%): 369.4 (100) [M + 2H]2+. HRMS calcd for C38H77N10O4 737.6123 ([M + H]+), mass found m/z: 737.6107.
O); δ: 1542 (N–H); 1466 (C–H) cm−1. MS (ES+): m/z (%): 369.4 (100) [M + 2H]2+. HRMS calcd for C38H77N10O4 737.6123 ([M + H]+), mass found m/z: 737.6107.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O); δ: 1539 (N–H); 1466 (C–H) cm−1. MS (ES+): m/z (%): 369.4 (100) [M + 2H]2+. HRMS calcd for C38H77N10O4 737.6123 ([M + H]+), mass found m/z: 737.6122.
O); δ: 1539 (N–H); 1466 (C–H) cm−1. MS (ES+): m/z (%): 369.4 (100) [M + 2H]2+. HRMS calcd for C38H77N10O4 737.6123 ([M + H]+), mass found m/z: 737.6122.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O); δ: 1541 (N–H); 1468 (C–H) cm−1. MS (ES+): m/z (%): 376.4 (100) [M + 2H]2+. HRMS calcd for C39H79N10O4 751.6280 ([M + H]+), mass found m/z: 751.6280.
O); δ: 1541 (N–H); 1468 (C–H) cm−1. MS (ES+): m/z (%): 376.4 (100) [M + 2H]2+. HRMS calcd for C39H79N10O4 751.6280 ([M + H]+), mass found m/z: 751.6280.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O); δ: 1539 (N–H); 1467 (C–H) cm−1. MS (ES+): m/z (%): 383.5 (100) [M + 2H]2+. HRMS calcd for C40H81N10O4 765.6436 ([M + H]+), mass found m/z: 765.6406.
O); δ: 1539 (N–H); 1467 (C–H) cm−1. MS (ES+): m/z (%): 383.5 (100) [M + 2H]2+. HRMS calcd for C40H81N10O4 765.6436 ([M + H]+), mass found m/z: 765.6406.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O); δ: 1536 (N–H); 1454 (C–H) cm−1. MS (ES+): m/z (%): 280.1 (100) [M + 3H]3+, 419.8 (50) [M + 2H]2+. HRMS calcd for C40H81N14O5 837.6508 ([M + H]+), mass found m/z: 837.6547.
O); δ: 1536 (N–H); 1454 (C–H) cm−1. MS (ES+): m/z (%): 280.1 (100) [M + 3H]3+, 419.8 (50) [M + 2H]2+. HRMS calcd for C40H81N14O5 837.6508 ([M + H]+), mass found m/z: 837.6547.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O); δ: 1538 (N–H); 1466 (C–H) cm−1. MS (ES+): m/z (%): 289.4 (100) [M + 3H]3+, 433.5 (30) [M + 2H]2+. HRMS calcd for C42H85N14O5 865.6822 ([M + H]+), mass found m/z: 865.6822.
O); δ: 1538 (N–H); 1466 (C–H) cm−1. MS (ES+): m/z (%): 289.4 (100) [M + 3H]3+, 433.5 (30) [M + 2H]2+. HRMS calcd for C42H85N14O5 865.6822 ([M + H]+), mass found m/z: 865.6822.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O); δ: 1542 (N–H); 1454 (C–H) cm−1. MS (ES+): m/z (%): 298.71 (100) [M + 3H]3+, 447.5 (35) [M + 2H]2+. HRMS calcd for C44H89N14O5 893.7135 ([M + H]+), mass found m/z: 893.7135.
O); δ: 1542 (N–H); 1454 (C–H) cm−1. MS (ES+): m/z (%): 298.71 (100) [M + 3H]3+, 447.5 (35) [M + 2H]2+. HRMS calcd for C44H89N14O5 893.7135 ([M + H]+), mass found m/z: 893.7135.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O); δ: 1541 (N–H); 1462 (C–H) cm−1. MS (ES+): m/z (%): 303.1 (100) [M + 3H]3+. HRMS calcd for C45H90N14O5 907.7291 ([M + H]+), mass found m/z: 907.7288.
O); δ: 1541 (N–H); 1462 (C–H) cm−1. MS (ES+): m/z (%): 303.1 (100) [M + 3H]3+. HRMS calcd for C45H90N14O5 907.7291 ([M + H]+), mass found m/z: 907.7288.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O); δ: 1539 (N–H); 1466 (C–H) cm−1. MS (ES+): m/z (%): 317.5 (100) [M + 3H]3+, 475.5 (30) [M + 2H]2+. HRMS calcd for C48H97N14O5 949.7761 ([M + H]+), mass found m/z: 949.7733.
O); δ: 1539 (N–H); 1466 (C–H) cm−1. MS (ES+): m/z (%): 317.5 (100) [M + 3H]3+, 475.5 (30) [M + 2H]2+. HRMS calcd for C48H97N14O5 949.7761 ([M + H]+), mass found m/z: 949.7733.