Effect of clustered peptide binding on DNA condensation†
Jennifer
Haley
,
Paul
Kabiru
and
Yan
Geng
*
Department of Chemistry, University of Georgia, Athens, GA 30602, USA. E-mail: ygeng@chem.uga.edu
First published on 25th September 2009
Abstract
DNA condensation in-vitro has been studied as a model system to reveal common principles underlying gene packaging in biology, and as the critical first step towards the development of non-viral gene delivery vectors. In this study, we use a bio-inspired approach, where small DNA-bindingpeptides are controllably clustered by an amphiphilic block copolymer scaffold, to reveal the effect of clustered peptide binding on the energetics, size, shape and physical properties of DNA condensation in-vitro. This provides insights into the general architectural effect of gene-binding proteins on DNA condensation process. Moreover, the versatility afforded by regulating the clustering density and composition of peptides may provide a novel design platform for gene delivery applications in the future.
Introduction
Condensation of long strands of DNA into compact, ordered structures is an important biological process for gene protection, storage and replication. This phenomenon has attracted tremendous interest to a broad spectrum of scientific communities. DNA condensation in-vitro has been pursued as a model system to study the phase transition phenomena of polyelectrolytes,1 to reveal common principles underlying gene packaging in biology,2,3 and as the critical first step towards the development of non-viral gene delivery vectors.4,5 Historically, toroids—where loops of DNA double helix pack in hexagonal arrays—have attracted the most attention and are considered as the predominant in-vitroDNA condensate structure.6–9 Occasionally, metastable rod-like DNA condensates have also been discovered and are attracting increasing interest in recent years.10,11A wide variety of materials have been explored as DNA condensing agents, ranging from the original small natural amines (e.g.spermidine and spermine)6 and multivalent cations (e.g. Co(NH3)63+)7 to much more complex materials, such as lipids,12 crowding agents,13dendrimers,14peptides,15,16polyamines,17–20 and their corresponding block copolymers .21,22 For small natural amines and multivalent cations, the mechanism and pathway of their DNA condensation have been vigorously studied and are fairly well understood, providing invaluable foundation for later studies.8,23 However, other than certain viruses, most DNA condensation process in biological systems, especially in bacteria and eukaryocyte cells, all involves much more complex DNA interactions with large molecules of proteins. Thus, relevant biological information that can be revealed by simple small condensing agents is rather limited. Moreover, simple small agents are unlikely to provide sufficient stabilization and protection of DNA for practical applications. However, as condensing materials become more complex, elucidation of their complexation process with DNA becomes increasingly difficult. Polymeric condensing agents, for example, have attracted significant attention in recent years, due to their superior ability in compacting and stabilizing DNA, and to their chemical flexibility in functionalization towards improving gene delivery efficiency.17–22 However, interactions between long polymer chains and DNA strands are much more complicated,24 and the innate polydispersity of synthetic polymers further complicates the DNA complexation process. Lack of systematic understanding and precise control about their DNA condensation process represents a severe drawback.
We have recently developed a bio-inspired combinative self-assembly approach that can efficiently condense and package DNA into nanoparticles.25 Small oligopeptides that emulate the active nucleotide binding site of DNA compaction proteins can provide direct insights into DNA–protein interactions, and when compared to large whole proteins, have the advantages of high efficiency in functionality, less antigenicity, flexibility, and precision in the sequence design.15 When grafted onto the hydrophobic segment of a block copolymer scaffold, a clustered spatial arrangement of the peptides is created towards DNA binding, Scheme 1. Synthetic polymers have been used as scaffolds in the past to create multivalent ligands with controlled density, to probe the mechanism of receptor clustering at cell surface and cell signaling pathways.26 In this report, we elucidate the effect of controlled peptide clustering on the energetics, size, shape, as well as physical properties of DNA condensation in-vitro. This provides insights into the general architectural effect of gene-binding proteins on DNA condensation and packaging process. Moreover, the versatility afforded by regulating the clustering density and composition of the peptides may provide a novel design platform for gene delivery applications in the future.
 | ||
| Scheme 1 Combinative self-assembly of the block copolymer –peptide clustered hybrid with DNA into toroid and rod condensates. | ||
Experimental
Materials
λ-DNA in 10 mM Tris buffer was purchased from New England Biolabs (Ipswich, MA). All chemicals and solvents were purchased from Sigma-Aldrich (St. Louis, MO). Oligopetides were synthesized by the standard Fmoc solid phase peptide synthesis procedure, using HOBT, HBTU and DIPEA couplings, followed by N-capping with acetylation and C-capping with amidation. Each peptide was analyzed by ESI-MS and 1H NMR before grafting. Amphiphilic block copolymer PEG93-b-PBD14, where the subscripts denote the average number of repeating units, was prepared by the sequential living anionic polymerization of 1,3-butadiene and ethylene oxide.27Grafting of model gene-binding KWKnpeptides onto PEG-b-PBD
For grafting purposes, cysteine that contains a thiol group was attached to KWKnoligopeptides as the linker terminus. CKWKn were grafted to PEG93-b-PBD14 according to a modular procedure published elsewhere, utilizing the free radical addition of the thiol group onto the double bonds of PBD.25,28 The grafting scheme and the representative NMR and GPC spectra of the polymer–peptide hybrid are available as ESI S1 and S2.† The grafting density, i.e. the percentage of the PBD double bonds grafted with peptides, can be tuned by changing the molar ratio between the reacting thiol groups and double bonds, and the peptides are expected to be randomly linked along the PBD chain.28EtBr displacement assay
λ-DNA in Tris buffer (20 μg mL−1) was incubated with EtBr (0.8 μg mL−1) for 1 h prior to analysis. The fluorescent intensity of the DNA·EtBr complex was measured using a Jobin Yvon FluoroMax-3 Fluorimeter (excitation: 520 nm, emission: 590 nm). Measured concentrated KWKn or their polymer clustered hybrids was then titrated into the DNA·EtBr solution, and the corresponding fluorescence at different lysine/phosphate (N/P) ratios was determined.Circular dichroism analysis
CD spectra were recorded using Jasco J-715 spectropolarimeter, at the far-UV region (200–320 nm) and with a scanning speed of 50 nm min−1. A total of four scans were accumulated, and temperature was maintained at 25 °C. DNA complexes with KWKn or their polymer clustered hybrids in Tris buffer were set the concentration of 50 μg ml−1 and N/P = 0.5 for the CD studies. Weak background absorbance from the buffer and condensing agents were directly subtracted from the measurements.TEM Imaging
λ-DNA complexes with KWKn and their polymer clustered hybrids were prepared by mixing an equal volume of DNA (10 μg mL−1) with the condensing agents at desired N/P in 1× TE buffer (10 mM Tris-Cl, 1 mM EDTA, pH 7.0). The complex solution was then vortexed for 30 s and allowed to equilibrate at room temperature for 2 h. The complex sample was then deposited onto the glow discharged formvar coated copper grids and stained with 2% uranyl acetate for 1 min. The grids were blotted and then air-dried for TEM imaging on a 200 kV Tecnai 20 transmission electron microscope at a magnification of 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000×.
000×.
DNA melting study
Melting profiles of native λ-DNA and the DNA complexes with KWKn or their polymer clustered hybrids (50 μg mL−1, N/P = 0.5) were obtained by monitoring their absorbance at 260 nm with a Cary 100 UV-Vis spectrophotometer. Samples were heated from 30 to 95 °C with a heating rate of 1 °C min−1. Weak background absorbance from the buffer and condensing agents were directly subtracted from the measurements.DNaseI degradation assay
DNaseI (1 unit) in 10× digestion buffer (100 mM Tris-HCl, 25 mM MgCl2, 5 mM CaCl2, pH 7.6) was added to 0.02 ml, 10 μg mL−1DNA and DNA complex samples (0.2 μg DNA). The samples were incubated at 37 °C for 15 min, followed by inactivation with 4 μL of 25 mM EDTA at room temperature for 10 min. Finally, 7.5 μL of 100 mg mL−1heparin was added and incubated at room temperature for 2 h to release DNA for gel electrophoresis analysis (0.8% agarose gel, 1× TAE running buffer, 0.5 μg mL−1ethidium bromide, 100 V, 1 h).Results and discussion
Model gene-binding oligopeptides, KWKn (K = lysine; W = tryptophan), with different numbers of lysine residues (n = 2 or 4) were used for this study, and they were controllably grafted onto the hydrophobic polybutadiene segment of an amphiphilic poly(ethylene glycol)-block-polybutadiene (PEG93-b-PBD14) block copolymer scaffold at different grafting densities, i.e. either with four peptide grafts or eight peptide grafts, via an established modular peptide grafting route.28 The peptide grafted polymer hybrids are designated as PP series, where PP12 and PP24 represent the polymer–peptide hybrids with four and eight KWK2 grafted, respectively, and PP20 and PP40 represent the polymer–peptide hybrids with four and eight KWK4 grafted, respectively. Literature shows that KWKnpeptides bind to DNA through two kinds of interactions: electrostatic neutralization between the positively charged amino group of lysine (N) and the negatively charged phosphate DNA backbone (P), and the hydrophobic intercalation of aromatic tryptophan within the DNAbase pairs.29,30 Such attractions between KWKnpeptides and DNA are expected to be the driving force for the complexation of the polymer–peptide hybrids with DNA, as control experiments reveal negligible interactions between DNA and the neutral block copolymer scaffold alone.25Five or more lysine residues are generally required in an oligopeptide sequence to condense DNA.15,16 With only three lysine residues, free KWK2 alone exhibited fairly low DNA binding affinity from the ethidium bromide (EB) displacement assay , Fig. 1(a). In the EB displacement assay , binding of an agent to DNA would displace the intercalated EB and subsequently quench the fluorescence caused by the EB·DNA complex. Fig. 1(a) shows that free KWK2 can only weakly quench fluorescence over a wide range of N/P values. Even in large excess, at N/P = 12, only 40% of quenching (I/I0 ∼ 0.6) could be achieved by free KWK2. However, when KWK2 was clustered into proximity by the polymer PEO93-b-PBD14 scaffold, PP12 and PP24 quenched the fluorescence much more efficiently at the same stoichiometric N/P of free KWK2, and both were able to achieve nearly complete quenching (I/I0 ≤ 0.3), Fig. 1(a). At higher grafting density, where more oligopeptides were clustered into closer proximity along the polymer scaffold, PP24—with eight peptides grafted—demonstrated more enhanced DNA binding than PP12–with four peptides grafted. It seems that the clustered oligopeptide array gathered by the polymer scaffold can recognize the DNA double helix in a positive cooperative manner and thus can significantly strengthen the DNA binding. The surrounding overall hydrophobic environment generated by the PBD polymers may also contribute to the strengthened DNA binding effect.
 | ||
| Fig. 1 Characterization of the binding and conformational change of λ-DNA complexation with KWK2, PP12 and PP24. (a) EB displacement assay on DNA binding; (b) DNA conformation by CD analysis. | ||
Conformational changes in the DNA double helix induced by the binding of KWK2, PP12 and PP24 were monitored by circular dichroism (CD), Fig. 1(b). The CD spectrum of native λ-DNA shows a typical B-form conformation, which is composed of four major peaks in the UV-Vis region: a negative 210 nm peak, a positive 220 nm peak, a negative 245 nm peak and a positive 280 nm peak.31 Transition of the DNA conformation from the B-form to the less compact C-form, which is characterized by a decrease in the intensity of the positive 280 nm peak, is commonly found in condensed DNA systems, such as in virus heads and nucleosomes.31,32 With free KWK2, negligible intensity change at the 280 nm peak was observed and the B-form DNA conformation largely remained intact. PP12 and PP24, on the other hand, provoked much more significant change in the 280 nm peak, indicating a partial B-to-C transition has occurred in such polymer–peptide clustered systems, and the higher the clustering density, the more dramatic the conformational change. It seems that the clustered peptide–DNA binding can cooperatively distort and loosen the DNA double helix into the less compact C form, which facilitates DNA condensation.
Transmission electron microscopy (TEM) analysis reveals distinctively different DNA complexation phenomena between free KWK2 and its polymer clustered PP12 and PP24, Fig. 2. No DNA compaction, but rather exclusively extended DNA bundles, was observed for free KWK2, even in large excess of N/P of 12. The bundles are 10–20 nm thick and each contains 5–10 λ-DNA strands, considering the individual ds-DNA is ∼2 nm in width. With just three lysines, free KWK2 can not compact DNA, but its triple valency is able to bridge different DNA strands together. In sharp contrast, when KWK2 was clustered by the polymer scaffold, PP12 and PP24 were observed to condense DNA into toroidal structures, and the clustering density exhibited a strong effect on the condensation process. With low clustering density, PP12 gave rise to ill-defined nucleation loops with an average large diameter of 80 nm at the same stoichiometric N/P 12. However, no subsequent winding of DNA strands around the nucleation loops, i.e. toroid growth, was fostered, as floppy DNA strands surrounding the loops are clearly visible from the TEM images. As the peptide clustering density doubles in PP24, the toroid loop size was notably reduced and the subsequent toroid growth efficiently promoted. TEM analysis shows well-defined DNA toroids that have the average diameter of 50 nm and thickness of 20 nm at N/P = 12 (definition of diameter and thickness of a toroid is available in ESI S3† ).
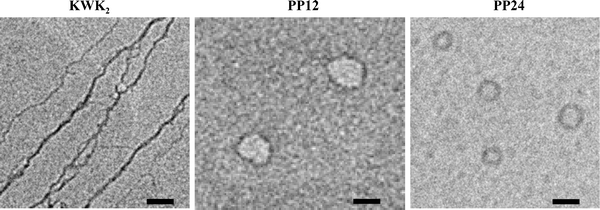 | ||
| Fig. 2 Representative TEM micrographs of λ-DNA condensates by KWK2, PP12 and PP24 at N/P = 12. Scale bar is 50 nm. | ||
It is well known that the formation of DNA toroid condensates proceeds through two stages—the initial nucleation loop stage, followed by growth.33 Looping of a semiflexible DNA chain is a spontaneous, diffusion-limited process, and the ease and size largely depend on the flexibility and bending energy of the DNA chain, as well as the ability of the condensing agents to anneal and stabilize the loop.34 The cooperative, clustered binding of KWK2 seems to be able to significantly lower the bending energy of DNA and promote the formation of nucleation loops. With low grafting density, PP12, however, can not sufficiently reduce the DNA helix–helix association energy to foster the further winding of DNA strands around the loop. As the clustering density increases, PP24 can not only overcome the extra strain associated with the smaller nucleation loops, but is also sufficient to promote the further growth of toroids. It appears that for oligopeptides with weak DNA affinity, clustered binding shifts the DNA complexation process from intermolecular bridging to intramolecular toroidal compaction, and the clustering density has a strong impact on the energetics and dimension of the DNA condensation process.
To reveal the effect of clustered binding on DNA condensation for peptides with strong DNA affinity, KWK4 with five lysine residues and its corresponding polymer clustered hybrids, PP20—with four peptides grafted, and PP40—with eight peptides grafted, were studied respectively. Incorporation of more lysine residues is well known to enhance the binding between the peptide and DNA.16 Indeed, EBassay shows that KWK4 quenched the fluorescence much more efficiently than KWK2 and reached near complete binding as N/P increased to 10, Fig. 3(a). CD analysis also shows that the binding of KWK4 to DNA notably reduced the intensity of the 280 nm peak and induced a partial B-to-C conformational change in λ-DNA, Fig. 3(b). Like KWK2, clustering by the polymer scaffold nonetheless enhanced the DNA binding and provoked more pronounced B-to-C conformational change, and the higher the clustering density, the stronger the effect.
 | ||
| Fig. 3 Characterization of the binding and conformational change of λ-DNA complexation with KWK4, PP20 and PP40. (a) EB displacement assay on DNA binding; (b) DNA conformation by CD analysis. | ||
TEM studies confirmed that free KWK4 is able to condense DNA into compact structures, Fig. 4. Intertwined aggregates of toroids were observed as the primary DNA condensate structure. It appears that at initial low N/P values, KWK4 can simultaneously promote both intermolecular DNA aggregation and the formation of toroid nucleation loops, which progressively grew into intertwined full toroids. When KWK4 was clustered by the polymer scaffold, PP20 produced discrete toroids without aggregation. Distinct nucleation loops with an average diameter of 70 nm were observed at N/P = 3, and higher N/P values further reduced the toroid loop size and promoted toroid growth. At N/P = 12, dispersed, well-defined toroids with average diameter of 45 nm and thickness of 15 nm were found as the exclusive DNA condensate structure. Compared to free KWK4, which does not seem to differentiate intramolecular DNA compaction from the intermolecular DNA association process, the polymer–peptide clustered PP20 must be able to lower the bending energy of DNA chains much more efficiently, so that it exclusively favors the intramolecular toroidal DNA condensation route. With higher clustering density in PP40, the difference in DNA condensation process becomes even more dramatic from the free KWK4. PP40 produced much smaller toroid loops with average diameter of 50 nm at N/P = 3, and fostered fast toroid growth to 25 nm in thickness at N/P = 7. Starting from N/P = 10, rod-like DNA condensate structures began to emerge, and upon reaching N/P = 12, a significant population (∼50%) of well-defined rods with average length of 200 nm and width of 15 nm were observed. The rods were quite stable in solution and there was no apparent population change with time. This is surprising, considering studies with small condensing agents in literature suggest that DNA rod condensates are generally unstable, and would quickly convert to toroids with time. Controlling the morphology of DNA condensates between toroids and rods has been quite difficult,11 and only in the presence of an alcoholsolvent that destabilizes the DNA double helix, or with special bacterial chromatinproteins that can induce pronounced kinks in the DNA double helix, has higher population of rods been reported.10,35 Here, the finding of a significant population of stable rods induced by PP40 suggests that at sufficiently high density, the cooperative, clustered binding can sharply bend the DNA double helix into rod-forming kinks, as well as help stabilize the rods once formed. The entanglement nature of the polymer scaffold may also contribute to the stabilization of the rod DNA condensates. Thus, in the event of oligopeptides with strong DNA affinity, such as KWK4, we show that peptide clustering by a polymer scaffold can alter the DNA condensation pathway from intertwined toroid aggregates to discrete toroid or rod DNA condensates. Controlled density plays a vital role in the clustering effect, which not only determines the size and dimension of DNA condensate structure, but also the shape transition from toroids to rods.
 | ||
| Fig. 4 Representative TEM micrographs of λ-DNA condensates by KWK4, PP20 and PP40 at increasing N/P from 3 to 12. Scale bar is 50 nm. | ||
To evaluate the effect of the polymer scaffolded peptide clustering on physical properties of the resultant DNA condensates, melting and nuclease degradation studies were carried out to analyze their thermal and biological stability, respectively, Fig. 5. In the melting study, dissociation of double-stranded DNA into single strands was monitored by an increase in the absorbance of 260 nm, due to the disruption of hydrogen bonds between base pairs with raising temperature (i.e. hyperchromic effect). Fig. 5(I) shows that compared to naked DNA, condensation with KWK4 notably shifted the DNA dissociation curve to higher Tm, the temperature at which 50% ds-DNA dissociates, and reduced the degree of change in absorbance, indicating enhanced DNA stabilization against double-strand breakage. When clustered by the polymer scaffold, PP20 and PP40 further stabilized DNA in comparison to free KWK4, by shifting the DNA dissociation curve to even higher temperatures and more reduced absorbance changes, and the higher the clustering density, the more significant the enhancement. Even at the upper limit of DNA melting studies in aqueous solution, i.e. 95 °C, which is just below the boiling point of water, the majority of the ds-DNA remained intact.
 | ||
| Fig. 5 Stability of DNA complexes against thermal and bio-degradation. (I) (A) Melting profiles and (B) Table of estimated Tm of native λ-DNA and its complexes with KWK4, PP20 and PP40; (II) DNaseI protection assay . (−) before treatment, (+) after treatment. | ||
DNA is also prone to nuclease degradation in biofluids, which represents a major challenge for gene delivery. To assess their resistance against DNase degradation, naked λ-DNA, DNA condensates with KWK4 and the polymer–peptide clustered PP40, were incubated with DNaseI for 30 min, and the integrity of the DNA before and after the treatment was analyzed by agarose gel electrophoresis, Fig. 5(II). For naked λ-DNA, no intact DNA band could be detected after DNaseI treatment, indicating that the DNA has been completely degraded into small fragments that are beyond the detection limit. For DNA–KWK4 condensates, even at high N/P = 12, only a faint intact DNA band was observed after DNase treatment. Comparison between the DNA band intensity before and after the DNase treatment shows that only a small fraction of DNA was preserved, and KWK4 itself does not provide sufficient protection for DNA. However, the polymer–peptide clustered PP40 demonstrated much more enhanced DNA protection against nuclease degradation. As more PP40 was used in DNA condensation, the intensity of the intact DNA band after treatment continuously increased with N/P, and at N/P = 12, the DNA band before and after DNase treatment was measured to be nearly the same, suggesting that the integrity of the DNA has been largely preserved. We speculate that the superior protection of the block copolymer –peptide clustered hybrids originates from two aspects—highly efficient DNA compaction by the clustered peptides inside the core, and the surrounding dense, stealthy PEG shell that prevents the deposition and degradation of the nuclease. Apparently, the polymer–peptide clustered hybrids here have inherited all the general advantages in DNA stabilization and protection that are associated with polymeric systems, and the PEG shell can be further conjugated with a wide variety of functional groups to foster specific targeting, endosomal release and nuclear transport for future gene delivery applications.
Conclusions
In this study, we demonstrate that the peptide clustering can controllably alter the pathway and morphology of DNA condensation in vitro. Moreover, such peptide clustering by block copolymer scaffolds also significantly improves the DNA stability against breakage and DNase bio-degradation. The study here has comprehensively elucidated the general architectural effect of the clustered peptide binding on DNA condensation, as well as having provided a versatile new approach to tailor and optimize synthetic gene delivery vector design.Acknowledgements
The authors would like to thank Dr Jeff Urbauer (Biochemistry Department, UGA) and Dr Robert Scott (Chemistry Department, UGA) for their assistance with the EtBr displacement assay and the use of the UV transilluminator. We also thank Dr Robert Phillips (Chemistry Department, UGA) and Dr Vinny Manoharan (Physics Department, Harvard University) for useful discussions. This work was supported by the UGA Research Foundation and Georgia Cancer Coalition.References
- G. S. Manning, Q. Rev. Biophys., 1978, 11, 103–178 CAS.
- S. M. Klimenko, T. I. Tikchonenko and V. M. Andreev, J. Mol. Biol., 1967, 23, 523–533 CrossRef CAS.
- M. Cerritelli, N. Cheng, A. Rosenberg, C. McPherson, F. Booy and A. Steven, Cell, 1997, 91, 271–280 CrossRef CAS.
- D. Luo and W. M. Saltzman, Nat. Biotechnol., 2000, 18, 33–37 CrossRef CAS.
- M. A. Mintzer and E. E. Simanek, Chem. Rev., 2009, 109, 259–302 CrossRef CAS.
- L. C. Gosule and J. A. Schellman, Nature, 1976, 259, 333–335 CAS.
- J. Widom and R. L. Baldwin, J. Mol. Biol., 1980, 144, 431–453 CrossRef CAS.
- C. C. Conwell, I. D. Vilfan and N. V. Hud, Proc. Natl. Acad. Sci. U. S. A., 2003, 100, 9296–9301 CrossRef CAS.
- N. V. Hud, K. H. Downing and R. Balhorn, Proc. Natl. Acad. Sci. U. S. A., 1995, 92, 3581–3585 CrossRef CAS.
- P. G. Arscott, C. Ma, J. R. Wenner and V. A. Bloomfield, Biopolymers, 1995, 36, 345–364 CrossRef CAS.
- I. D. Vilfan, C. C. Conwell, T. Sarkar and N. V. Hud, Biochemistry, 2006, 45, 8174–8183 CrossRef CAS.
- J. O. Radler, I. Koltover, T. Salditt and C. R. Safinya, Science, 1997, 275, 810 CrossRef CAS.
- U. K. Laemmli, Proc. Natl. Acad. Sci. U. S. A., 1975, 72, 4288–4292 CrossRef CAS.
- J. Haensler and F. C. J. Szoka, Bioconjugate Chem., 1993, 4, 372–379 CrossRef CAS.
- L. C. Smith, J. Duguid, M. S. Wadhwa, M. J. Logan, C. H. Tung, V. Edwards and J. T. Sparrow, Adv. Drug Delivery Rev., 1998, 30, 115–131 CrossRef CAS.
- M. S. Wadhwa, W. T. Collard, R. C. Adami, D. L. McKenzie and K. G. Rice, Bioconjugate Chem., 1997, 8, 81–88 CrossRef CAS.
- E. Wagner, M. Zenke, M. Cotten, H. Beug and M. L. Birnstiel, Proc. Natl. Acad. Sci. U. S. A., 1990, 87, 3410–3414 CrossRef CAS.
- Y. H. Choi, F. Liu, J. S. Park and S. W. Kim, Bioconjugate Chem., 1998, 9, 708–718 CrossRef CAS.
- K. A. Mislick, J. D. Baldeschwieler, J. F. Kayyem and T. J. Meade, Bioconjugate Chem., 1995, 6, 512–515 CrossRef CAS.
- W. Zauner, M. Ogris and E. Wagner, Adv. Drug Delivery Rev., 1998, 30, 97–113 CrossRef CAS.
- K. Kataoka, H. Togawa, A. Harada, K. Yasugi, T. Matsumoto and S. Katayose, Macromolecules, 1996, 29, 8556–8557 CrossRef CAS.
- Y. Kakizawa and K. Kataoka, Adv. Drug Delivery Rev., 2002, 54, 203–222 CrossRef CAS.
- V. A. Bloomfield, Biopolymers, 1997, 44, 269–282 CrossRef CAS.
- I. Nayvelt, T. Thomas and T. J. Thomas, Biomacromolecules, 2007, 8, 477–484 CrossRef CAS.
- J. Haley, X. Li, N. Marshall, J. Locklin and Y. Geng, Mol. BioSyst., 2008, 4, 515–517 RSC.
- C. W. Cairo, J. E. Gestwicki, M. Kanai and L. L. Kiessling, J. Am. Chem. Soc., 2002, 124, 1615–1619 CrossRef.
- S. Förster and E. Kramer, Macromolecules, 1999, 32, 2783–2785 CrossRef.
- Y. Geng, D. E. Discher, J. Justynska and H. Schlaad, Angew. Chem., Int. Ed., 2006, 45, 7578–7581 CrossRef CAS.
- D. Pörschke and J. Ronnenberg, Biophys. Chem., 1981, 13, 283–290 CrossRef CAS.
- D. P. Mascotti and T. M. Lohman, Biochemistry, 1993, 32, 10568–10579 CrossRef CAS.
- W. A. Baase and W. C. J. Johnson, Nucleic Acids Res., 1979, 6, 797–814.
- C. Böttcher, C. Endisch, J. H. Fuhrhop, C. Catterall and M. Eaton, J. Am. Chem. Soc., 1998, 120, 12–17 CrossRef.
- N. V. Hud and I. D. Vilfan, Annu. Rev. Biophys. Biomol. Struct., 2005, 34, 295–318 CrossRef CAS.
- S. Jun, J. Bechhoefer and B.-Y. Ha, Europhys. Lett., 2003, 64, 420–426 CrossRef CAS.
- T. Sarkar, I. Vitoc, I. Mukerji and N. V. Hud, Nucleic Acids Res., 2007, 35, 951 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available: NMR and GPC of the peptide grafted polymer hybrids; illustration of the diameter and thickness of a toroid. See DOI: 10.1039/b908873c |
| This journal is © The Royal Society of Chemistry 2010 |
